Abstract
Extracellular ATP and UTP nucleotides increase the proliferation and engraftment potential of normal human hematopoietic stem cells via the engagement of purinergic receptors (P2Rs). In the present study, we show that ATP and UTP have strikingly opposite effects on human acute myeloblastic leukemia (AML) cells. Leukemic cells express P2Rs. ATP-stimulated leukemic cells, but not normal CD34+ cells, undergo down-regulation of genes involved in cell proliferation and migration, whereas cell-cycle inhibitors are up-regulated. Functionally, ATP induced the inhibition of proliferation and accumulation of AML cells, but not of normal cells, in the G0 phase of the cell cycle. Exposure to ATP or UTP inhibited AML-cell migration in vitro. In vivo, xenotransplantation experiments demonstrated that the homing and engraftment capacity of AML blasts and CD34+CD38− cells to immunodeficient mice BM was significantly inhibited by pretreatment with nucleotides. P2R-expression analysis and pharmacologic profiling suggested that the inhibition of proliferation by ATP was mediated by the down-regulation of the P2X7R, which is up-regulated on untreated blasts, whereas the inhibition of chemotaxis was mainly mediated via P2Y2R and P2Y4R subtypes. We conclude that, unlike normal cells, P2R signaling inhibits leukemic cells and therefore its pharmacologic modulation may represent a novel therapeutic strategy.
Introduction
Acute myeloid leukemia (AML) is a clonal disorder characterized by the accumulation of myeloid progenitor cells in the BM. Leukemic blasts show aberrant differentiation patterns that hinder the physiologic maturation process of myeloid cells, eventually leading to hematopoietic failure.1 Most of the efforts made over the years to develop therapeutic approaches against AML have proven to be inadequate, with the disease relapsing in many of the patients. Therefore, although they are able to kill the bulk of AML blasts, current therapies are ultimately incapable of eliminating the leukemic stem cells (LSCs) responsible for the disease.2 In 1994, Dick et al identified LSCs in the CD34+CD38− fraction of AML blasts through transplantation experiments in a xenogenic murine model.3 LSCs retain stem-cell properties, including self-renewal and extensive proliferation, and represent the ideal target for AML therapy. More recently, several investigations have demonstrated the heterogeneity of the normal and the LSC compartment and have questioned the expression of CD34 as a marker of “stemness.”4,5
Extracellular nucleotides, mainly ATP, UTP, ADP, and UDP, are key players in several biologic activities, including modulation of tumor cell function. Several reports have shown the ability of extracellular nucleotides to regulate cell proliferation, migration, and death, depending on the expression of the purinergic receptor (P2R) subtype and the type/concentration of the nucleotide(s) released or present in the extracellular milieu.6,7 The P2R family is subdivided in 2 subgroups: the P2XR family, which includes receptors that, upon activation, act as membrane channels regulating the flux of ions into the cells,8 and the P2YR group,9 which encompasses the G-protein–coupled membrane receptors.10-12
We reported previously that normal human hematopoietic stem cells (HSCs) and mesenchymal stromal cells express several P2Rs and that, in synergy with several hematopoietic cytokines, extracellular nucleotides can increase the clonogenic activity and the number of human BM-repopulating CD34+ cells in immunodeficient mice.13,14 Moreover, we showed that extracellular UTP modulates HSC migration and homing in synergy with the chemotactic peptide CXCL12 via the activation of Rho gGTPase transduction pathway.15
The expression of P2XRs in neoplastic cells, particularly the P2X7R subtype, has been well documented in chronic B-lymphocytic leukemia (B-CLL), prostate cancer, neuroblastoma, and thyroid papillary cancer.16-18 The expression of P2X7R has also been found to be elevated in acute and chronic myelogenous leukemia and in myelodysplastic syndromes.19
Chong et al reported the differential expression of P2XRs in BM cells from pediatric AML patients. Particularly, P2X1R, P2X4R, P2X5R, and P2X7R were overexpressed compared with normal cells, whereas P2X2R, P2X3R, and P2X6R were not expressed or only slightly expressed in these cells.20 However, despite the suggestion of the potential contribution of P2R expression to the malignant phenotype, neither study addressed its function or biologic significance in leukemia.
In the present study, we confirm that AML cells express several functional P2Rs. In contrast to what was observed in normal human HSCs, we found that in AML cells, P2R stimulation induced a significant inhibition of both proliferation and migration in vitro. Extracellular nucleotides were also capable of inhibiting the homing and the engraftment of both AML blasts and purified LSCs when transplanted into immunodeficient mice. Our data on P2R signaling in normal and AML cells may help to define novel molecular pathways that deregulate leukemic cells while stimulating normal HSCs.
Methods
Reagents
ATP and UTP were purchased from Sigma-Aldrich. P2Y2R/P2Y4R subtype agonist INS973 and P2Y2R/P2Y6R subtype agonist INS415 were kindly provided by Inspire Pharmaceutical (Durham, NC). KN62, a P2X7 receptor antagonist, was purchased from Tocris Bioscience. The concentrations of study reagents were chosen based on previous findings.13-16
Cells
The human leukemia cell lines U937 and OCI-AML3 (DSMZ) were cultured in RPMI 1640 medium supplemented with 10% FBS and maintained at 37°C and 5% CO2.
Primary leukemic cells were obtained from the peripheral blood (PB) of 35 AML patients at diagnosis (before treatment; supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Mononuclear cells (MNCs) were separated from AML samples by Ficoll-Hypaque centrifugation (Amersham). The percentage of leukemic blasts in the final cell preparation was always > 90%. Leukemic stem/progenitor cell purification and phenotypic analyses were performed as described previously.21 Aliquots of sorted CD38−CD34+ cells were reanalyzed by FACScan (BD Biosciences) to assess their purities, which were found to be 99.2% ± 0.8%.
PBMCs were enriched by Ficoll-Hypaque (Amersham Bioscience) from leukapheresis products from healthy stem cell donors receiving recombinant G-CSF (Lenograstim; Sanofi-Aventis).
MNCs were processed with a MiniMACs high-gradient magnetic separation column (Miltenyi Biotec) to obtain highly purified CD34+ cells (mean purity, 94% ± 5%).14 The research was approved by the ethics committee of the University Hospital and each individual gave written informed consent in accordance with the Declaration of Helsinki.
Cytosolic Ca2+ concentration measurements
Cells were seeded onto glass coverslips and loaded with the Ca2+ indicator Fura-2 AM (4μM) for 20 minutes at 37°C in a standard saline solution as described previously.13-15,22 Intracellular Ca2+ concentration changes were measured with a PerkinElmer fluorometer at an excitation of 340-380 nm and emission at 510 nm.22 The Ca2+ concentration was calculated using FL WinLab 3.0 software (PerkinElmer).
