Abstract
Acute myeloid leukemia (AML) is commonly associated with alterations in transcription factors because of altered expression or gene mutations. These changes might induce leukemia-specific patterns of histone modifications. We used chromatin-immunoprecipitation on microarray to analyze histone 3 lysine 9 trimethylation (H3K9me3) patterns in primary AML (n = 108), acute lymphoid leukemia (n = 28), CD34+ cells (n = 21) and white blood cells (n = 15) specimens. Hundreds of promoter regions in AML showed significant alterations in H3K9me3 levels. H3K9me3 deregulation in AML occurred preferentially as a decrease in H3K9me3 levels at core promoter regions. The altered genomic regions showed an overrepresentation of cis-binding sites for ETS and cyclic adenosine monophosphate response elements (CREs) for transcription factors of the CREB/CREM/ATF1 family. The decrease in H3K9me3 levels at CREs was associated with increased CRE-driven promoter activity in AML blasts in vivo. AML-specific H3K9me3 patterns were not associated with known cytogenetic abnormalities. But a signature derived from H3K9me3 patterns predicted event-free survival in AML patients. When the H3K9me3 signature was combined with established clinical prognostic markers, it outperformed prognosis prediction based on clinical parameters alone. These findings demonstrate widespread changes of H3K9me3 levels at gene promoters in AML. Signatures of histone modification patterns are associated with patient prognosis in AML.
Introduction
Acute leukemias are associated with transcription factor mutations that often cause specific phenotypes and predict patients' therapy response and prognosis.1,2 The fine tuning of transcription factor activity is crucial for cell fate decisions in normal hematopoiesis.3,4 Importantly, small alterations in transcription factor activity levels can induce leukemia in mouse models.5 Although of potential importance in leukemia development, these small changes in transcription factor activity are not detectable in clinical patient samples. Transcription factors recruit histone-modifying enzymes and alter chromatin structure.6 In contrast to the difficulty in assessing transcription factor activity in clinical samples in vivo, their effects on chromatin modifications are readily accessible by analysis of histone modifications. Indeed, in several cancers including leukemia, epigenetic changes such as posttranslational modifications of histone residues have been recognized at single-gene loci. For example, histone H3 lysine 9 trimethylation (H3K9me3) recruits heterochromatin protein 1 and is associated with heterochromatin.7 In contrast, histone H3 acetylation is generally regarded as a mark for chromatin primed to undergo transcription or for transcriptionally active chromatin.8
Epigenetic changes are thought to play an important role in leukemia pathogenesis.9 The origins of these changes in patients and their association with specific transcription factors have remained obscure in most instances. Nonetheless, the potential importance of these changes is highlighted by the promising activity of several drugs that target epigenetic alterations in leukemia.10 Recent technical advances, such as the hybridization of chromatin-immunoprecipitation on microarray (ChIP-Chip) or deep sequencing, have been applied to cell lines and cells from healthy donors.11,12 In addition, the CALM-AF10 fusion protein has been shown to reduce H3K79 methylation on a global level.13 However, it is still unknown whether and which global changes exist in the epigenome of patients with acute myeloid leukemia (AML). Here, we applied ChIP-Chip to identify global changes of the epigenome in primary blast cells derived from patients with acute leukemia. The well-characterized posttranslational modification histone 3 lysine 9 trimethylation (H3K9me3) was analyzed by ChIP-Chip in a large cohort of AML patients and controls. Leukemia-specific chromatin modification patterns were discovered. The global patterns of epigenome changes allowed disease classification and distinguished between normal hematopoiesis, acute lymphoid leukemia (ALL), and AML. Histone modification changes were associated with specific transcription factor binding sites. This association was further supported by luciferase assays that showed consistent changes in promoter activities. From a clinical perspective, global changes of H3K9me3 were associated with AML patients' prognosis and served as an independent prognostic parameter for event-free survival (EFS) in AML.
Methods
Patient material
Blasts from patients with AML and ALL were obtained at the time of diagnosis (or, in a few cases, at first relapse; Table 1). Informed consent was obtained from all patients in accordance with the Declaration of Helsinki, and the study was approved by the Ethics Committee of the University of Muenster. Two batches of experiments were performed and analyzed separately (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). One group of specimens in the final analysis contained ALL (n = 28) and AML samples (n = 38), and the other one contained AML samples (n = 70), CD34+ progenitor cells (n = 21), and white blood cells (WBCs; n = 15) as controls. Median blast percentage in bone marrow at the time of diagnosis was 76%. Usually, more than 90% of cells were blast cells after enrichment by density centrifugation. Blasts were stored in liquid nitrogen until used and fixed in 1% formaldehyde during thawing. AML patients were treated with standard cytarabine- and daunorubicin-based induction therapy (2 cycles) usually followed by 3 cycles of consolidation therapy with high-dose cytarabine. Older patients (> 60 years) received a second cycle of induction therapy only in the case of blast persistence after cycle 1. Consolidation therapy in older patients consisted of 2 cycles of high-dose cytarabine (1 g/m2). Treatment of patients with acute promyelocytic leukemia (APL) also included all-trans retinoic acid. The ALL patients were treated within the German Childhood Acute Lymphoblastic Leukemia study group protocols. Leukapheresis products were obtained from granulocyte colony-stimulating factor–stimulated patients undergoing harvest for autologous transplantation for a nonleukemia disease. CD34+ progenitor cells were purified with anti-CD34 antibodies by magnetic separation (MACS; Miltenyi Biotec) from leukapheresis products. Purity of CD34+ cells was usually more than 85%. WBCs were obtained from the non-CD34 fraction of the leukapheresis products. After isolation, WBC fractions and CD34+ progenitor cells were fixed with 1% formaldehyde.
