Abstract
The ability of natural killer (NK) cells to kill malignant or infected cells depends on the integration of signals from different families of cell surface receptors, including cytokine receptors. How such signals then regulate NK-cell cytotoxicity is incompletely understood. Here we analyzed an endogenous inhibitor of protein phosphatase 2A (PP2A) activity called SET, and its role in regulating human NK-cell cytotoxicity and its mechanism of action in human NK cells. RNAi-mediated suppression of SET down-modulates NK-cell cytotoxicity, whereas ectopic overexpression of SET enhances cytotoxicity. SET knockdown inhibits both mRNA and protein granzyme B expression, as well as perforin expression, whereas SET overexpression enhances granzyme B expression. Treatment of NK cells with the PP2A activator 1,9-dideoxy-forskolin also inhibits both granzyme B expression and cytotoxicity. In addition, pretreatment with the PP2A inhibitor okadaic acid rescues declining granzyme B mRNA levels in SET knockdown cells. Down-modulation of SET expression or activation of PP2A also decreases human NK-cell antibody-dependent cellular cytotoxicity. Finally, the induction of granzyme B gene expression by interleukin-2 and interleukin-15 is inhibited by SET knockdown. These data provide evidence that granzyme B gene expression and therefore human NK-cell cytotoxicity can be regulated by the PP2A-SET interplay.
Introduction
Human natural killer (NK) cells are CD56+CD3− large granular lymphocytes that participate in immune responses against infection or malignant transformation through production of cytokines and chemokines, and via spontaneous and antibodyantibody-dependent cellular cytotoxicity (ADCC) of target cells that lack matching major histocompatibility complex.1,2
Human NK cells rapidly kill pathogen infected or tumor cells via different mechanisms, including Fas-L, TRAIL, and granule-exocytosis dependent pathways.3,4 The secretion of cytotoxic granules is the main mechanism by which NK cells lyse target cell populations and depends on the expression of perforin and granzyme molecules. The pore-forming protein perforin allows the entry of granzymes into target cells where these proteases induce cell death. Among granzymes, granzyme A and granzyme B are the most abundant. Granzyme B plays a critical and nonredundant role in the rapid killing of susceptible target cells by both human and mouse NK cells.5-7 It has been shown that interleukin-2 (IL-2) stimulation of primary NK cells induces an enhancement of granzyme B expression by inducing its transcription.8,9
The SET (I2PP2A, IGAAD, and TAF1b) protein plays a critical role in the regulation of normal and cancer signal transduction.10 Indeed, SET is a potent inhibitor of the major cellular serine threonine phosphatase protein phosphatase 2A (PP2A).11,12 In addition, it has also been described as an inhibitor of the tumor suppressor NM23-H1, a granzyme A DNAse-activated factor,13 and a negative regulator of histone acetylation.14 PP2A regulates a variety of cellular processes and signal transduction pathways.10,15,16 The role of PP2A in the expansion and activation of NK cells is not fully defined. In vivo administration of the PP2A inhibitor cytostatin in mice inhibits pulmonary metastasis of B16 melanoma, possibly via the up-regulation of cytokines that are important for NK-cell development and/or activation.17 In vitro, primary human NK cells treated with the PP2A inhibitor okadaic acid have enhanced spontaneous cellular cytotoxicity.18 Recently, we reported the SET-PP2A interplay as a novel pathway that regulates monokine-induced interferon-γ (IFN-γ) production by human NK cells.19 Indeed, monokines, such as IL-15, IL-12, and/or IL-18, can induce both mRNA and protein expression of SET. SET functionally inactivates PP2A in NK cells. Subsequently, we discovered that endogenous protein SET modulates human NK-cell IFN-γ production at least in part via PP2A after monokine stimulation. In the current study, we assess the role of the PP2A-SET interplay in the regulation of human NK-cell cytotoxicity.
Methods
Cells lines and NK-cell preparations
The human IL-2–dependent NK cell-line NK-92 (gift of Dr H. Klingemann, Rush Cancer Center, Chicago, IL) was maintained in culture in RPMI 1640 medium (Invitrogen), supplemented with 20% heat-inactivated fetal bovine serum (FBS; Invitrogen), 2mM l-glutamine, and 150 IU/mL rhIL-2 (Hoffman-LaRoche). The amphotropic-packaging cell line 293T was maintained in culture in Dulbecco modified Eagle medium (Invitrogen)/10% FBS medium and grown for 16 to 18 hours to 80% confluence before transfection by calcium phosphate-DNA precipitation (Profection system, Promega). Human NK cells were isolated from peripheral blood leukopacks of healthy persons (American Red Cross) by incubation for 30 minutes with RosetteSep NK-cell antibody cocktail (Stem Cell Technologies), followed by Ficoll-Hypaque density gradient centrifugation. NK-cell preparations containing more than 98% CD56+ NK cells were obtained by positive selection using CD56 MicroBeads and MACS Separation Columns from Miltenyi (Miltenyi Biotec), as determined by direct immunofluorescence using an anti-CD56 phycoerythrin (PE)-conjugated monoclonal antibody (Immunotech). All work with human materials was approved by the Institutional Review Board of the Ohio State University Comprehensive Cancer Center.