Real-time qRT-PCR
Total RNA was isolated using the RNeasy kit (QIAGEN) according to the manufacturer's instructions. Real-time quantitative RT-PCR (qRT-PCR) was performed on an ABI PRISM 7900 Sequence Detector (PerkinElmer) using SDS2.3 software. qRT-PCR data were analyzed using the 2−ΔΔCt method.23 The relative level of a specific mRNA for each P2R was calculated by subtracting cycle threshold (Ct) values of the control gene (GAPDH) from the Ct values of the specific gene. Universal human RNA (Stratagene; Agilent Technologies) was used as reference and taken as a value of 1.
The P2X1R Assay ID is Hs 00175686_m1; P2X2R Assay ID: Hs00247255_m1; P2X3R Assay ID: Hs 00175689_m1; P2X4R Assay ID: Hs00602442_m1; P2X5R Assay ID: Hs 00531938-m1; P2X6R Assay ID: Hs01003997_m1; P2X7R Assay ID: Hs00175721_m1; P2Y1R Assay ID: Hs 00704965_s1; P2Y2R Assay ID: Hs00175732_m1; P2Y4R Assay ID: Hs00267404_s1; P2Y6R Assay ID: Hs00602548_m1; P2RY11R Assay ID: Hs01038858_m1; P2Y12R Assay ID: Hs00224470_m1; P2Y13R Assay ID: Hs03043902_s1; P2Y14R Assay ID: Hs00208434_m1; and GAPDH Assay ID: Hs00266705_g1.
RNA extraction and microarray data analysis
Leukemic cells obtained from 12 AML patients (3 M0/M1: numbers 6, 10, and 19; 3 M2: numbers 5, 20, and 28; 3 M4: numbers 15, 21, and 31; and 3 M5: 18, 23, and 24 in supplemental Table 1) and normal CD34+ cells purified from the PB of 3 healthy donors were cultured in DMEM at a concentration of 1 × 106/mL for 24 hours with or without 1mM ATP. Total RNA from 106 ATP-treated and ATP-untreated cells was extracted using the RNeasy Micro Kit (QIAGEN). Disposable RNA chips (Agilent RNA 6000 Nano LabChip kit; Agilent Technologies) were used to determine the purity/integrity of RNA samples using an Agilent 2100 Bioanalyzer.24 A NanoDrop ND-1000 spectrophotometer was used to evaluate the RNA sample concentration, and 260/280 and 260/230 nm ratios were considered to verify the purity of total RNA. ATP-treated and control CD34+-derived RNAs originating from 3 healthy donors were pooled to obtain at least 1 μg per sample. One-cycle target labeling assays, as well as the Affymetrix HG-U133 Plus2 GeneChip array hybridization, staining, and scanning, were performed starting from 1 μg of total RNA using standard Affymetrix protocols.24 All of the data have been deposited in the Gene Expression Omnibus MIAME compliant public database, at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE30903
The GeneChip Operating Software absolute analysis algorithm was used to determine the amount of a transcript mRNA (signal), whereas the comparison analysis algorithm was used to compare gene expression levels between 2 samples (the signal-log ratio). Present genes were selected as the sequences showing the signal average ≥ 50 in at least in 1 sample. Differentially expressed genes were selected as the sequences showing a signal-log ratio average ≥ 0.5 or ≤ −0.5 in the pairwise comparisons between 1mM ATP and untreated control cells. Microarray data were analyzed with the 2-tailed Student t test for comparison of signal averages in paired samples; P < .05 was considered significant. The gene list passing this filter was selected as “changing genes.”
DAVID Version 6.7 software (National Institute of Allergy and Infectious Diseases, http://david.abcc.ncifcrf.gov/) was used to examine the selected lists of genes to identify overrepresentation of functional classes accordingly with gene-ontology classification.
Progenitor cell assays
Leukemic colony-forming cells (CFU-Ls) and normal CFUs were cultured in methylcellulose-based medium.14 Leukemic cells (10 × 104) were cultured in serum-free medium (MethoCult SFBIT H4236; StemCell Technologies) supplemented with cytokines: 50 ng/mL of SCF, 50 ng/mL of IL-3, and 10 ng/mL of GM-CSF (all from Thermo Fisher Scientific) with or without nucleotides. Normal and leukemic CFUs were scored after 10-12 days of incubation at 37°C in a fully humidified 5% CO2 atmosphere. Results are shown as the fold change of the number of CFUs compared with untreated conditions.
Proliferation assay
CellTiter 96AQueous One Solution Cell Proliferation Assay (Promega) was used to detect the effects of nucleotides on normal and leukemic cell proliferation. Culture medium (5 × 104 cells/100 μL) supplemented with cytokines (SCF, IL-3, and GM-CSF) was seeded into a 96-well microplate. After overnight incubation at 37°C in humidified 5% CO2 atmosphere, 1mM ATP was added to the culture. After 48 hours of incubation, CellTiter 96AQueous One Solution reagent was added to each well and the microplate was incubated for 4 hours at 37°C in 5% humidified CO2. The optical density value was measured with an ELISA plate reader (Multiskan Ex; Thermo Electron Corporation) at a wavelength of 492 nm. Each variant group was performed in triplicate wells. In selected experiments, the ATP blocking agent apyrase (1 U/mL) or denaturated apyrase (Sigma-Aldrich) was added to the culture medium.
Migration assay
Migration of leukemic cells was tested using the Transwell assay (diameter 6.5 mm, pore size 8 μm; Costar; Corning).15 Leukemic cells were incubated overnight in the presence of ATP or UTP; extracellular nucleotides were then washed out before the migration assay. Briefly, 100 μL of IMDM plus 10% FBS containing 1 × 105 pretreated cells was added to the upper chamber, and 600 μL of medium with or without 150 ng/mL of CXCL-12 (Biodesign International) was added to the bottom chamber.
After overnight incubation at 37°C in 5% humidified CO2 atmosphere, inserts (upper chambers) were removed and cells transmigrated into lower chamber were recovered and counted. The number of migrating cells was counted with an inverted microscope (Nikon) using a 5× magnification. Results are shown as the fold change of the percentage of migration compared with the untreated condition.
Cell-cycle and apoptosis analysis
Cells were resuspended in complete medium at a concentration of 1 × 106/mL, and primed for 24 hours with a cocktail of cytokines (GM-CSF, SCF, and IL-3; Thermo Fisher Scientific) before culture in the presence of extracellular nucleotides. Changes in the cell-cycle distribution were evaluated using the Acridine Orange technique, as described previously.25 The percentage of cells in the G0/G1, S, and G2M phases was determined by measuring simultaneously the DNA and RNA total cellular content. The percentage of apoptotic cells was measured based on the decreased stainability of apoptotic elements in DNA green fluorescence (sub-G1 peak on DNA-frequency histograms) coupled with a higher RNA red fluorescence (which is associated with chromatin condensation).25 Cell-cycle distribution was analyzed using ModFit LT 3.0 software (Verity Software House). Induction of apoptosis was also assessed by measuring annexin V binding to externalized phosphatidylserine.25
Xenotransplantation experiments
Experiments involving mice were performed by the Institute for Cancer Research and Treatment (Candiolo, Turin, Italy) and all procedures were carried in accordance with national and international laws and policies. NOD.cg-PrkdcscidIl2rgtm1Wjll (NSG) mice were obtained from The Jackson Laboratory, and bred and maintained under defined flora conditions in individually ventilated, sterile, microisolated cages. After sublethal irradiation (240 cGy from a cesium 137 source), female mice between 4 and 5 weeks of age underwent transplantation with human primary leukemia cells by lateral tail vein injection.