ChIP-Chip procedure
Chromatin-IP was carried out as described previously with slight modifications.14 In brief, formaldehyde fixed cells were neutralized with glycine and rinsed with phosphate-buffered saline. Lysates were prepared from 2 × 107 frozen cells, and chromatin was sonicated to an average DNA length of 500 bp. Lysates were precleared and a total of 0.5 mg of protein was used for immunoprecipitation. Immunoprecipitations were performed sequentially for antiacetylated histone H3, anti-HDAC1, and anti-H3K9me3 (all antibodies obtained from Upstate Biotechnology). Antibody quality and suitability for ChIP-Chip were demonstrated previously.14 The antiacetylated histone H3 data and the anti-HDAC1 data will be presented elsewhere. The sequential ChIPs did not affect recovery, and results are as determined in extensive preceding experiments. Nonetheless, this procedure cannot recover bivalently labeled histones. Chromatin-IPs were amplified using ligation-mediated polymerase chain reaction and labeled using Cy3 coupled to incorporated amino-allyl-uridine triphosphate. Common reference DNA consisting of a mixture of genomic DNA from AML patients was prepared, amplified, and labeled in a similar way with Cy5. Hybridization was performed onto custom promoter oligonucleotide arrays with 31 000 independent genomic oligonucleotides covering the promoter regions of more than 10 000 genes. Hybridizations were performed for 18 hours, and slides were scanned using an Axon 4000B scanner. Array data were deposited at Gene Expression Omnibus (National Center for Biotechnology Information) as GSE20452.
Transient transfection and luciferase assays
To study whether the H3K9me3 pattern is associated with corresponding changes in the activity of a cyclic adenosine monophosphate (cAMP) response element (CRE)–controlled promoter, primary CD34+ cells or primary AML blasts were cultured in 6-well plates and transiently transfected with a CRE-controlled Photinus pyralis luciferase reporter gene construct (pGL4.29; Promega) or empty-vector control (5 μg/well) using the Amaxa-Kit (Invitrogen) according to the manufacturer's instructions. Cells were cotransfected with an SV40 enhancer and early promoter element-driven Renilla reniformis luciferase vector (pRL-SV40; 1 μg/well) for normalization of the transfection efficiency. Cells were cocultured in either the presence or absence of forskolin (10−5M), a direct activator of the adenylyl cyclase, to identify effects of a stimulation of the cAMP pathway. Transfected cells were harvested and analyzed after 6 hours. Luciferase assays were performed with the dual-reporter luciferase assay kit (Promega). For all cotransfection studies, firefly luciferase activity was normalized using Renilla luciferase activity.
Bioinformatics
Only arrays of sufficient quality were included in the analyses. The ChIP-Chip channel data were normalized with the variance stabilizing normalization method15 separately for each of the datasets resulting from batch effects. After unsupervised filtering, differences in H3K9me3 levels were analyzed as described in the supplemental data. The false discovery rate (Benjamini-Hochberg corrected) was calculated to adjust for multiple testing. Heatmaps and plots of the large variance principle components were used to visualize the results. In addition, we applied support vector machines to classify samples based on their H3K9me3 data. Accuracy of support vector machines was assessed by leave-one-out cross validation.
We used the Gene Set Analysis (GSA) method by Efron and Tibshirani16 to study the association between histone modifications and the presence of specific transcription factor binding sites. First, we searched for binding sites within the DNA sequences surrounding the oligonucleotides spotted on the arrays based on weight matrices collected in the TRANSFAC database. Subsequently, GSA was used to test whether the oligonucleotides with a putative binding site for a specific transcription factor showed stronger differences in intensity values between AML and CD34+ (AML and ALL) than the rest of the oligonucleotides.
To analyze whether histone modifications were related to EFS in AML patient samples, we developed a risk group predictor according to the method by Bair and Tibshirani17 as implemented in BRB-Array Tools.18 The 3 largest variance principle components of all probes that were significantly correlated with EFS in probe-wise regression models (P < .0005) were computed. The linear predictor of a Cox regression model with these 3 principle components was then used as a continuous risk score. A dichotomized version of the risk predictor was constructed by choosing a threshold such that 40% of the patients (with small risk scores) in the training set fall into the low-risk group and the other 60% into the high-risk group. Test samples were classified based on this threshold. Such classifiers were built with (1) H3K9me3 only, (2) the covariate age, NPM1/FLT3-ITD status, and karyotype, and (3) H3K9me3 plus the covariates as explanatory variables. The classifiers were assessed via leave-one-out cross validation on samples from the AML versus CD34+ dataset. In addition, we used samples from the AML versus ALL dataset to assess classifiers trained on the AML versus CD34+ dataset. Details about the construction and assessment of the classifiers, the training and test samples, and the other bioinformatics methods are given in the supplemental data.
The mRNA gene expression arrays and the consecutive analyses were described previously.19
Results
Identification of histone modification patterns in primary patient specimens
To identify global changes of the histone modification patterns in acute leukemia, we analyzed primary patient samples using ChIP-Chip. The experiments were performed in 2 batches: first, 70 AML, 21 CD34+, and 15 WBCs were analyzed and in a second batch 38 AML and 28 ALL samples. Chromatin with H3K9me3 was immunoprecipitated from primary acute leukemia blasts enriched by density centrifugation and from control samples (patient information in Table 1). After hybridization and scanning of the amplified and labeled chromatin-IPs onto oligonucleotide promoter microarrays, data were normalized and controlled for quality before the final datasets were obtained (supplemental Figure 1).