Intracellular staining by flow cytomety
PSUPER and pSUPER.retro-shSET NK-92 cells were fixed, permeabilized, and washed using a BD Cytofix/Cytoperm kit (BD Biosciences) and stained with an allophycocyanin-conjugated mouse monoclonal antibody to human granzyme B (clone GB12) or IgG1 allophycocyanin isotype control antibody from Invitrogen. Samples were acquired using a FACSCalibur (BD Biosciences) and analyzed with FlowJo software v7.6.1 (TreeStar).
Retroviral and lentiviral infection of the NK-92 cell line and primary human NK cells
The generation of pSUPER.retro-shSET retroviral vector was previously described.19 To generate pNaldini-GFP-SET, the human SET cDNA was obtained from MigRI-FLAG SET12 by polymerase chain reaction (PCR), using a 5′ primer containing BamHI restriction site, the ATG codon, and the nucleotides encoding for the FLAG epitope, and a downstream primer that includes the last nucleotides of SET cDNA linked to a BamHI restriction site. After BamHI digestion, the FLAG-SET product was cloned in the BamHI digested pNaldini-CMV-IRES-EGFP.20 For infection, the NK-92 cell line and primary NK cells were first incubated overnight in the presence of 300 IU/mL IL-2 and then followed by previously published standards.21 Briefly, infectious supernatant from pSUPER, pSUPER.retro-shSET, pNaldini, and pNaldini-GFP-SET retrovirally transfected 293 T cells were collected after 48 hours and used for 3 cycles of infections. On infection, NK-92 cells and CD56+ NK cells were cultured for several days and then sorted (FACSVantage and FACSAria II cytometer, BD Biosciences) for green fluorescent protein (GFP) expression. GFP+CD56+ primary NK cells were used for experimentation 18 hours after sorting and NK-92 cells were used 48 hours after IL-2 starvation. Expression of SET mRNA and protein was confirmed by real-time reverse-transcription (RT)–PCR and Western blot. To generate NK-92 cells expressing CD16, Phoenix cells were transfected with PINCO or PINCO-CD16 retrovirus, and their supernatants were used for infection of NK92 cells, as previously described.22 CD16 surface expression was confirmed by flow cytometry using a biotinylated anti-CD16 antibody and streptavidin-PE conjugate (BD Biosciences), an allophycocyanin-Cy7–conjugated anti-CD16 antibody (BD Biosciences), or a PE-conjugated anti-CD16 antibody (Immunotech).
Cell culture conditions
Before cytokine stimulation, NK-92 cells were cultured in IL-2–free medium containing 10% FBS for 36 hours. Cells were next incubated in medium plus 10% FBS at 37°C (1 × 106/mL) for the indicated times with the addition of IL-15 (100 ng/mL, kindly provided by Amgen) or IL-2. Where noted, the PP2A activator 1,9-dideoxy-forskolin (Sigma-Aldrich), the PP2A inhibitor okadaic acid (Calbiochem), or dimethyl sulfoxide (DMSO) vehicle control was added for 24 hours at 37°C.
Western blot analysis
Cells were harvested, washed once with ice-cold phosphate-buffered saline and lysed (108 cells/mL RIPA buffer: 0.15M NaCl, 1% NP-40, 0.1% sodium dodecyl sulfate, 50mM Tris [pH 8.0], supplemented with protease and phosphatase inhibitors, 1mM phenylmethylsulfonylfluoride, 1mM Na3VO4, 50mM NaF, 10mM β-glycerol-phosphate, 1mM ethylenediaminetetraacetic acid, and a protease inhibitor cocktail tablet from Roche Applied Science) as described.23 Alternatively, cells were directly lysed in Laemmli buffer (2 × 105 cells/20 μL). Western blotting was performed according to previously published protocols,24 and antibody-reactive proteins were detected with horseradish peroxidase-labeled sheep anti–rabbit, mouse, and/or goat Ig sera and enhanced chemiluminescence (GE Healthcare). Proteins were analyzed in 4% to 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Bio-Rad) using reducing conditions. Monoclonal and polyclonal antibodies used were: rabbit polyclonal anti-SET (I2PP2A) from Globozymes, the anti-actin from Santa Cruz Biotechnology, rabbit polyclonal anti-granzyme B antibody from Cell Signaling Technology, monoclonal anti-perforin antibody from Abcam, and monoclonal anti-GRB2 antibody from Transduction Laboratories.
Real-time RT-PCR
Total mRNA was extracted using RNeasy Mini kits (QIAGEN), and cDNA was generated according to the manufacturer's recommendations (Invitrogen). Real-time RT-PCR reactions were performed with the primer/probe set specific for the human SET and granzyme B transcripts, and 18S as control as previously described.19,22 Reactions were performed using an ABI 7900 HT sequence detector (TaqMan; PE Applied Biosystems), and data were analyzed with the Sequence Detector Version 2.3 software to establish the PCR cycle at which the fluorescence exceeded a set threshold, CT, for each sample. Data were analyzed according to the comparative CT method using the internal control 18S transcript levels to normalize differences in sample loading and preparation. Results represent the n-fold difference of transcript levels in a particular sample compared with calibrator cDNA (cDNA samples of unstimulated, DMSO vehicle-treated or control vector-infected NK-92 or primary NK cells). Results are expressed as the mean plus or minus SEM of triplicate reaction wells.