For induction of AML, 10-40 × 106 MNCs from the PB or BM of newly diagnosed AML patients were injected into mice. To evaluate human leukemia cell homing, mice were killed 16 hours thereafter. Nucleated cells, obtained from long-bone BM, were stained analyzed by flow cytometry for detection of human CD45+ cells. Eight weeks after transplantation, human long-term engraftment was evaluated by cytometry analyzing BM cells after bone flushing. Cells were incubated with rat and mouse IgG (Sigma-Aldrich) and then stained with the anti–human CD45 leukocyte common antigen (clone 2D1; BD Biosciences). After RBC lysis, cell suspensions were evaluated with a FACSCalibur (BD Biosciences) using analysis gates designed to exclude dead cells, platelets, and debris. After the acquisition of at least 100 000 cells per sample, analyses were considered as informative when adequate numbers of events (ie, > 50, typically 100-200) were collected in the enumeration gates. The percentages of stained cells were determined and compared with appropriate negative controls. 7-amino actinomycin D (Sigma-Aldrich) was used to enumerate viable, apoptotic, and dead cells. Positive staining was defined as being greater than nonspecific background staining.
Statistical analysis
Results are expressed as means ± SEM of at least 3 different experiments. Statistical comparisons were performed using the t test ANOVA. Two-sided P < .05 was considered statistically significant.
Results
P2Rs are expressed on AML cells
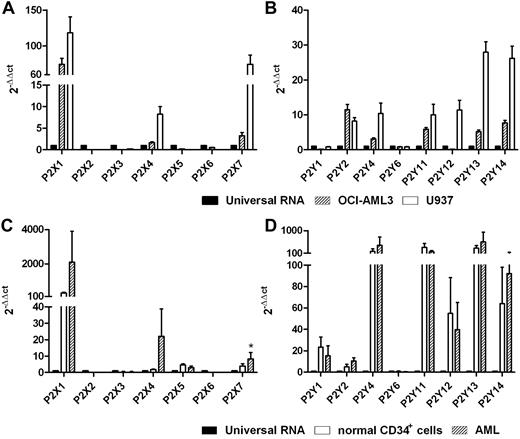
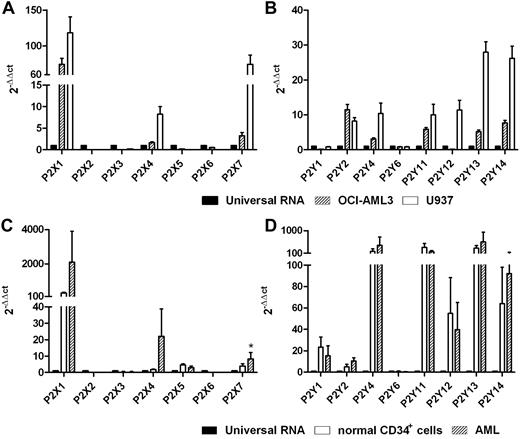
The expression levels of 15 cloned human P2XR and P2YR subtypes were analyzed in U937 and OCI-AML3 cell lines and in primary blasts from 15 leukemia patients. As shown in Figure 1A-B, OCI-AML3 and U937 cells expressed the P2X1R, P2X4R, and P2X7R subtypes. U937 cells expressed all P2YR subtypes, whereas P2Y1R and P2Y12R were not detected in OCI-AML3 cells. Primary leukemic cells expressed mRNAs of P2X1R, P2X4R, P2X5R, and P2X7R, whereas P2X2R, P2X3R, and P2X6R were not detected. The P2X7R mRNA level was significantly higher in AML cells than in normal CD34+ cells (P < .05). Like normal CD34+ cells, leukemic cells also expressed mRNA for all P2YR subtypes cloned so far, although we found low levels of P2Y6R (Figure 1C-D).
Human AML cells express P2Rs. mRNA expression of P2R subtypes was evaluated by qRT-PCR. Histograms indicate the expression level of all P2Rs cloned thus far in the U937 and OCI-AML3 cell lines (A-B) and in primary blasts (C-D). Universal human RNA was used as a reference and taken as a value of 1. *P < .05.
Human AML cells express P2Rs. mRNA expression of P2R subtypes was evaluated by qRT-PCR. Histograms indicate the expression level of all P2Rs cloned thus far in the U937 and OCI-AML3 cell lines (A-B) and in primary blasts (C-D). Universal human RNA was used as a reference and taken as a value of 1. *P < .05.
Functional expression of P2Rs in AML cells
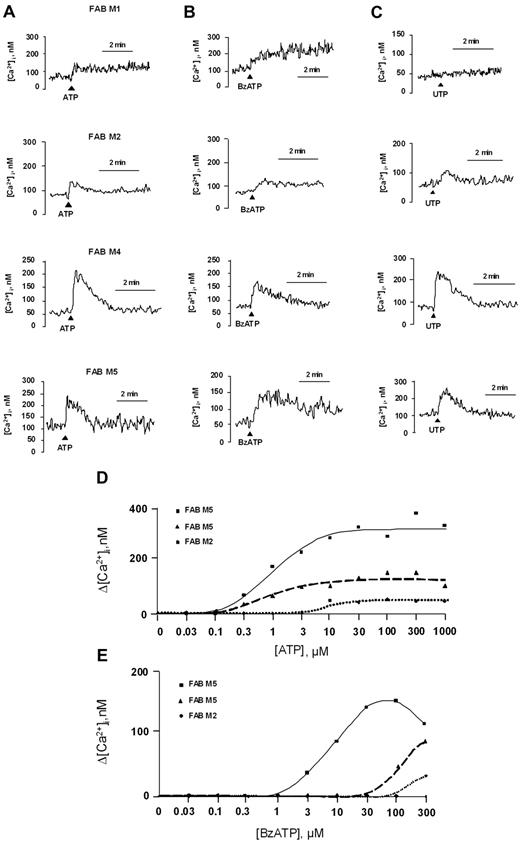
Modulation of Ca2+ signaling is the hallmark of all P2R function. Therefore, AML cells were loaded with the fluorescent Ca2+ indicator Fura-2 AM and stimulated with the P2R agonists ATP, BzATP, and UTP. Ca2+ changes in the cytoplasm were monitored by fluorometric analysis. Cells from all tested subjects (n = 22) were responsive to ATP, although with Ca2+ peaks of very different amplitude; ΔCa2+ (ie, the difference between maximum Ca2+ concentration induced by the stimulus and basal Ca2+ level) ranged from 36-334nM. Figure 2 shows that stimulation with 1mM ATP triggered a fast Ca2+ increase in AML cells belonging to FAB M1, M2, M4, and M5. Interestingly, a 300μM concentration of BzATP, the potent P2X7R pharmacologic agonist, was sufficient to evoke a measurable Ca2+ increase in cells from AML patients that was identical to that caused by ATP.