Profound differences of H3K9me3 histone modification pattern occur in AML promoter regions
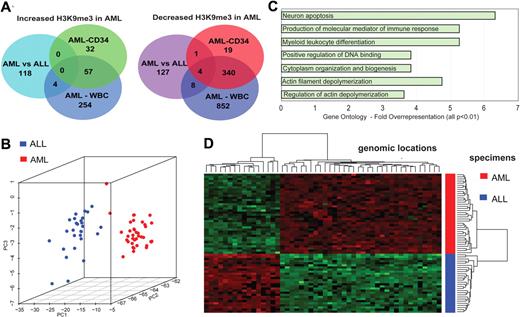
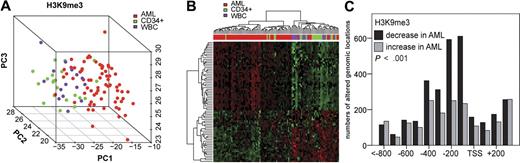
We analyzed differences in global histone modification patterns between AML-ALL as well as between AML-CD34+ progenitor cells and between AML and WBCs (Figure 1A). Class comparison procedures were used with Benjamini-Hochberg correction for multiple testing. At a corrected P < .05, hundreds of promoter regions were identified that differed in H3K9me3 levels between AML blasts, ALL blasts, and controls. The AML versus ALL analysis identified 290 genomic locations that differed in H3K9me3 levels (supplemental Table 1). A more than 2-fold difference was seen for 262 genomic loci with 122 showing an increase of H3K9me3 levels in AML. A Gene Ontology analysis revealed several groups of genes whose promoters showed changes in H3K9me3 levels (Figure 1C). Some of the enriched gene ontology groups, such as immune response and myeloid differentiation, were rather expected, but others were unexpected. According to distinct patterns of H3K9me3 levels at gene promoters, AML and ALL were clearly separated in 2 different groups. Figure 1B depicts a principle component analysis (PCA). Each dot represents one patient sample (38 AMLs and 28 ALLs). A heatmap of hierarchically clustered genomic regions differing in H3K9me3 patterns further showed the differences between AML and ALL specimens (Figure 1D). Alterations were found across all chromosomes (data not shown). Histone modification patterns were not related to patient age (data not shown). Overall, these findings indicate specific patterns of H3K9me3 deposition in AML versus ALL blasts.
ChIP-Chip profiling identifies leukemia-specific loci with altered H3K9me3 loci in leukemia. (A) The Venn diagrams depict the number of genomic loci that were significantly altered in the indicated analyses. Genomic loci included here had a Benjamini-Hochberg corrected P < .05 and were at least 2-fold altered between 2 analyzed groups. The numbers in the overlaps indicate genomic loci that were altered in the same direction in at least 2 analyses. A detailed list of all genomic loci altered in H3K9me3 levels is provided in the supplemental data. (B) PCA identifies distinct patterns of H3K9me3 distribution in AML compared with ALL samples. PC1 is the first principal component, and PC2 and PC3 are the second and third principle components, respectively. (C) Gene Ontology analysis of promoter regions with altered H3K9me3 levels. Among others, regulation of DNA binding and myeloid leukocyte differentiation was overrepresented, indicating that functionally relevant genes were altered in H3K9me3 levels between AML and ALL specimens. (D) Genomic regions differing in H3K9me3 patterns were hierarchically clustered and are indicated in this heatmap. Green represents higher H3K9me3 levels; and red, decreased H3K9me3 levels.
ChIP-Chip profiling identifies leukemia-specific loci with altered H3K9me3 loci in leukemia. (A) The Venn diagrams depict the number of genomic loci that were significantly altered in the indicated analyses. Genomic loci included here had a Benjamini-Hochberg corrected P < .05 and were at least 2-fold altered between 2 analyzed groups. The numbers in the overlaps indicate genomic loci that were altered in the same direction in at least 2 analyses. A detailed list of all genomic loci altered in H3K9me3 levels is provided in the supplemental data. (B) PCA identifies distinct patterns of H3K9me3 distribution in AML compared with ALL samples. PC1 is the first principal component, and PC2 and PC3 are the second and third principle components, respectively. (C) Gene Ontology analysis of promoter regions with altered H3K9me3 levels. Among others, regulation of DNA binding and myeloid leukocyte differentiation was overrepresented, indicating that functionally relevant genes were altered in H3K9me3 levels between AML and ALL specimens. (D) Genomic regions differing in H3K9me3 patterns were hierarchically clustered and are indicated in this heatmap. Green represents higher H3K9me3 levels; and red, decreased H3K9me3 levels.
The global histone H3K9me3 code pattern differs between AML blasts and CD34+ progenitors
Next, we analyzed differences between normal CD34+ hematopoietic progenitor cells and AML specimens in more detail (Figure 2) using similar algorithms. More than 2000 genomic loci were identified that differed in their H3K9me3 intensities between CD34+ (n = 21) progenitors and AML blasts (n = 70) (supplemental Table 2). A total of 453 promoters showed a more than 2-fold change in H3K9me3 levels (Figure 1A). H3K9me3 pattern clearly distinguished between AML and blood or progenitor cells from healthy donors. Separation between CD34+ cells and WBCs was clearly less prominent as visualized in a PCA (Figure 2A). In addition, gene promoters altered at least 2-fold in H3K9me3 levels between CD34+ cells, and AML showed a high overlap with those altered between WBCs and AML blasts (Figure 1A). Hierarchical cluster analysis of genomic regions differing in H3K9me3 revealed specific alterations of chromatin modification patterns according to AML, CD34+ progenitor, or WBC group (Figure 2B). Next, we analyzed whether differences in H3K9me3 levels at gene promoters could be used to distinguish specimen origin. For this purpose, we used support vector machine algorithms with leave-one-out cross-validation. Classification was correct in 100% of the cases (66 of 66) in AML versus ALL. Correct classification of CD34+ and AML samples was predicted in 77% (70 of 91) based on differences in H3K9me3 patterns. Altogether, these data indicated distinct patterns of histone modification between leukemia and normal blood cells. Interestingly, most of the changes occurred in the core promoter regions of the genes; and in many cases, promoters from AML blasts had preferentially lost the H3K9me3 mark in the core promoter region. Figure 2C depicts the altered regions and indicates that the decrease in H3K9me3 levels was centered in the core promoter regions between −300 and the transcriptional start site (TSS; P = .001).
Chromatin modification changes comparing CD34+ hematopoietic progenitors with AML blasts. (A) PCA identifies distinct patterns of H3K9me3 in AML versus CD34+ progenitors and WBCs. (B) Genomic regions differing in H3K9me3 patterns were hierarchically clustered and are indicated in this heatmap. Green represents higher H3K9me3 levels; and red, decreased H3K9me3 levels. (C) Number of genomic locations altered in H3K9me3 levels between AML and CD34+ specimens with regard to their distance from the TSS. A high number of core promoter regions (−400 to TSS) showed decreased H3K9me3 levels in AML.