Cytotoxicity assays
Before the assay, primary human NK effector cells were cultured in IL-2–free medium containing 10% FBS for 18 hours and NK-92 effector cells for 36 hours. When indicated, 1,9-dideoxy-forskolin was used at the indicated times and doses. K562 or P815 coated with an anti–mouse lymphocyte rabbit antibody (Accurate Chemical and Scientific) was used as target in a 4-hour 51Cr release assay.25 A constant number of target cells and serial dilution of effectors cells were used in triplicate. Spontaneous release was always less than 10%.
Statistics
Comparisons between 2 groups were performed using the 2-sided paired t test or linear mixed model to account for the correlation among replicates from the same donor. A P value less than .05 was considered statistically significant.
Results
Role of SET in the regulation of NK cell-mediated cytotoxicity in primary human NK cells
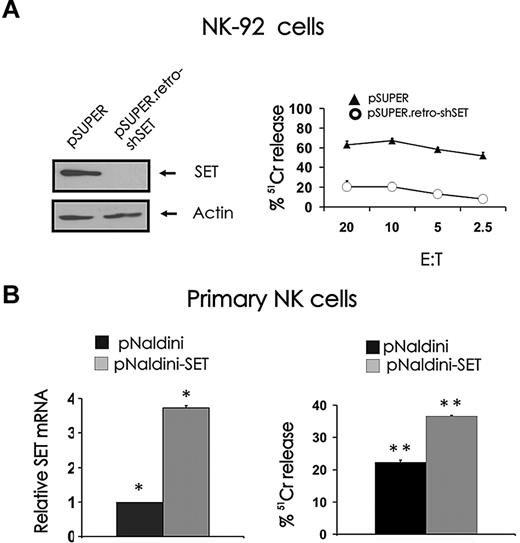
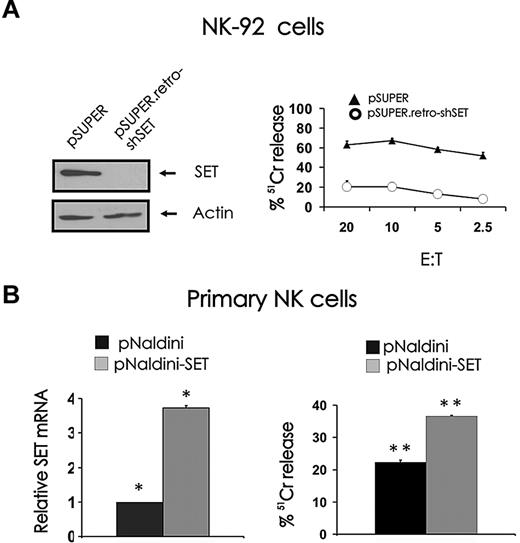
In our study characterizing the role of the PP2A inhibitor SET in the regulation of human NK-cell IFN-γ expression, we performed an experiment with the NK-92 cell line, which suggested that SET may have a role in NK-cell cytotoxicity.19 Indeed, down-modulation of SET by RNA interference using pSUPER.retro-shSET carrying short hairpin RNA corresponding to nucleotides 120 to 138 of human SET diminishes spontaneous cytotoxicity of NK-92 cells against K562 tumor cells (Figure 1 right).19 To determine whether the PP2A inhibitor SET could serve as a positive regulator of NK-cell cytotoxicity, we overexpressed SET in primary human CD56+ NK cells using pNaldini lentivirus vector coexpressing SET and GFP cDNAs. The ability of the vector to induce the expression of SET protein was first validated in 293T and NK-92 cell lines by Western blot (data not shown). GFP-sorted primary human NK cells infected with pNaldini-GFP-SET were tested for SET mRNA overexpression by real-time RT-PCR (Figure 1B left) and used as effectors to kill tumor K562 target cells in a 51Cr release assay (Figure 1B right). Because of the low number of primary NK cells recovered after GFP sorting, a single effector/target ratio was used for the assay. In 3 separate experiments, primary NK cells that overexpressed SET killed K562 cells with 34.6% plus or minus 4.6% higher efficiency compared with primary NK cells infected with pNaldini control vector (P < .03).
Effect of SET knockdown or overexpression on NK cell–mediated cytotoxicity. (A) NK-92 cells retrovirally infected using the pSUPER or pSUPER.retro-shSET vector were starved from IL-2 for 48 hours, analyzed for SET protein expression by Western blot (left), and used as effector cells for NK-cell cytotoxicity assays19 (right). Technical limitations precluded infection of primary human NK cells using the pSUPER or pSUPER.retro-shSET vector. (B) Primary human CD56+ NK cells were infected using pNaldini-GFP and pNaldini vector encoding both GFP and SET cDNAs, and next FACS sorted for GFP. Sorted primary human CD56+GFP+ NK cells were analyzed for SET transcript by real-time RT-PCR (left; n = 4, *P < .01) and used as effectors for NK-cell cytotoxicity assay (right; n = 3, **P < .03). Spontaneous cytotoxicity against K562 cells was tested in a 4-hour 51Cr release assay. This experiment is representative of 3 performed with similar results. Errors bars represent plus or minus SE.