ATP, UTP, and BzATP trigger intracellular Ca2+ concentration increases in AML cells. Cells from AML patients were resuspended in a standard saline solution and loaded with the Ca2+ indicator Fura-2 AM, as reported in “Methods.” Cells were then rinsed and resuspended in the same saline solution. Stimulation was performed with 1mM ATP (A), 300μM BzATP (B), or 1mM UTP (C) on AML cells from the same patients. Closed triangles indicate nucleotide additions. One representative experiment is shown. Cells were also stimulated with increasing doses of ATP (D) or BzATP (E). Cells from the same patients were used for the ATP and BzATP stimulations.
ATP, UTP, and BzATP trigger intracellular Ca2+ concentration increases in AML cells. Cells from AML patients were resuspended in a standard saline solution and loaded with the Ca2+ indicator Fura-2 AM, as reported in “Methods.” Cells were then rinsed and resuspended in the same saline solution. Stimulation was performed with 1mM ATP (A), 300μM BzATP (B), or 1mM UTP (C) on AML cells from the same patients. Closed triangles indicate nucleotide additions. One representative experiment is shown. Cells were also stimulated with increasing doses of ATP (D) or BzATP (E). Cells from the same patients were used for the ATP and BzATP stimulations.
We performed a dose-dependent curve for ATP and BzATP in 3 AML patients (2 FAB M5 and 1 FAB M2). Data are reported in Figure 2D (ATP) and 2E (BzATP). The 3 patients were responsive to ATP and BzATP, although with different kinetics of the Ca2+ signal. Cells that were more responsive to ATP (Figure 2D FAB M5) were also more responsive to BzATP (Figure 2E FAB M5). AML cells were also responsive to UTP (8 of 10 patients; Figure 2C).
These results demonstrate that AML cells express functional P2Rs and that ATP and, to a lesser extent, UTP stimulate P2R signaling.
GEP of ATP-treated blast cells
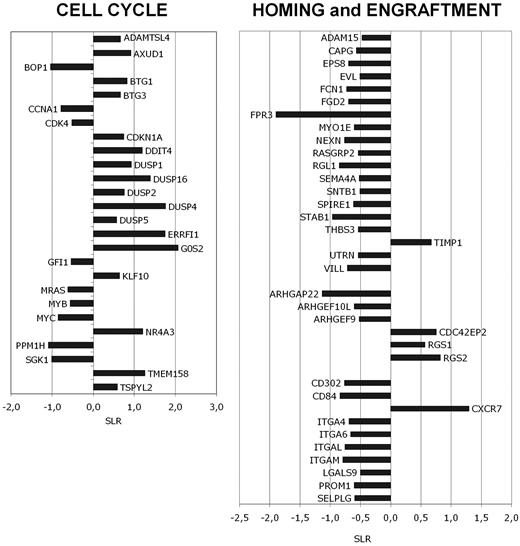
Previous experiments have shown that ATP induces the most relevant changes in Ca+ signaling of all AML samples studied. Therefore, we assessed the transcriptome profile of ATP-treated blasts by gene-expression profiling (GEP) using Affymetrix HG-U133 Plus 2 GeneChip array. Using the filtering procedure described in “Methods,” we identified several genes significantly modulated by ATP treatment (692 probe sets increased and 520 probe sets decreased in 1mM ATP vs control). DAVID 6.7 analysis showed that the “negative regulation of cell proliferation,” “negative regulation of cell cycle,” and “cell cycle arrest” GO categories are mainly represented in the gene list of probe sets increased in ATP-treated versus control cells (supplemental Table 2A). Moreover, GO categories such as “negative regulation of cell-matrix adhesion” and “negative regulation of cell-substrate adhesion” were significantly up-regulated by ATP treatment (supplemental Table 2A). Conversely, GO categories down-regulated by ATP included “positive regulation of cell proliferation” and “cell cycle.” Many genes belonging to the GO category “leukocyte adhesion” were significantly down-regulated by ATP treatment (supplemental Table 2B). The study of differentially expressed genes showed that ATP-treated cells were characterized by the preferential expression of growth-arrest genes and cell-cycle inhibitors (CDKN1A/p21 waf-1, G0S2, BTG1, and BTG3; Figure 3A). ATP treatment down-regulated the expression cell-cycle related genes such as cyclins and CDKs (CCNA1 and CDK4; Figure 3A) and transcription factors involved in cell proliferation (MYB, MYC, and GFI1; Figure 3A). Interestingly, the MAPK pathway was inhibited as the MRAS gene was down-regulated, whereas many MAPK pathway inhibitors (DUSPs) were up-regulated. Furthermore, ATP treatment induced the expression of genes involved in the negative regulation of cell motility (eg, TIMP1) and inhibitors of GTPase activity (eg, CDC42EP2, RGS1, and RGS2)26 (Figure 3B). Consistently, ATP decreased many activators of cell motility, such as Rho-GTPase regulators (eg, ARHGEF9, ARHGEF10L, and FGD2),26,27 and molecules involved in cellular migration, such as matrix degradation enzymes (eg, ADAM15) and cytoskeleton proteins (eg, NEXN, THBS3, EVL, FCN1, MYO1E, SNTB1, and UTRN; Figure 3B). Many integrins (eg, ITGA4, ITGA6, ITGAL, and ITGAM) and adhesion molecules (eg, SELPLG, LGALS9, PROM1, and STAB1)28 involved in HSC homing and engraftment were down-regulated in ATP-treated cells (Figure 3B). Several genes whose expression if required for the induction of programmed cell death (eg, BNIP3, BNIP3L, CRADD, CASP5, and TNFRSF21) were preferentially expressed in ATP-treated cells, whereas apoptosis inhibitors were down-regulated after ATP treatment (data not shown).
GEP of ATP-treated cells. Microarray analysis was performed after 24 hours of incubation with 1mM ATP. Treated and untreated AML cells were obtained from 12 AML patients (3 for M0/M1, 3 for M2, 3 for M4, and 3 for M5). Changes in gene expression are reported on the x-axis as the signal-log ratio (SLR) in the pairwise comparisons ATP versus control cells. (A) Cell-cycle regulators. (B) Migration and cell adhesion molecules.
GEP of ATP-treated cells. Microarray analysis was performed after 24 hours of incubation with 1mM ATP. Treated and untreated AML cells were obtained from 12 AML patients (3 for M0/M1, 3 for M2, 3 for M4, and 3 for M5). Changes in gene expression are reported on the x-axis as the signal-log ratio (SLR) in the pairwise comparisons ATP versus control cells. (A) Cell-cycle regulators. (B) Migration and cell adhesion molecules.