Chromatin modification changes comparing CD34+ hematopoietic progenitors with AML blasts. (A) PCA identifies distinct patterns of H3K9me3 in AML versus CD34+ progenitors and WBCs. (B) Genomic regions differing in H3K9me3 patterns were hierarchically clustered and are indicated in this heatmap. Green represents higher H3K9me3 levels; and red, decreased H3K9me3 levels. (C) Number of genomic locations altered in H3K9me3 levels between AML and CD34+ specimens with regard to their distance from the TSS. A high number of core promoter regions (−400 to TSS) showed decreased H3K9me3 levels in AML.
Histone modification patterns are associated with specific transcription factor binding sites
Transcription factor complexes induce altered chromatin patterns at the genomic regions to which they bind. The aberrant function of transcription factor complexes at specific genomic regions is thought to be crucial for leukemia pathogenesis.
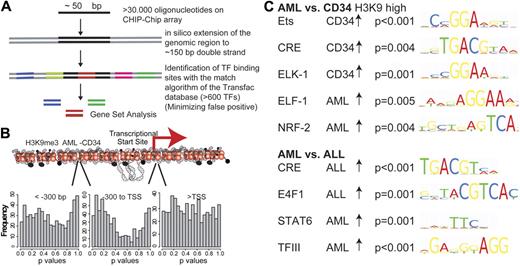
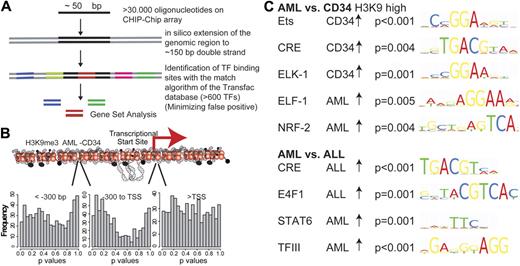
The H3K9me3 alterations in AML are probably the result of locus-specific transcription factor-associated enzymatic activity. We therefore analyzed the association between H3K9me3 level changes and transcription factor binding sites using a novel approach. We searched for transcription factor binding sites in 150-bp genomic sequences that surround the genomic locations of all oligonucleotides present on the microarray (Figure 3). Association of transcription factor binding sites with histone modification changes was analyzed in the entire dataset (AML-CD34+) using GSA. Approximately 600 bona fide transcription factor binding sites (TRANSFAC database, Version 12.0, Biobase) were analyzed by GSA in different promoter regions16 (supplemental data). Because histone modification alterations occurred predominantly in the core promoter regions (Figure 2C), analyses were performed separately for the core promoter region as well as for regions further upstream and downstream. The number of resulting P values in each range was plotted for H3K9me3 in AML versus CD34+ cells (Figure 3B). A shift to lower P values (indicating more significantly associated transcription factor binding sites) was noted in the core promoter region. This finding suggested that transcription factor activity alterations in these regions were reflected by changes in H3K9me3 levels. These changes occurred most prominently in the core promoter region of affected genes.
Association of histone modifications with transcription factor binding sites of different specimen groups. (A) Association of transcription factor binding sites with histone modification changes was analyzed. Overall, 150 bp enclosing each oligonucleotide was screened with high stringency for approximately 600 bona fide transcription factor binding sites. GSA was used to identify associations between transcription factor binding sites and histone modification changes. (B) GSA analyses were performed independently for regions with different distances to the TSSs. Approximately 600 bona fide transcription factor binding sites (TRANSFAC database) were analyzed by GSA in the H3K9me3 dataset of AML versus CD34+ in different promoter regions, and the number of resulting P values in each range was plotted. A shift to lower P values indicates an association between the presence of specific transcription factor binding sites and changes in H3K9me3 levels in the core promoter region. (C) Among the overrepresented transcription factor binding sites, ETS and CRE binding elements were most prominently altered in different datasets. AML specimens consistently showed decreased levels of H3K9me3 at binding sites for most ETS factors and CREB.
Association of histone modifications with transcription factor binding sites of different specimen groups. (A) Association of transcription factor binding sites with histone modification changes was analyzed. Overall, 150 bp enclosing each oligonucleotide was screened with high stringency for approximately 600 bona fide transcription factor binding sites. GSA was used to identify associations between transcription factor binding sites and histone modification changes. (B) GSA analyses were performed independently for regions with different distances to the TSSs. Approximately 600 bona fide transcription factor binding sites (TRANSFAC database) were analyzed by GSA in the H3K9me3 dataset of AML versus CD34+ in different promoter regions, and the number of resulting P values in each range was plotted. A shift to lower P values indicates an association between the presence of specific transcription factor binding sites and changes in H3K9me3 levels in the core promoter region. (C) Among the overrepresented transcription factor binding sites, ETS and CRE binding elements were most prominently altered in different datasets. AML specimens consistently showed decreased levels of H3K9me3 at binding sites for most ETS factors and CREB.
Significant enrichment of specific transcription factor binding sites was observed (Figure 3). Several transcription factor binding sites were identified by comparing H3K9me3 methylation levels between AML and CD34+ cells (Figure 3C). H3K9 trimethylation at ETS-binding sites (P < .001) and at CRE sites (P = .004) was decreased in AML blasts compared with CD34+ cells. Using a different, de novo motif finding algorithm,20 we also identified an ETS-binding motif in the H3K9me3 dataset (P < .002, data not shown). Finally, the decreased, AML-specific H3K9 trimethylation was also recapitulated in the comparison between independent AML and ALL specimens. ETS-binding sites (P = .01) and CREs (P = .004) showed lower levels of H3K9 trimethylation in AML blasts compared with ALL blasts. These experiments indicated that ETS sites as well as CRE elements were consistently altered in AML compared with all other specimens.