Effect of SET knockdown or overexpression on NK cell–mediated cytotoxicity. (A) NK-92 cells retrovirally infected using the pSUPER or pSUPER.retro-shSET vector were starved from IL-2 for 48 hours, analyzed for SET protein expression by Western blot (left), and used as effector cells for NK-cell cytotoxicity assays19 (right). Technical limitations precluded infection of primary human NK cells using the pSUPER or pSUPER.retro-shSET vector. (B) Primary human CD56+ NK cells were infected using pNaldini-GFP and pNaldini vector encoding both GFP and SET cDNAs, and next FACS sorted for GFP. Sorted primary human CD56+GFP+ NK cells were analyzed for SET transcript by real-time RT-PCR (left; n = 4, *P < .01) and used as effectors for NK-cell cytotoxicity assay (right; n = 3, **P < .03). Spontaneous cytotoxicity against K562 cells was tested in a 4-hour 51Cr release assay. This experiment is representative of 3 performed with similar results. Errors bars represent plus or minus SE.
The effect of SET on the regulation of granzyme B expression in NK cells
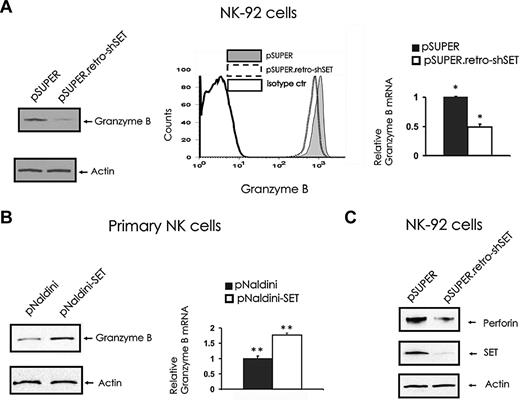
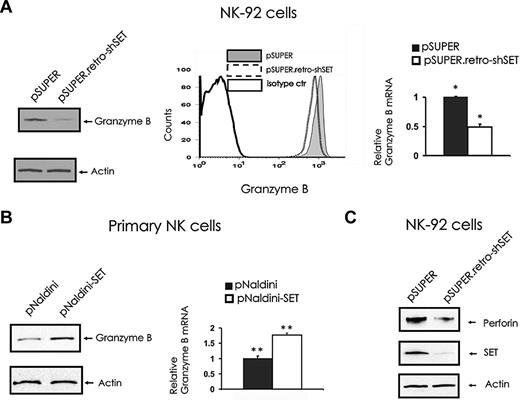
To elucidate the molecular mechanism by which SET regulates NK-cell cytotoxicity, we assessed the levels of lytic granule proteins in the NK-92 cells with SET knockdown. The analysis of granzyme B levels in pSUPER retro-shSET cells compared with pSUPER vector control cells indicated that SET knockdown down-modulates granzyme B expression at both protein (Figure 2A left and middle) and mRNA (Figure 2A right) levels. To confirm the role of SET in regulating granzyme B expression, we overexpressed SET in primary human CD56+ NK cells after infection with pNaldini-GFP-SET, and checked for mRNA and protein levels of granzyme B (Figure 2B). Importantly, primary NK cells overexpressing SET after infection with pNaldini-GFP-SET express significantly higher levels of granzyme B at both mRNA and protein levels compared with control vector-infected cells (P = .032). Of interest, the NK-92 cell line with SET knockdown after infection with pSUPER retro-shSET also showed down regulation of the perforin protein compared with the same cells infected with the pSUPER vector control (Figure 2C). Notably, fluorescence-activated cell sorter (FACS) analysis of NKp30, NKp46, and NKG2D did not reveal any changes in their surface expression in the presence or absence of SET knockdown (data not shown).
The effect of SET knockdown or overexpression on granzyme B and perforin expression. (A) pSUPER or pSUPER.retro-shSET NK-92 cells were starved from IL-2 for 48 hours and analyzed for granzyme B protein expression by Western blot (left), intracellular flow cytometry (middle), and granzyme B transcript by real-time RT-PCR (right; n = 6). *P < .01. (B) Primary human CD56+ NK cells sorted for GFP after lentivirus infection using pNaldini-GFP and pNaldini-GFP-SET vectors were cultured overnight without IL-2 and analyzed for granzyme B protein by Western blot (left) and granzyme B mRNA expression by real-time RT-PCR (right; n = 7). **P = .03. In data not shown, primary human NK cells were also retrovirally infected using PINCO-SET vector, and an 8-fold increase in granzyme B mRNA relative to identical cells infected with PINCO alone was noted. (C) pSUPER or pSUPER.retro-shSET NK-92 cells were starved from IL-2 for 48 hours and analyzed for perforin protein expression by Western blot. Each experiment is representative of at least 3 performed with similar results.