We also evaluated whether incubation with ATP influences the molecular profile of normal CD34+ cells purified from the PB of healthy donors by performing transcriptome analysis of treated (1mM ATP for 24 hours) and untreated cells. The GEP of ATP-treated CD34+ cells was not significantly modulated compared with control samples (supplemental Table 3A-B). These findings indicate that ATP exerts a profound modulatory effect on GEP of leukemic cells but not on normal cells.
ATP inhibits the proliferation and clonogenic efficiency of leukemic cells
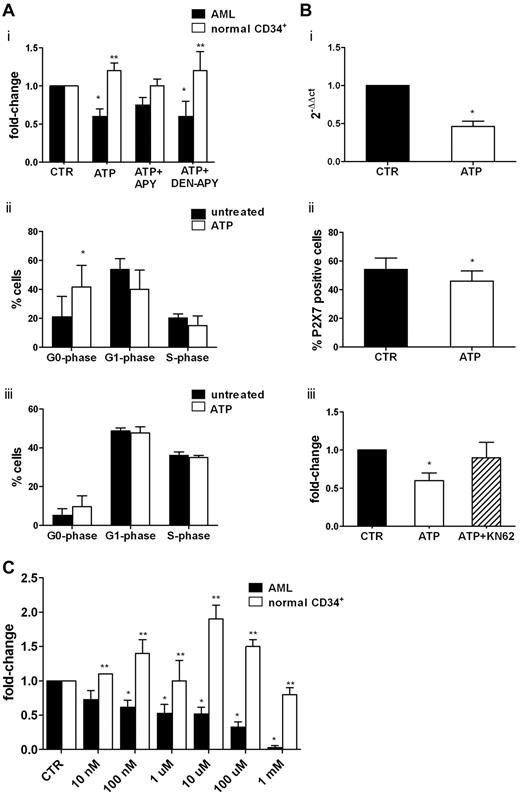
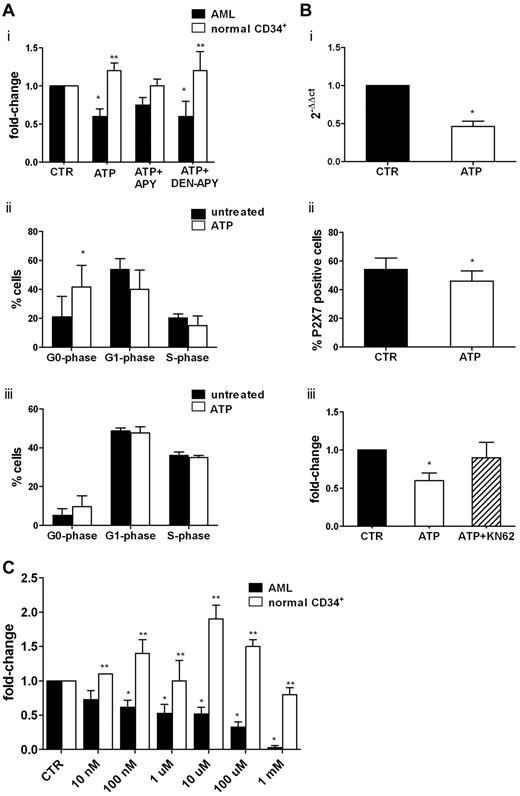
We then studied, at the functional level, the inhibitory activity of ATP on leukemic cells. Consistent with molecular studies, ATP significantly inhibited U937 and OCI-AML3 leukemic cell lines (supplemental Figure 1A) and primary AML cell proliferation (Figure 4Ai) with a change of 0.6 ± 0.1-fold for AML cells (P < .05). Moreover, apoptosis was induced by ATP in OCI-AML3 after 72 hours, with an increase from 9.4% ± 7% to 26% ± 14% (1mM; P = .05) and to 87.7% ± 10.8% (5mM, P = .003; supplemental Figure 1B). Conversely, normal CD34+ cells were not inhibited by ATP (Figure 4Ai).
ATP inhibits AML cell proliferation by repressing the overexpression of P2X7R. Freshly isolated leukemic blasts from 7 AML patients and highly purified CD34+ cells from 4 healthy donors were seeded at a density of 1 × 106/mL, primed with a cocktail of cytokines for 24 hours, and then treated or not with extracellular nucleotides. (Ai) Cell proliferation was measured by CellTiter 96 Aqueous One Solution (Promega) as described in “Methods.” The inhibitory activity of ATP was reverted by the blocking agent apyrase and restored when denaturated apyrase was used. The percentage of proliferating cells in control samples was 60% ± 23%. Conversely, CD34+ cell proliferation was not inhibited by ATP. Shown are the results of cell-cycle analysis of 4 AML samples (ii) and CD34+ cells from 4 healthy donors (iii). Results are expressed as the percentage of cells in different phases of the cell cycle. (B) Leukemic blasts from 10 patients were incubated for 24 hours in the presence or absence of 1mM ATP. The expression of the P2X7R was studied by real-time PCR using untreated cells as a control (i) and by flow cytometry (ii). (iii) Pretreatment with KN-62, a specific P2X7R antagonist, counteracted the inhibitory effect of ATP on AML cells proliferation. (C) Leukemic cells from 15 patients and CD34+ cells from 7 healthy donors were plated in methylcellulose in the presence of cytokines and increasing concentrations of ATP. The mean number of colonies in AML and healthy control samples was 103 ± 36 and 306 ± 68, respectively. *P < .05 comparing ATP-treated samples with control samples; **P < .05 comparing leukemic cells with normal CD34+ cells.
ATP inhibits AML cell proliferation by repressing the overexpression of P2X7R. Freshly isolated leukemic blasts from 7 AML patients and highly purified CD34+ cells from 4 healthy donors were seeded at a density of 1 × 106/mL, primed with a cocktail of cytokines for 24 hours, and then treated or not with extracellular nucleotides. (Ai) Cell proliferation was measured by CellTiter 96 Aqueous One Solution (Promega) as described in “Methods.” The inhibitory activity of ATP was reverted by the blocking agent apyrase and restored when denaturated apyrase was used. The percentage of proliferating cells in control samples was 60% ± 23%. Conversely, CD34+ cell proliferation was not inhibited by ATP. Shown are the results of cell-cycle analysis of 4 AML samples (ii) and CD34+ cells from 4 healthy donors (iii). Results are expressed as the percentage of cells in different phases of the cell cycle. (B) Leukemic blasts from 10 patients were incubated for 24 hours in the presence or absence of 1mM ATP. The expression of the P2X7R was studied by real-time PCR using untreated cells as a control (i) and by flow cytometry (ii). (iii) Pretreatment with KN-62, a specific P2X7R antagonist, counteracted the inhibitory effect of ATP on AML cells proliferation. (C) Leukemic cells from 15 patients and CD34+ cells from 7 healthy donors were plated in methylcellulose in the presence of cytokines and increasing concentrations of ATP. The mean number of colonies in AML and healthy control samples was 103 ± 36 and 306 ± 68, respectively. *P < .05 comparing ATP-treated samples with control samples; **P < .05 comparing leukemic cells with normal CD34+ cells.