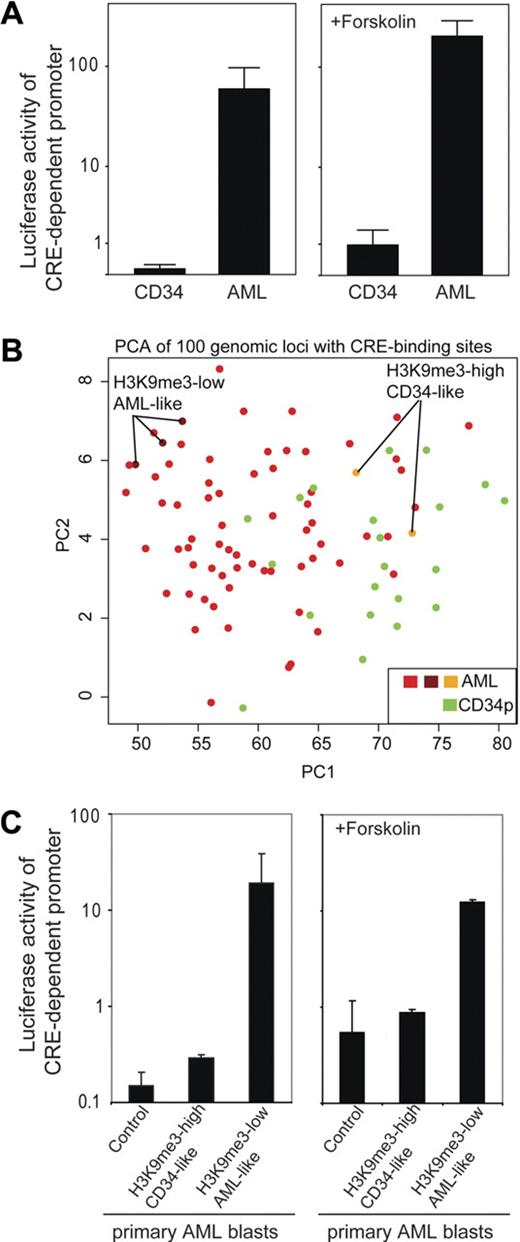
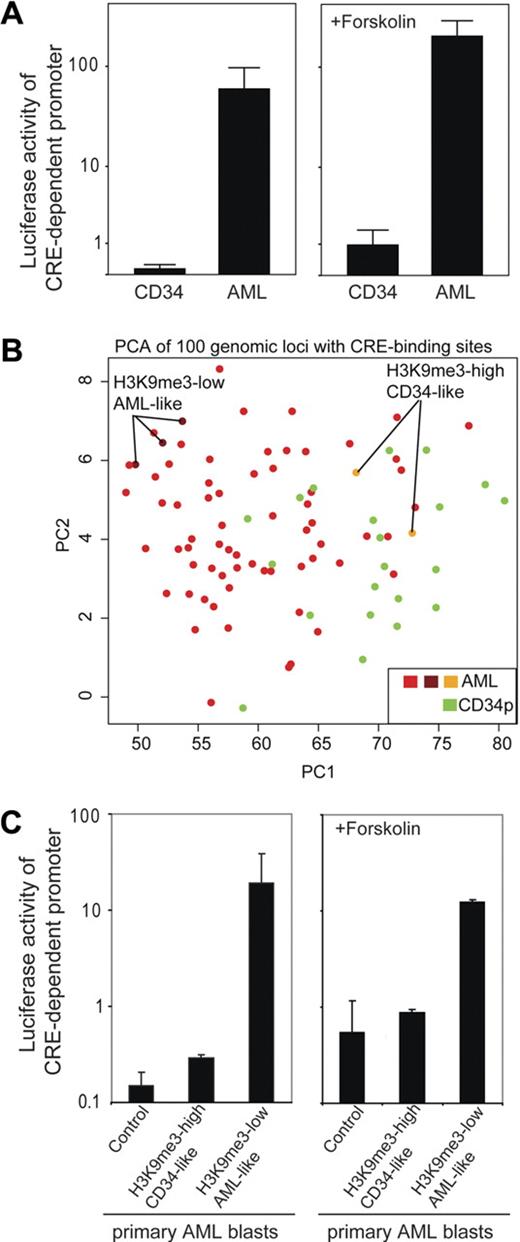
Given that altered H3K9me3 levels at gene promoters could be a result of transcription factor activity, we performed promoter assays to follow up on our findings. Cells were transfected with a CRE-controlled luciferase reporter gene construct, and promoter activity was analyzed. Transfection efficiency as determined in separate experiments using enhanced green fluorescent protein expression vector ranged between 0.8% and 7.5% (data not shown). In all subsequent experiments, a renilla expression vector was used for standardization, and only samples with sufficient renilla luciferase activity were included into the final analysis. Repeated analyses of cells from the same patient showed approximately 2-fold variation (data not shown). In a first experiment, we transiently transfected either CD34+ progenitor cells or primary AML blasts and analyzed CRE-driven promoter activity. Intriguingly, CRE-dependent promoter activity was significantly (P < .01) lower in CD34+ progenitor cells compared with AML cells (Figure 4A left). The higher H3K9me3 levels in CD34+ progenitor cells were in accordance with a lower transcriptional activity of the CRE-controlled promoter in CD34+ cells. The addition of forskolin, an activator of the cAMP-signaling transduction pathway, resulted in an increase of CRE-controlled promoter activity but did not change the observed differences between AML and CD34+ specimens (Figure 4A right). In this limited number of patients, no clear association between CRE-dependent luciferase activity and clinical paramters (eg, French-American-British type, age, or cytogenetics) was found (data not shown). To verify the influence of H3K9me3 pattern on CRE-activity, we separated AML samples because of their H3K9me3 signatures at CREs in vivo (Figure 4B). Next, we tested whether differences of CRE-associated H3K9me3 levels in vivo were associated with CRE-controlled promoter activity. We chose AML specimens either with a CD34+-like H3K9me3 pattern (high H3K9me3 associated with CREs) or a strong AML-like pattern (low H3K9me3 associated with CREs). Interestingly, CRE-driven promoter activity mirrored the estimated activity obtained from its H3K9me3 signature (Figure 4C). Selected AML samples with an H3K9me3 signature similar to CD34+ progenitor cells had very low CRE-activity, whereas AML samples with a very weak H3K9me3 pattern revealed largely higher CRE-driven promoter activity.