The effect of SET knockdown or overexpression on granzyme B and perforin expression. (A) pSUPER or pSUPER.retro-shSET NK-92 cells were starved from IL-2 for 48 hours and analyzed for granzyme B protein expression by Western blot (left), intracellular flow cytometry (middle), and granzyme B transcript by real-time RT-PCR (right; n = 6). *P < .01. (B) Primary human CD56+ NK cells sorted for GFP after lentivirus infection using pNaldini-GFP and pNaldini-GFP-SET vectors were cultured overnight without IL-2 and analyzed for granzyme B protein by Western blot (left) and granzyme B mRNA expression by real-time RT-PCR (right; n = 7). **P = .03. In data not shown, primary human NK cells were also retrovirally infected using PINCO-SET vector, and an 8-fold increase in granzyme B mRNA relative to identical cells infected with PINCO alone was noted. (C) pSUPER or pSUPER.retro-shSET NK-92 cells were starved from IL-2 for 48 hours and analyzed for perforin protein expression by Western blot. Each experiment is representative of at least 3 performed with similar results.
Evidence that SET regulates cytotoxicity and granzyme B expression by inhibiting the negative regulatory effects of endogenous PP2A
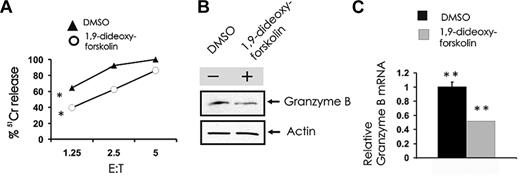
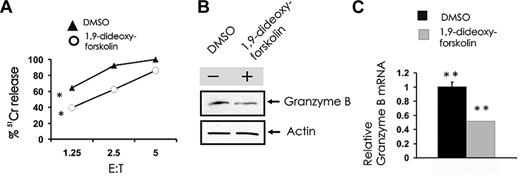
SET functionally inhibits the serine-threonine phosphatase PP2A in human NK cells,19 and we postulated that this in turn regulates granzyme B expression. To test our hypothesis, we used the known activator of PP2A, 1,9-dideoxy-forskolin. Primary CD56+ NK cells were treated with vehicle or a dose of 1,9-dideoxy-forskolin that activates PP2A in NK cells,19 which were then used as effectors in a cytotoxicity assay and were also tested for the expression of granzyme B; 1,9-dideoxy-forskolin was able to significantly inhibit NK-cell cytotoxicity against K562 cells (Figure 3A) and also inhibited granzyme B expression in primary human NK cells at the protein (Figure 3B) and mRNA levels (Figure 3C).
Pharmacologic induction of PP2A activity inhibits spontaneous NK cell–mediated cytotoxicity and granzyme B expression. (A) Primary human NK cells were first incubated (18 hours) with medium containing either DMSO vehicle control or 40μM 1,9-dideoxy-forskolin and then used as effector cells in a 51Cr release assay using K562 tumor cell targets (n = 4). *P < .03. Cell pellets from DMSO or 1,9-dideoxy-forskolin-treated cells were collected for quantification of granzyme B protein by Western blot (B) and granzyme B transcript by real-time RT-PCR (C; n = 3). **P < .03. Each experiment is representative of at least 3 performed with similar results.
Pharmacologic induction of PP2A activity inhibits spontaneous NK cell–mediated cytotoxicity and granzyme B expression. (A) Primary human NK cells were first incubated (18 hours) with medium containing either DMSO vehicle control or 40μM 1,9-dideoxy-forskolin and then used as effector cells in a 51Cr release assay using K562 tumor cell targets (n = 4). *P < .03. Cell pellets from DMSO or 1,9-dideoxy-forskolin-treated cells were collected for quantification of granzyme B protein by Western blot (B) and granzyme B transcript by real-time RT-PCR (C; n = 3). **P < .03. Each experiment is representative of at least 3 performed with similar results.
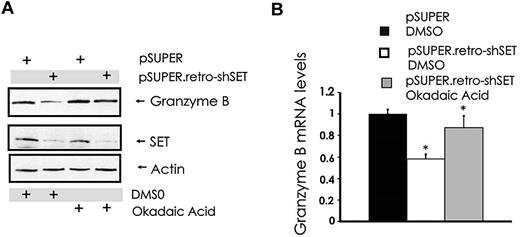
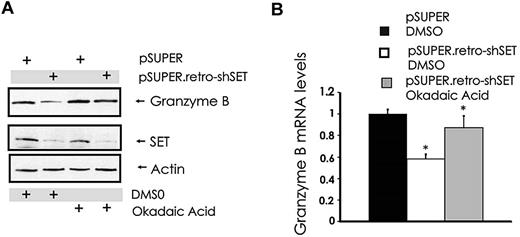
Our attempts to overexpress or down-modulate the catalytic subunit PP2Ac in NK cells with standard gene transfer techniques were unsuccessful (data not shown). However, similar to the results obtained with 1,9-dideoxy-forskolin, NK cells showed diminished cytotoxicity and granzyme B expression using another activator of PP2A called FTY720, at doses that activate PP2A in NK cells (data not shown). To further strengthen the functional relationship between the SET-PP2A interplay in the regulation of granzyme B expression, we treated pSUPER.retro-shSET-expressing and pSUPER-expressing NK-92 cells with the PP2A serine/threonine phosphatase inhibitor okadaic acid and used them to analyze the changes in granzyme B expression. Despite down-regulation of SET in SET knockdown cells, granzyme B protein and mRNA levels were rescued by the pretreatment of the same NK cells with PP2A inhibitor okadaic acid (Figure 4A-B). Thus, the ability of SET to regulate granzyme B expression in human NK cells appears to be mediated via the direct effect of SET on PP2A.