The inhibitory activity of ATP was reversed by its inhibitor, apyrase, and restored when denaturated apyrase was used. The inhibitory effect of ATP was mainly because of a cell-cycle arrest in the G0 phase (percentage of untreated G0-phase cells = 21 ± 14.2% vs 41.7% ± 17.8% of ATP-treated cells; P < .05). We also found a decrease of G1- and S-phase cells (Figure 4Aii). Conversely, we did not find any effect of ATP on CD34+ cell cycling (Figure 4Aiii). Apoptosis was significantly increased in 2 of 7 AML samples by 1mM ATP (data not shown), whereas incubation with 5mM ATP increased programmed cell death in all specimens from 15.4% ± 7.0% to 34.6% ± 19.4 (P < .05).
Consistent with the GEP analysis, normal CD34+ cells exposed to 1 or 5mM ATP for 48 hours did not show growth decrease, cell-cycle changes, or induction of apoptosis (data not shown).
To confirm the functional role of P2X7R signaling in mediating ATP effects on cell proliferation, we assessed the expression of the receptor at the molecular (Figure 4Bi) and protein (Figure 4Bii) levels before and after ATP treatment. Incubation with ATP induced the significant (P < .05) down-regulation of P2X7R mRNA and protein on cell membrane (the percentage of P2X7R positive cells in untreated samples was 54% ± 8% vs 46% ± 7% in ATP-treated cells; P < .01). The addition of the P2X7R antagonist KN62 to the culture restored the proliferation of AML cells to levels comparable to control samples (0.9 ± 0.2-fold; Figure 4Biii).
We next investigated whether P2R ligation with the ligands ATP and UTP could affect the clonogenic activity of leukemic cells. As shown in Figure 4C, CFU-Ls were significantly inhibited in a dose-dependent manner by the addition of a wide range of ATP concentrations, with the change ranging from 0.62 ± 0.10-fold (100nM ATP; P < .05) to 0.33 ± 0.08-fold (100μM ATP; P < .0001). At 1mM ATP, the clonogenic activity was almost abolished. A striking opposite effect was observed on normal CD34+ cells, thus confirming previous data.14 Conversely, we did not find an inhibitory effect of UTP on CFU-L growth (data not shown).
Whereas they support the GEP data, these results are in sharp contrast to the remarkable stimulatory activity of extracellular nucleotides on normal HSCs.14 Our data also indicate that ATP-mediated inhibition may be due to down-regulation of the P2X7R, which was overexpressed in AML cells at baseline (Figure 1).
Inhibitory effect of triphosphate nucleotides on the migration of AML cells
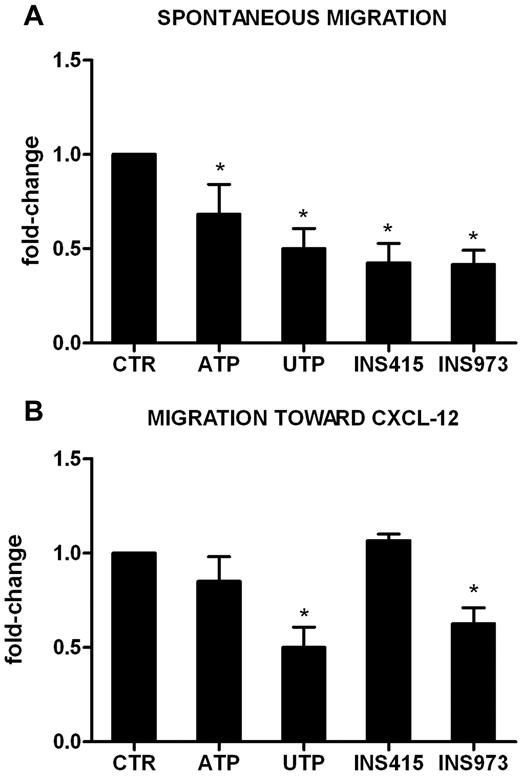
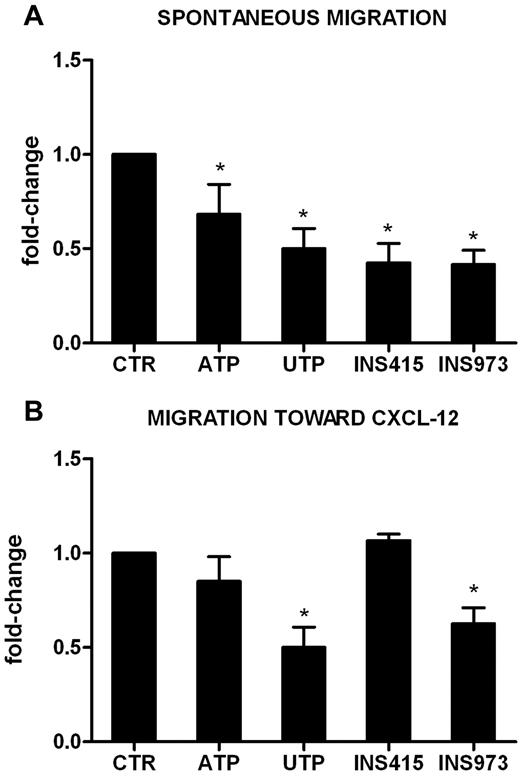
We reported previously15 that UTP is a potent inducer of HSC migration in vitro and in vivo. Conversely, pretreatment of U937 cells (supplemental Figure 1B) and primary leukemic cells (Figure 5) with UTP inhibited their spontaneous migration (Figure 5A) and the migration toward the chemoattractant agent CXCL-12 (0.5 ± 0.1-fold change, P < .01 for AML in Figure 5B). The inhibitory activity of UTP was not mediated by the modulation of the expression of cell membrane or intracytoplasmic CXCR4 (data not shown). In addition, ATP exerted a significant inhibitory effect on fresh AML cell spontaneous migration (0.7 ± 0.2-fold change, P < .05; Figure 5A).
UTP inhibits the migration of leukemic cells via specific P2Y receptors. Leukemic cells from 5 patients (106/mL) were preincubated for 24 hours in the presence of ATP, UTP, INS973 (a P2Y2R/P2Y4R agonist), or INS415 (aP2Y2R/P2Y6R agonist). Migration of leukemic cells was then tested using Transwell assays, as described in “Methods.” Transmigrated cells were counted under the microscope, whereas untreated cells were used as a reference. (A) Spontaneous migration of AML cells. The percentage of migrating cells in control samples was 15% ± 4%. (B) Migration of AML cells toward the CXCL-12 chemokine. The percentage of migrating cells in control samples was 22% ± 5%. *P < .05.