Increased CREB-dependent promoter activity in AML blasts in vivo is associated with H3K9me3 levels at CRE-binding sites. (A) Primary CD34+ (n = 3) and AML blasts (n = 10) were transiently transfected with a CRE-driven reporter gene construct (or empty vector as a control; data not shown) and cultured for 6 hours before luciferase activity was determined. Transfection efficiency was normalized to renilla luciferase activity. To analyze the effect of stimulation of the cAMP pathway, forskolin was added after transfection and samples were analyzed accordingly (diagram on the right). In the absence (P < .01) and presence (P < .01) of forskolin, CRE-driven reporter activity was significantly higher in AML blasts. Indicated are mean ± SEM (Mann-Whitney U test). (B) PCA is shown that groups AML and CD34+ progenitor cells according to their H3K9me3 levels at genomic loci with CREs. Indicated are also AML specimens with a more “CD34-like” higher H3K9me3 level at CRE sites and those with “true AML-like” lower H3K9me3 levels at CRE sites. (C) Luciferase reporter assays were performed to identify differences in CRE-driven promoter activities within AML samples. AML samples were selected because of their H3K9me signature either close to the CD34+ pattern or to the AML pattern and analyzed for CRE-dependent promoter activity in transient transfection assays. The samples used for analysis are indicated in Figure 4B. “Control” depicts AML blasts (with AML-like H3K9me3 level) transfected with a promoter-less luciferase construct. AML patients with a CD34-like H3K9me3 pattern showed only minimal CREB-dependent promoter activity, whereas AML blasts with an “AML-like” H3K9me3 pattern showed more than 60-fold higher CREB-dependent promoter activity. Similar analyses carried out in the presence of forskolin to stimulate the cAMP-response pathway are indicated on the right. Indicated are mean ± SD.
Increased CREB-dependent promoter activity in AML blasts in vivo is associated with H3K9me3 levels at CRE-binding sites. (A) Primary CD34+ (n = 3) and AML blasts (n = 10) were transiently transfected with a CRE-driven reporter gene construct (or empty vector as a control; data not shown) and cultured for 6 hours before luciferase activity was determined. Transfection efficiency was normalized to renilla luciferase activity. To analyze the effect of stimulation of the cAMP pathway, forskolin was added after transfection and samples were analyzed accordingly (diagram on the right). In the absence (P < .01) and presence (P < .01) of forskolin, CRE-driven reporter activity was significantly higher in AML blasts. Indicated are mean ± SEM (Mann-Whitney U test). (B) PCA is shown that groups AML and CD34+ progenitor cells according to their H3K9me3 levels at genomic loci with CREs. Indicated are also AML specimens with a more “CD34-like” higher H3K9me3 level at CRE sites and those with “true AML-like” lower H3K9me3 levels at CRE sites. (C) Luciferase reporter assays were performed to identify differences in CRE-driven promoter activities within AML samples. AML samples were selected because of their H3K9me signature either close to the CD34+ pattern or to the AML pattern and analyzed for CRE-dependent promoter activity in transient transfection assays. The samples used for analysis are indicated in Figure 4B. “Control” depicts AML blasts (with AML-like H3K9me3 level) transfected with a promoter-less luciferase construct. AML patients with a CD34-like H3K9me3 pattern showed only minimal CREB-dependent promoter activity, whereas AML blasts with an “AML-like” H3K9me3 pattern showed more than 60-fold higher CREB-dependent promoter activity. Similar analyses carried out in the presence of forskolin to stimulate the cAMP-response pathway are indicated on the right. Indicated are mean ± SD.
H3K9me3 patterns predict event-free survival in AML
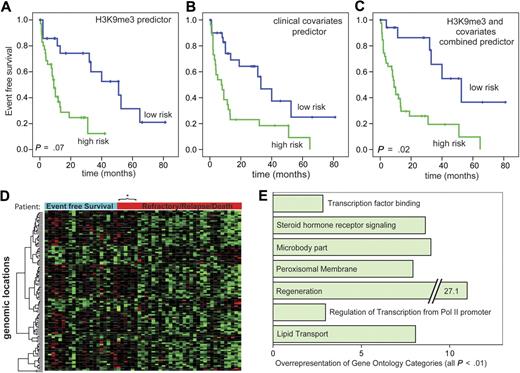
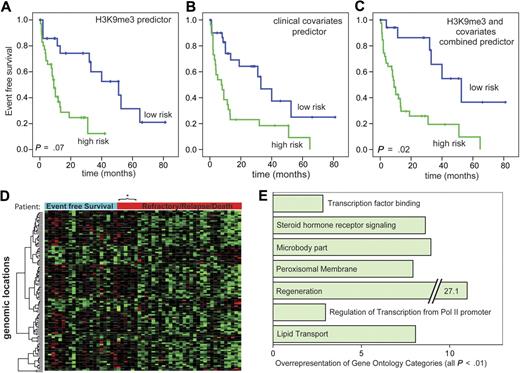
We next analyzed whether specific chromatin modifications are associated with survival in AML at the time of primary diagnosis.17 This analysis included all AML patients at the time of primary diagnosis from the AML versus the CD34+ dataset except those with APL. Median EFS in this group was 13 months with 33% estimated EFS after 3 years. More detailed patient information in the cohorts used for survival analyses can be found in the Supplemental data. For H3K9me3 levels, 91 genomic loci were identified where changes in H3K9me3 levels were significantly associated with patient prognosis at a univariate significance level of P < .001 (supplemental Table 3). A predictor of survival risk groups was developed based on the H3K9me3 signature and assessed by leave-one-out cross-validation (Figure 5A). The permutation P value of the log-rank test statistic between the 2 predicted risk groups based on 100 permutations was .07. Next, a predictor was built based on known clinical parameters that are important for survival in AML. This predictor, based on age, FLT3/NPM1, and karyotype, successfully distinguished between patients with good versus poor prognosis (P < .003, log-rank test; Figure 5B). Finally, we analyzed whether the clinical predictor could be improved by addition of information from the H3K9me3 dataset. Indeed, the combined information from clinical variables and the H3K9me3 signature significantly improved the prediction of prognosis (Figure 5C; P = .02 for statistical test with 100 permutations). These results indicated that the H3K9me3 signature possesses independent power to predict patient prognosis.