Modulation of PP2A overrides SET-mediated regulation of granzyme B expression in human NK cells. pSUPER-infected and pSUPER.retro-shSET-infected NK-92 cells were treated for 15 hours with vehicle (DMSO) or the PP2A inhibitor okadaic acid (10nM). Cell pellets were collected and assayed for granzyme B protein by Western blot (A) and granzyme B mRNA by real-time RT-PCR (B; n = 4). *P < .02. Each experiment is representative of at least 3 performed with similar results.
Modulation of PP2A overrides SET-mediated regulation of granzyme B expression in human NK cells. pSUPER-infected and pSUPER.retro-shSET-infected NK-92 cells were treated for 15 hours with vehicle (DMSO) or the PP2A inhibitor okadaic acid (10nM). Cell pellets were collected and assayed for granzyme B protein by Western blot (A) and granzyme B mRNA by real-time RT-PCR (B; n = 4). *P < .02. Each experiment is representative of at least 3 performed with similar results.
SET regulation of granzyme B expression in IL-2 and IL-15 activated NK cells
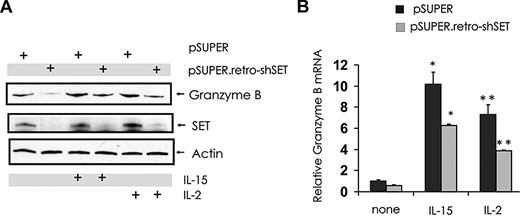
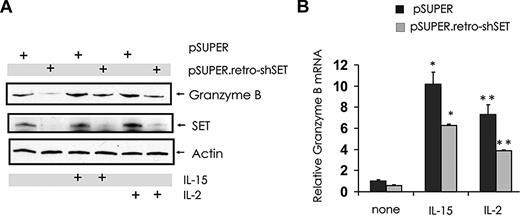
Stimulation of NK cells with activating cytokines, such as IL-2 and IL-15, induces granzyme B expression9 (Figure 5A-B) and also induces expression of SET19 (Figure 5A). To determine whether SET has a regulatory role in the induction of granzyme B expression mediated by IL-2 or IL-15, NK-92 cells with SET knockdown were stimulated with IL-2 or IL-15 and analyzed for granzyme B expression (Figure 5A-B). Both IL-2 and IL-15 were able to induce granzyme B expression in NK-92 cells, and this induction was inhibited by SET knockdown at both mRNA and protein levels.
Effect of SET down-modulation on granzyme B expression in IL-15– or IL-2–activated human NK cells. pSUPER-infected and pSUPER.retro-shSET-infected NK-92 cells starved from IL-2 were left untreated or stimulated with IL-15 (100 ng/mL) or IL-2 (15 ng/mL) for 18 hours. Cells were analyzed for granzyme B, SET and actin protein levels by Western blot (A), and granzyme B transcript level by real-time RT-PCR (B; n = 4). *,**P < .01. Each experiment is representative of at least 3 performed with similar results.
Effect of SET down-modulation on granzyme B expression in IL-15– or IL-2–activated human NK cells. pSUPER-infected and pSUPER.retro-shSET-infected NK-92 cells starved from IL-2 were left untreated or stimulated with IL-15 (100 ng/mL) or IL-2 (15 ng/mL) for 18 hours. Cells were analyzed for granzyme B, SET and actin protein levels by Western blot (A), and granzyme B transcript level by real-time RT-PCR (B; n = 4). *,**P < .01. Each experiment is representative of at least 3 performed with similar results.
Role of SET in regulating CD16-mediated antibody-dependent cellular cytotoxicity in human NK cells
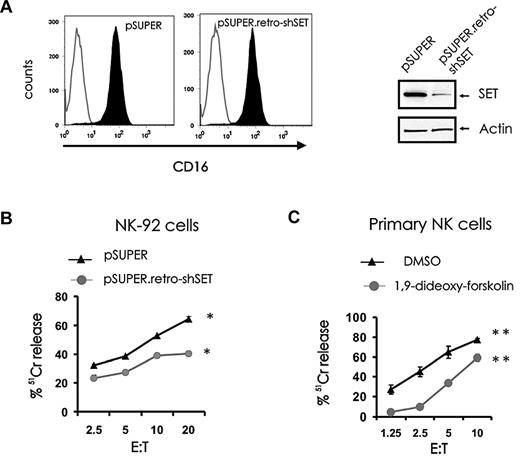
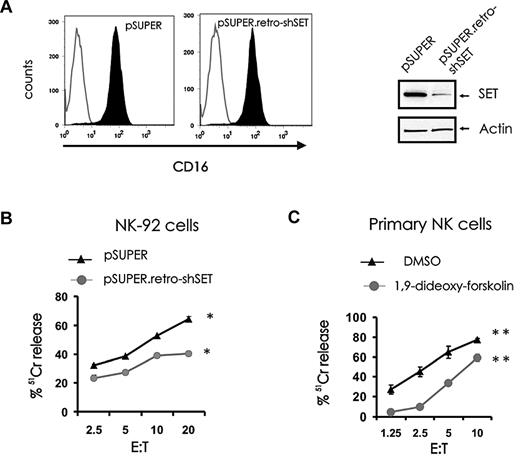
In addition to natural cytotoxicity, NK cells mediate ADCC through its FcγRIIIA receptor, CD16. To determine whether modulation of SET expression could influence ADCC, the NK-92 cell line with low levels of endogenous SET was retrovirally infected to express CD16 α-chain and then infected with pSUPER.retro-shSET or with pSUPER. After confirming CD16 expression by FACS on sorted cells (Figure 6A left) and SET expression by Western blot (Figure 6A right), pSUPER.retro-shSET-CD16+ cells were used as effectors in an ADCC assay and compared with pSUPER-CD16+ cells (Figure 6B). Knockdown of SET expression in CD16+ NK-92 cells significantly inhibited ADCC. Parallel experiments were performed to determine whether activation of PP2A could affect ADCC. Primary NK cells were treated with the PP2A activator 1,9-dideoxy-forskolin and used as effectors for ADCC in 51Cr release assay (Figure 6C). Treatment of primary CD56+ NK cells with 1,9-dideoxy-forskolin inhibited ADCC. In addition, inhibition of ADCC was also observed after treatment with the PP2A activator FTY720 (data not shown).