UTP inhibits the migration of leukemic cells via specific P2Y receptors. Leukemic cells from 5 patients (106/mL) were preincubated for 24 hours in the presence of ATP, UTP, INS973 (a P2Y2R/P2Y4R agonist), or INS415 (aP2Y2R/P2Y6R agonist). Migration of leukemic cells was then tested using Transwell assays, as described in “Methods.” Transmigrated cells were counted under the microscope, whereas untreated cells were used as a reference. (A) Spontaneous migration of AML cells. The percentage of migrating cells in control samples was 15% ± 4%. (B) Migration of AML cells toward the CXCL-12 chemokine. The percentage of migrating cells in control samples was 22% ± 5%. *P < .05.
Because P2YRs, which are widely expressed on AML cells (Figure 1), mediate signals that regulate cell motility,29,30 and because UTP is known to be a high-affinity ligand for the P2Y2R, P2Y4R, and P2Y6R subtypes, we hypothesized that the observed inhibition may be because of the engagement of these receptors. Therefore, we evaluated the migration capacity of AML cells after pretreatment with the P2Y2R and P2Y6R agonist INS415 and with the P2Y2R and P2Y4R agonist INS973. As shown in Figure 5, INS973 exerted an inhibitory effect on AML cell migration, both in the presence (Figure 5B) and the absence (Figure 5A) of CXCL-12. This effect was identical to that of UTP, whereas INS415 only reduced the spontaneous migration of leukemic cells (Figure 5A). These results suggest that the ligation of selected P2YRs may be the main mechanism by which ATP and UTP regulate AML cell motility.
Subsequently, using FN-coated culture wells, we found that the treatment of leukemia cells with ATP and UTP did not induce significant modulation of their adhesion capacity and the phenotypic expression of CD49e and CD49d integrins (data not shown).
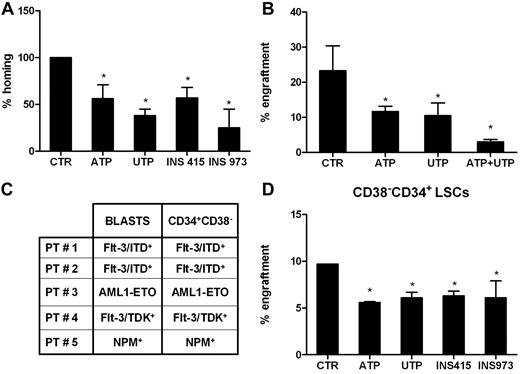
Extracellular nucleotides inhibit homing and engraftment of AML cells in immunodeficient mice
NSG mice were sublethally irradiated and IV injected with human leukemic cells incubated overnight with or without ATP, UTP, or their analogs (Figure 6).
Extracellular nucleotides inhibit homing and engraftment of AML cells in immunodeficient mice. Low-density mononuclear cells (10 × 106) from the PB or the BM of 5 newly diagnosed AML patients were injected into sublethally irradiated NSG-immunodeficient mice after overnight incubation with or without nucleotides or P2YR analogs. Mice were killed 16 hours later to evaluate the homing of human cells. Nucleated cells obtained from the BM of long bones were stained and analyzed by flow cytometry for the detection of human CD45+ cells, as described in “Methods.” The homing capacity of AML cells to the BM was significantly inhibited by ATP (A). Similarly, when the long-term engraftment of leukemic cells pretreated with nucleotides was analyzed, we found a significant inhibition by ATP and by UTP (B). Furthermore, CD34+CD38− cells from 5 AML patients at diagnosis were sorted and analyzed for the presence of the same molecular marker found in the unselected blast population. The results shown in panel C confirmed the leukemic origin of the study cell populations. Purified CD34+CD38− LSCs preincubated with or without nucleotides were injected into NSG-immunodeficient mice, and a significant inhibition of long-term engraftment of leukemic hematopoiesis was observed under all the tested experimental conditions (D). *P < .05.
Extracellular nucleotides inhibit homing and engraftment of AML cells in immunodeficient mice. Low-density mononuclear cells (10 × 106) from the PB or the BM of 5 newly diagnosed AML patients were injected into sublethally irradiated NSG-immunodeficient mice after overnight incubation with or without nucleotides or P2YR analogs. Mice were killed 16 hours later to evaluate the homing of human cells. Nucleated cells obtained from the BM of long bones were stained and analyzed by flow cytometry for the detection of human CD45+ cells, as described in “Methods.” The homing capacity of AML cells to the BM was significantly inhibited by ATP (A). Similarly, when the long-term engraftment of leukemic cells pretreated with nucleotides was analyzed, we found a significant inhibition by ATP and by UTP (B). Furthermore, CD34+CD38− cells from 5 AML patients at diagnosis were sorted and analyzed for the presence of the same molecular marker found in the unselected blast population. The results shown in panel C confirmed the leukemic origin of the study cell populations. Purified CD34+CD38− LSCs preincubated with or without nucleotides were injected into NSG-immunodeficient mice, and a significant inhibition of long-term engraftment of leukemic hematopoiesis was observed under all the tested experimental conditions (D). *P < .05.
The homing capacity of AML cells to the BM was significantly inhibited by ATP (mean decrease, 44%; P < .05), UTP (mean decrease, 72%; P < .05), INS415 (43% inhibition; P < .05), and INS973 (78% inhibition; P < .01; Figure 6A). By comparison, incubation with UTP had double the homing ability of normal HSCs.15 When we analyzed the long-term engraftment of leukemic cells pretreated with nucleotides, we found a median 53% inhibition by ATP and 51% inhibition by UTP (engraftment: control, 23.3% ± 7.1%; ATP, 11.7% ± 1.5%; UTP, 10.5% ± 3.6%; P < .05). Moreover, the combination of the 2 nucleotides showed a synergistic effect (Figure 6B), thus supporting the finding that ATP and UTP may have different mechanisms of action to induce the inhibition of leukemic cell engraftment.
Finally, we assessed whether the inhibitory activity of nucleotide signaling could be observed on highly purified LSCs. To this end, CD34+CD38− cells from 5 AML patients at diagnosis were sorted and analyzed for the presence of the same molecular marker found in the unselected blast population. The results shown in Figure 6C confirmed the leukemic origin of the study cell populations. Afterward, purified CD34+CD38− cells preincubated with or without nucleotides were injected into sublethally irradiated immunodeficient mice, and caused a significant inhibition of long-term engraftment of leukemic hematopoiesis under all of the tested experimental conditions (Figure 6D).