A H3K9me3 chromatin signature predicts EFS in AML patients. (A) EFS analyses were performed according to Bair and Tibshirani.17 A predictor was built based on 91 genomic regions with significant association (P < .001) with EFS in 57 AML patients. Overall, 35 patients were predicted to be at high risk and 22 to be at low risk. The predictor separated well between patients with good and poor prognosis. Mean EFS was 43.5 months (low risk) versus 12.4 months (high risk). A permutation test analysis identified the quality of the predictor to be based on chance to be less than 10% (P = .07, permutation test). (B) A clinical predictor for EFS was built based on karyotype, age, and FLT3/NPM1 mutation status, which are the strongest clinical predictors for survival in AML patients. For this predictor, 26 and 31 patients were predicted to be at high and low risk, respectively. As expected, this predictor also separated well between surviving and nonsurviving patients. Mean EFS was 42.2 versus 17.5 months. (C) A combined predictor was built from H3K9me3 and the clinical covariates. For this predictor, 39 and 18 patients were predicted to be at high and low risk, respectively. This predictor predicted the outcome better than the clinical predictor (P = .02, permutation test). The EFS times of the predicted groups were 53.9 versus 17.2 months for the low-risk and the high-risk groups, respectively. (D) A heatmap of genomic loci whose H3K9me3 level were associated with EFS. The columns represent patient specimens that were ordered from the left to the right according to their EFS status and the time of censoring or event. Patients marked with the blue bar “Event Free Survival” were alive without relapse at the time of analysis. The bar in red indicates patients with relapse, refractory disease, or death. These were arranged with the longest EFS time on the left. Gene loci in rows were hierarchically clustered for H3K9me3 levels (red indicates low-level H3K9me3; and green, high-level H3K9me3). Two large clusters are visible with the group of good prognosis H3K9me3 patterns extending to 6 additional patients in the other group. *Each of these patients experienced an event more than 3 years after initial diagnosis indicating a generally more favorable prognosis. (E) Gene Ontology analysis of genes whose level of H3K9 trimethylation was closely associated with patient survival in the combined predictor. Indicated are Gene Ontology categories with at least 2.5-fold overrepresentation. All changes were statistically significant with P < .01.
A H3K9me3 chromatin signature predicts EFS in AML patients. (A) EFS analyses were performed according to Bair and Tibshirani.17 A predictor was built based on 91 genomic regions with significant association (P < .001) with EFS in 57 AML patients. Overall, 35 patients were predicted to be at high risk and 22 to be at low risk. The predictor separated well between patients with good and poor prognosis. Mean EFS was 43.5 months (low risk) versus 12.4 months (high risk). A permutation test analysis identified the quality of the predictor to be based on chance to be less than 10% (P = .07, permutation test). (B) A clinical predictor for EFS was built based on karyotype, age, and FLT3/NPM1 mutation status, which are the strongest clinical predictors for survival in AML patients. For this predictor, 26 and 31 patients were predicted to be at high and low risk, respectively. As expected, this predictor also separated well between surviving and nonsurviving patients. Mean EFS was 42.2 versus 17.5 months. (C) A combined predictor was built from H3K9me3 and the clinical covariates. For this predictor, 39 and 18 patients were predicted to be at high and low risk, respectively. This predictor predicted the outcome better than the clinical predictor (P = .02, permutation test). The EFS times of the predicted groups were 53.9 versus 17.2 months for the low-risk and the high-risk groups, respectively. (D) A heatmap of genomic loci whose H3K9me3 level were associated with EFS. The columns represent patient specimens that were ordered from the left to the right according to their EFS status and the time of censoring or event. Patients marked with the blue bar “Event Free Survival” were alive without relapse at the time of analysis. The bar in red indicates patients with relapse, refractory disease, or death. These were arranged with the longest EFS time on the left. Gene loci in rows were hierarchically clustered for H3K9me3 levels (red indicates low-level H3K9me3; and green, high-level H3K9me3). Two large clusters are visible with the group of good prognosis H3K9me3 patterns extending to 6 additional patients in the other group. *Each of these patients experienced an event more than 3 years after initial diagnosis indicating a generally more favorable prognosis. (E) Gene Ontology analysis of genes whose level of H3K9 trimethylation was closely associated with patient survival in the combined predictor. Indicated are Gene Ontology categories with at least 2.5-fold overrepresentation. All changes were statistically significant with P < .01.
These findings were validated using the AML specimens from the second batch of experiments as an independent test set. The test set contained 29 AML patients (no patients with APL) with defined genetic abnormalities inv(16) (n = 8), t(8;21) (n = 8), and complex karyotype (n = 13). In this group of patients, median EFS was 9 months with 37% being estimated to experience EFS after 3 years. We analyzed whether patients' prognosis prediction was improved by the combined predictor (H3K9me3 + clinical variables) versus the clinical predictor based on the clinical variables karyotype, age, NPM1, and FLT3-ITD mutation. The clinical risk predictor classified all samples with complex karyotype as high risk (n = 13) and the remaining samples as low risk (n = 16), whereas the combined model correctly predicted 2 additional core-binding factor leukemia patients, one each with inv(16) and t(8;21), to have a poor prognosis. In line with the results from the first batch of experiments, the risk group predictions of the combined model achieved a higher hazard ratio (8.9 vs 6.3) in a Cox regression model and so outperformed the clinical predictor. Nonetheless, karyotype remained the major explanatory factor for EFS in this dataset.
A heatmap of the H3K9me3 signature used for risk prediction indicated 2 major clusters of patient samples (Figure 5D). A Gene Ontology analysis showed that differing H3K9me3 levels at promoters of genes involved in gene regulation were overrepresented. Among other promoters overrepresented were those controlling genes involved in regeneration, transcription factor binding, and regulation of transcription from polymerase II–dependent promoters were overrepresented (Figure 5E).
Discussion
A detailed understanding of epigenetic alterations is important to further elucidate the pathogenesis of cancers. This might be particularly true in AML, where only a limited number of genetic mutations have been discovered.21 Epigenetic alterations are increasingly recognized as important contributors to human cancer pathogenesis.22 However, no global scale analyses of epigenome alterations exist for large numbers of clinical specimens in AML. We have demonstrated in our study that it is feasible to analyze primary patient specimens in sufficient numbers to be clinically relevant. Such studies offer new avenues for tackling the importance of global histone modification patterns in human cancer.