Effect of SET modulation or pharmacologic induction of PP2A activity on ADCC. pSUPER-infected and pSUPER.retro-shSET-infected NK-92 cells were infected with PINCO-CD16α chain virus and sorted for coexpression of GFP and CD16. After sorting, CD16 membrane expression was confirmed by FACS analysis (A, left), and down-modulation of SET was confirmed by immunoblotting (A, right). (B) ADCC of pSUPER-infected and pSUPER.retro-shSET–infected NK-92 cells expressing CD16 (n = 4, *P < .02) and (C) ADCC of primary CD56+ NK cells incubated in DMSO vehicle control or 1,9-dideoxy-forskolin (n = 5, **P < .01). Effector cells were assayed against P815 antibody-coated target cells in a 3-hour 51Cr release assay. Each experiment is representative of at least 3 performed with similar results.
Effect of SET modulation or pharmacologic induction of PP2A activity on ADCC. pSUPER-infected and pSUPER.retro-shSET-infected NK-92 cells were infected with PINCO-CD16α chain virus and sorted for coexpression of GFP and CD16. After sorting, CD16 membrane expression was confirmed by FACS analysis (A, left), and down-modulation of SET was confirmed by immunoblotting (A, right). (B) ADCC of pSUPER-infected and pSUPER.retro-shSET–infected NK-92 cells expressing CD16 (n = 4, *P < .02) and (C) ADCC of primary CD56+ NK cells incubated in DMSO vehicle control or 1,9-dideoxy-forskolin (n = 5, **P < .01). Effector cells were assayed against P815 antibody-coated target cells in a 3-hour 51Cr release assay. Each experiment is representative of at least 3 performed with similar results.
Discussion
Cellular cytotoxicity and production of cytokines are 2 primary functions of NK cells. NK cells kill pathogen-infected cells or malignant cells via spontaneous or natural cytotoxicity mediated by a variety of activating and inhibitory receptors, or through an antibody-dependent process of cellular cytotoxicity mediated by the interaction of the NK cell's Fc receptor CD16 with antibody coated target cells.2,26 A complete understanding of the molecular pathways that regulate spontaneous and antibody-dependent cellular cytotoxicity of NK cells is of fundamental importance to understanding how the body controls or fails to control cancer and infectious diseases.
We have recently described a novel pathway regulating IFN-γ production in human NK cells.19 We showed that SET expression is up-regulated by monokines IL-12, IL-15, and IL-18 acting on human NK cells, and that SET inhibits PP2A and thereby positively regulates the expression of human NK cell IFN-γ production. SET knockdown or pharmacologic activation of PP2A diminished ERK1/2, p65RelA, STAT4, and STAT5 activity in monokine-stimulated NK cells, potentially contributing to the reduction in IFN-γ gene expression.19 In support of our earlier study, Vesosky et al have identified the SET-PP2A interplay as being one possible mechanism by which CD8 T cells from aged mice produce enhanced levels of IFN-γ in response to Mycobacterium tuberculosis after IL-12 and IL-18 stimulation.27
In the current study, we hypothesized that SET could regulate NK cell–mediated cytotoxic activity via its regulation of PP2A. Performing loss- or gain-of-function experiments, we confirmed our preliminary evidence that modulation of SET altered cytotoxic activity of the NK-92 cell line. In particular, we pursued gain-of-function experiments using primary human CD56+ NK cells, thereby identifying SET as a novel positive regulator of NK cell–mediated lytic activity.