Discussion
In the present study, we investigated whether and to what extent P2Rs and their ligands are involved in the regulation of AML cells. We have shown that AML blasts express several P2Rs. Although different samples respond differently to ATP and UTP stimulation (reflecting the variability intrinsic to the group of AMLs), all of the tested samples appeared to be responsive to purinergic signaling, as demonstrated by intracellular calcium mobilization. Downstream, GEP analysis demonstrated that ATP induced, in leukemic cells but not in normal HSCs, the expression of cell-cycle inhibitors and negative modulators of cell motility, such as inhibitors of GTPase activity. Conversely, ATP inhibited the expression of cell-cycle–related genes, activators of cell motility, and adhesion molecules involved in homing and engraftment. At the functional level, in contrast to what has been found in human HSCs14,15 and in the present study and consistent with GEP results, the stimulation of P2Rs by extracellular ATP produced a significant inhibition of cell proliferation and colony formation. Previous studies demonstrated that extracellular ATP can induce apoptosis in several cell types.31 Our data suggest that in AML, a cell-cycle deregulation is more likely to be involved, because a significant increase of G0 cells was recorded in the presence of ATP, whereas apoptosis was induced only in few samples (although the induction of several proapoptotic genes at the transcriptional level was clearly detectable). Interestingly, AML blasts appear to be sensitive to the inhibitory activity of ATP only, whereas extracellular UTP does not affect colony growth in vitro. These findings suggest that, among extracellular nucleotides, ATP may play a key role in the leukemic microenvironment.
It has been shown that P2X7R stimulation by ATP supports cell proliferation in different cell models, including T lymphocytes, HEK293 cells ectopically expressing the receptor, and in several cancer types in which P2X7R subtype is expressed at high levels.16-18,32,33 The mechanism by which P2X7R influences cell proliferation is still unclear, although its ligation has been shown to increase the cytosolic and mitochondrial calcium concentration, thus increasing ATP production by the organelle.
Recently, Zhang et al showed higher expression of the P2X7R subtype in AML samples compared with normal controls.19 Interestingly, the remission rate in high-P2X7R-expressing patients was lower than that in either non-P2X7R-expressing or low-P2X7R-expressing patients. Moreover, Chong et al suggested that the “cooperation” between specific P2XRs subtypes may contribute to pediatric leukemia.20 The highest expression of P2X7R was detected in patients undergoing relapse, suggesting a role for this receptor in promoting AML proliferation. Overall, our results demonstrate that P2X7R is overexpressed in AML cells and its engagement mediates cell proliferation. In addition, ATP exerts its antiproliferative activity on leukemic blasts by repressing the overexpression of P2X7R, thus counteracting its proliferative function. The pivotal role of P2X7R in ATP-mediated inhibition of cell growth may explain the lack of proliferative effects of UTP, which does not bind to this receptor.
Several studies have demonstrated the capability of ATP to suppress the growth, in vitro and in vivo, of several cell lines, including prostate cancer cells, colon adenocarcinoma cells, murine fibroblasts, pancreatic adenocarcinoma cells, melanoma cells, and bladder cancer cells.34-36 Furthermore, extracellular ATP also suppressed the proliferation and stimulated the differentiation of human HL-60 acute promyelocytic leukemia cells.37 Several clinical applications of extracellular ATP and adenosine have been reported in many fields, including oncology,38-42 and several mechanisms have been proposed as being responsible for the effects of ATP in cancer.43-46 In the present study, 1mM ATP decreased AML proliferation with lower effects on apoptotic death, which was readily appreciable at 5mM perhaps because of the unregulated opening of membrane P2X pores (data not shown). No effects were seen on normal CD34+ cells. Interestingly, similar to AML, Adinolfi et al showed that P2X7R overexpression in B-CLL cells was correlated with the severity of the disease and with decreased survival.16 Indeed, as opposed to indolent variants, evolutive B-CLL stimulation by extracellular ATP (1mM) induced antiproliferative effects.16 In the present study, we have shown that P2X7R mRNA and protein levels are diminished in AML cells after treatment with ATP. These findings suggest that P2X7R stimulation by extracellular ATP may drive a negative feedback loop regulating the expression of P2X7R itself and thus counteracting cell growth.
We have also shown that extracellular nucleotides can inhibit the motility of AML cells in vitro. In particular, the migration assays showed that ATP inhibits AML spontaneous migration, whereas UTP also reduces AML migration in response to the chemokine CXCL-12. Further experiments with specific P2YR agonists indicated that the engagement of the P2Y2R and P2Y4R subtypes may be the main mechanism by which nucleotides stimulate motility. In vivo, BM homing and long-term engraftment also appeared to be inhibited when AML cells were treated with extracellular nucleotides or P2YR agonists before transplantation. Interestingly, extracellular ATP, UTP, and P2YR agonists significantly inhibited the long-term engraftment of a purified population of CD34+CD38− cells, indicating that purinergic signaling may be functionally operative at the LSC level. The observed decrease in long-term engraftment was likely because of the decreased homing after nucleotide treatment before transplantation. The finding that ATP effectively reduces the growth of AML cells in vitro and in vivo highlights the potential use of ATP in the treatment of leukemia.
In conclusion, our data indicate that extracellular nucleotides may be key players in the leukemic BM microenvironment, and therefore represent potential new therapeutic agents for the eradication of leukemia. Further experiments have been planned to define the optimal schedule of ATP administration in vivo to inhibit AML propagation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by the Regione Emilia-Romagna (Progetti di Ricerca Università-Regione Emilia Romagna, Progetto Medicina Rigenerativa, 2007; Innovative approaches to the diagnosis of inflammatory diseases, 2007; and Moniter); the Commission of European Communities 7th Framework Program HEALTH-F2-2007-202231; the Italian Ministry for University Education and Research (MIUR; PRIN 2006 and 2008); the National Research Council of Italy; the Italian Association for Cancer Research (AIRC; IG grant 5354); and by local funds from the University of Ferrara; Italian Association against Leukemia section of Bologna (BolognAil); “Special Program Molecular Clinical Oncology 5x1000” to the AIRC-Gruppo Italiano Malattie Mieloproliferative; Regione Piemonte Ricerca Sanitaria Finalizzata 2008 and 2009.
Authorship
Contribution: V.S., L.R., and M.R. performed the cell-culture experiments, assessed P2R expression, analyzed the data, and wrote the manuscript; R.Z., R.M., E.B., and S.S. performed the GEP experiments, analyzed the data, and wrote the manuscript; S.G., F.D.V., and D.F. evaluated the Ca2+ concentration, analyzed the data, and wrote the manuscript; W.P., L.C., and G.M. performed the in vivo assays, analyzed the data, and wrote the manuscript; A.T. and M.R.R. performed the cell-cycle and apoptosis assays, analyzed the data, and wrote the manuscript; S.F. and M.B. analyzed the data and wrote the manuscript; and R.M.L. designed the research, analyzed the data, wrote the manuscript, and gave the final approval for submission of the manuscript.
Conflict-of-interest disclosure: F.D.V. serves as a consultant for Cordex Pharma Inc (US) and Affectis Pharmaceuticals AG (Germany), companies involved in the development of P2 receptor-based drugs. The remaining authors declare no competing financial interests.
Correspondence: Dott Valentina Salvestrini, Department of Hematology and Oncological Sciences, L. & A. Seràgnoli Institute of Hematology, Via Massarenti, 9-40138 Bologna, Italy; e-mail: salvestrinivalentina@libero.it.