The multiple determinants of the histone code offer the cell an opportunity to regulate transcription patterns on multiple levels, a process we can only begin to understand by its genome-wide analyses in different physiologic and pathologic states. No significant association was found between H3K9me3 levels and gene expression levels (data not shown). This is in line with recent reports.23 Indeed, one recent report even described an increase of H3K9me3 at transcriptionally active promoters.24 At the core promoter region, AML specimens often showed decreased H3K9me3 levels compared with CD34+ hematopoietic progenitors and compared with ALL specimens. One possible explanation could be that an increase in transcriptional activity would lead to a decrease in nucleosome density. However, as stated above, the association between histone modifications and mRNA levels was weak. In addition, only a small fraction of genomic regions were significantly altered for H3K9me3 at the same time despite widespread mRNA expression.
Transcription factor alterations play an important role in AML pathogenesis. These are in some instances caused by genetic mutations but probably more often by regulatory mechanisms.25 Transcription factor activity is differentially regulated at specific binding sites by posttranslational modifications and protein-protein modifications as well as by transcriptional and translational control.26,27 Recently, it has been shown that the MLL-AF4 oncogene can specifically induce a H3K79me2 pattern in a murine leukemia model.28 Changes in H3K79me2 were also found in MLL-AF4 samples from ALL patients. Thus, the chromatin modification patterns that we describe here are also probably induced by disturbances in transcription factor activity. Thus, our data may offer a first comprehensive view on the functional consequences of altered transcription factor activity in AML. As a surrogate, ChIP-Chip analyses of resulting chromatin modifications might be usable as readout for the activity level of specific transcription factors in primary patient specimens.
AML-specific chromatin signatures were particularly associated with histone modification patterns at ETS and CRE-binding sites. ETS binding sites are characterized by a GGAA/T core motif and mediate DNA binding of a variety of transcription factors. Importantly, several of these factors, such as ERG-1, FLI-1, ELF-1, and others, play crucial roles in determining hematopoietic cell fate and shaping hematopoiesis. Deletion mutants of ETS factors lead to severe hematopoietic phenotypes. Importantly, several of these factors play important roles in leukemogenesis. In addition, ERG-1 expression levels are also of prognostic value in AML. Hence, it is not unexpected that H3K9me3 patterns in AML blasts appear to reflect altered ETS-factor activity. In contrast to ETS sites, the number of factors binding to CRE sites (TGACGTCA or variants thereof) is lower. In line with our findings, CREB has recently been shown to be an important player in hematopoiesis and leukemia.29 Furthermore, our CREB-luciferase experiments revealed both increased CRE-driven promoter activity in AML samples compared with CD34+ progenitor cells as well as different promoter activities within AML-samples associated with the H3K9me3 signature. Despite multiple genetic and epigenetic mutations that are found in human cancer, it appears that a few relevant pathways are consistently altered and act in a synergistic way. The prominent role of ETS factors and CREB/CREM/ATF transcription factors in the AML specific histone signature suggests that different pathogenetic events in AML might finally converge in deregulation of these transcription factor families.
Importantly, histone modification patterns predicted patients' prognosis. H3K9me3 patterns were closely associated with patients' EFS. In multivariate analyses, the H3K9 modification signature improved prognosis prediction independent from karyotype, age, and NPM1/FLT3 mutations. This might indicate that specific histone modifications are associated with certain disease features, and our data indicate that chromatin signatures can serve as biomarkers in cancer and leukemia. In this regard, it is interesting to mention that H3K9me3 patterns did not differ significantly between different cytogenetic subtypes (data not shown). In addition, no association was found between the H3K9me3 survival signature and the percentage of blasts in the bone marrow at the time of diagnosis (data not shown). The survival analyses were initially performed in a set of patients with low numbers of patients with balanced translocations. Analyses of the second set that contained samples from patients with t(8;21), t(15;17), inv(16), and complex karyotype also showed only weak differences in H3K9me3 levels between the cytogenetic groups. Most probably, H3K9me3 alterations do occur with regard to disease type (AML vs ALL vs CD34+ cells or WBCs). But we did not detect a clear association with cytogenetics. These findings are well recapitulated by the further finding that the prognostic power of H3K9me3 levels occurred independent of cytogenetic parameters or other established clinical features. Taken together, these analyses identify disease-specific epigenome changes in acute leukemia.
Our data suggest that specific patterns of H3K9me3 levels exist in AML. Alterations in H3K9me3 levels are probably the result of altered transcription factor activity. Importantly, H3K9me3 patterns are closely associated with patients' prognosis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank members of our laboratories for helpful discussions and the members of the AMLCG, Childhood Acute Lymphoblastic Leukemia, and Study Alliance Leukemia study groups for providing patient samples. Parts of the bioinformatics analyses were performed using BRB-ArrayTools developed by Dr Richard Simon and Amy Peng Lam.
This work was supported by the NGFN-Plus LeukemiaNet (GS010873), the José-Carreras Leukämiestiftung (R06/39f), the Deutsche Krebshilfe (Onconet2), Wilhelm-Sander Stiftung (2007.048.1), and the Interdisciplinary Center for Clinical Research (Mü2/018/07) at the University of Münster. F.I. was supported by a fellowship from the José-Carreras Leukämiestiftung. M.M. and Y.W. were supported by National Institutes of Health grants R01 CA068822 and U01 CA0114810 and Department of Defense grant W91XWH-06-1–0253.
National Institutes of Health
Authorship
Contribution: C.M.-T., H.-U.K., M.D., and H.S. designed research; C.M.-T., H.-U.K., C.D., A.H., F.I., L.T., N.T., S.A.-S., A.B., S.K., M.S., F.U.M., P.T., W.S., M.D., and H.S. performed research and analyzed the data; A.B., Y.W., M.M., C.T., G.E., P.S., and U.z.S. provided vital new reagents; and C.M.-T., H.-U.K., M.D., Y.W., M.M., W.E.B., and H.S. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A complete list of Study Alliance Leukemia participants appears in the online Appendix.
Correspondence: Carsten Müller-Tidow, Department of Medicine A, Hematology and Oncology, University of Münster, Domagkstr 3, 48129 Münster, Germany; e-mail: muellerc@uni-muenster.de.
References
Author notes
C.M.-T., H.-U.K., M.D., and H.S. contributed equally to this study.