Granule exocytosis is the main pathway used by NK cells for the elimination of virus-infected and malignant cells in both spontaneous and antibody-dependent cellular cytotoxicity. After target cell recognition, release of the granule contents including granzymes and perforin in the immunologic synapse formed between the killer cell and its target induces apoptosis. Granzyme B plays a fundamental role in NK-mediated apoptosis of tumor cell targets via exocytosis. Indeed, mouse NK cells lacking granzyme B do not kill,6 and down-modulation of granzyme B expression in human NK cells inhibits cytotoxicity.7 A characteristic of human NK cells is that they constitutively express granzyme B transcripts and protein and can perform cytotoxicity under resting conditions. Cytokine-mediated NK-cell activation results in up-regulation of granzyme B expression.9 In addition, we have recently shown that mRNA and protein levels of granzyme B are inhibited by treatment of NK cells with the anti-inflammatory cytokine transforming growth factor-β in a SMAD3-dependent manner.22
To explore the molecular mechanism by which SET regulates NK-cell cytotoxicity, we performed gain- and loss-of-function experiments in the human NK-92 cell line and in primary human NK cells, and assessed for granzyme B expression. We found that levels of SET expression positively regulate granzyme B expression. Regulation of granzyme B expression happens at both the mRNA and protein levels. Of interest, using the NK-92 cell line with SET knockdown, we show that SET also regulates the expression of another protein responsible for cytolysis (perforin), thus extending the role of SET to multiple aspects of granule exocytosis.
Regulation of granzyme B mRNA and protein expression by SET was also observed under conditions of NK cell activation. SET is induced by IL-2 and IL-15, suggesting that SET could have a role in cytokine-mediated enhancement of NK cytotoxic activity. In support of this notion, SET knockdown inhibited the induction of granzyme B expression normally induced by stimulation of NK cells with IL-2 or IL-15. However, we did not see an effect on the level of NK cytotoxic activity in the presence of IL-2 or IL-15 (not shown). This could be the result of compensatory regulatory mechanisms of granzyme B that are independent of SET after activation by IL-2 or IL-15, and/or from the multiple pathways of NK cytotoxic activity that are not regulated by SET yet are enhanced after activation with IL-2 or IL-15, such as expression of activating receptors NKG2D or NKp30.28-30 Nonetheless, given that we demonstrate a direct link between SET, PP2A, and granzyme B expression, a direct link between SET or PP2A and cytotoxicity in the absence of IL-2 or IL-15, and because granzyme B plays a nonredundant role in the killing of susceptible target cells by both human and mouse NK cells,5-7 our data do support a link between SET, PP2A, and granzyme B expression with an alteration in NK-cell cytotoxicity.
Whereas our data indicated that modulation of SET effects granzyme B expression and cytotoxic activity in resting NK cells, we cannot eliminate the possibility that the regulation of NK-cell lysis could be in part at the level of granzyme B mobilization. Indeed, we have shown that SET, possibly via PP2A, affects ERK activation in NK cells.19 We and others have previously demonstrated a role for ERK in regulating NK spontaneous and antibody-dependent cellular cytotoxicity.25,31,32 In particular, Wei et al showed that ERK is involved in the redistribution of granule components to the contact zone between NK and target cells.32
We have previously shown that SET affects PP2A activity in NK cells.19 Using a pharmacologic approach, we have shown here that, in primary resting human NK cells, activation of PP2A reduces granzyme B expression at both transcript and protein level and also reduces cytotoxic activity. This demonstrated that the effect of SET on cytotoxicity depends at least in part on its ability to functionally inhibit PP2A in NK cells. We think that PP2A modulates signaling molecule activation, which in turn alters the phosphorylation and activation of transcription factors, such as AP1 or nuclear factor-κB. These transcription factors then regulate NK-cell granzyme B transcription and protein expression.8,33
Finally, we show that the effect of the SET/PP2A interplay on NK-cell cytotoxicity is not only limited to spontaneous cytotoxicity but also includes ADCC. This can be explained by the fact that ADCC also depends on the release of granules containing granzyme B, and degranulation depends on molecules such as ERK that are affected by SET modulation or PP2A activation in NK cells.19,25
Collectively, our data indicate that SET is a molecular marker of NK-cell activation, and support a model were the SET-PP2A interplay regulates granzyme B expression and cytotoxicity in NK cells. It will be interesting to investigate the expression and role of SET after triggering NK-cell inhibitory killer immunoglobulin-like receptors, and after NK-cell treatment with the anti-inflammatory cytokine transforming growth factor-β, which suppresses NK-cell IFN-γ expression and NK cytotoxic activity.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Joshua Oaks for helpful discussion and Jeffrey Allard II for technical assistance.
This work was supported by the National Cancer Institute (grants CA16058, OSU-CCC; CA95426, M.A.C.; CA68458, M.A.C.; and CA095512, D.P.), the US Army, and CML Research Program (W81XWH-07-1-0270) (D.P.). D.P. is a Scholar of the Leukemia & Lymphoma Society.
National Institutes of Health
Authorship
Contribution: R.T. designed the study, performed research work, analyzed data, and wrote the manuscript; D.C., J.D.C., and L.C. performed experimental work and analyzed data; E.B. performed research work; H.M. performed cell sorting experiments; J.Z. performed statistical analysis; J.Y and D.P. discussed the data; and M.A.C. contributed to study design and wrote and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rossana Trotta, The Ohio State University Comprehensive Cancer Center, 884 OSU Biomedical Research Tower, 460 West 12th Ave, Columbus, OH 43210; e-mail: rossana.trotta@osumc.edu; and Michael A. Caligiuri, The Ohio State University Comprehensive Cancer Center, 886 OSU Biomedical Research Tower, 460 West 12th Ave, Columbus, OH 43210; e-mail: michael.caligiuri@osumc.edu.