Abstract
The prevalence, the prognostic effect, and interaction with other molecular markers of DNMT3A mutations was studied in 415 patients with acute myeloid leukemia (AML) younger than 60 years. We show mutations in DNMT3A in 96 of 415 patients with newly diagnosed AML (23.1%). Univariate Cox regression analysis showed that patients with DNMT3Amutant AML show significantly worse overall survival (OS; P = .022; hazard ratio [HR], 1.38; 95% confidence interval [CI], 1.04-1.81), and relapse-free survival (RFS; P = .005; HR, 1.52; 95% CI, 1.13-2.05) than DNMT3Awild-type AMLs. In a multivariable analysis, DNMT3A mutations express independent unfavorable prognostic value for OS (P = .003; HR, 1.82; 95% CI, 1.2-2.7) and RFS (P < .001; HR, 2.2; 95% CI, 1.4-3.3). In a composite genotypic subset of cytogenetic intermediate-risk AML without FLT3-ITD and NPM1 mutations, this association is particularly evident (OS: P = .013; HR, 2.09; 95% CI, 1.16-3.77; RFS: P = .001; HR, 2.65; 95% CI, 1.48-4.89). The effect of DNMT3A mutations in human AML remains elusive, because DNMT3Amutant AMLs did not express a methylation or gene expression signature that discriminates them from patients with DNMT3Awild-type AML. We conclude that DNMT3A mutation status is an important factor to consider for risk stratification of patients with AML.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous group of malignancies with different clinical behavior and different responses to therapy. AML can be classified according to distinct cytogenetic and genetic abnormalities, as well as on epigenetic differences that have been shown to be invaluable to guide risk assessment and choice of treatment.1-3 Although insight into the disease has improved in past years, the discovery and validation of new discriminative biomarkers remain of utmost value to improve outcome prediction, in particular for those patients who cannot be classified yet with the currently available biomarkers.
The effect of the association between DNA methylation and various types of human cancer has become a main research domain over the past decade. In conjunction with other epigenetic processes such as histone modifications and interfering RNA, DNA methylation represents an essential component of the transcriptional regulation machinery4 and of the repressive chromatin structure.5 According to their structure in mammals, 3 families of DNA methyltransferase enzymes have been identified.6,7 They are related to each other and probably diverged early in eukaryotic evolution.7 Mammalian DNA methyltransferases (DNMTs) catalyze the transfer of a methyl group onto the 5′-position of cytosine at CpG dinucleotides.8,9 DNMT3A and DNMT3B catalyze de novo DNA methylation, whereas DNMT1 is primarily responsible for maintenance methylation.4,7,10-12
Recently, a somatic change in DNMT3A was identified in human AML, resulting in Arg-to-His substitution at codon R882, located in the methyltransferase domain, causing loss of methylation activity.13 Recurrent DNMT3A mutations at multiple sites, including codon R882, were subsequently detected in a large cohort of patients with AML.14-16 DNMT3A mutations were reported to frequently occur in AML with a normal karyotype and associated with French-American-British (FAB) M5 morphology.14-16 An association with unique DNA methylation and gene expression profiles16 and unfavorable prognosis were suggested.15
In this study we investigated the distribution of DNMT3A mutations in a cohort of 415 AML cases. The relation with cytogenetic and molecular risk categories and the effect of DNMT3A mutations on treatment outcome were analyzed, and the association of DNMT3A mutations with gene methylation and gene expression patterns in AML was investigated. We provide evidence that the DNMT3A mutation status in AML is an important factor to consider for risk stratification of the disease.
Methods
Patients and patient samples and mutation analysis
BM aspirates of patients with AML were collected after written informed consent in accordance with the Declaration of Helsinki. All experiments described were approved by the Erasmus University Medical Center Institutional Review Board. Patients with AML were treated according to the HOVON (Dutch-Belgian Hematology-Oncology Cooperative Group) AML protocols HO04, HO04A, HO29, HO42, HO42A, and HO43 (http://www.hovon.nl). Molecular aberrations in FLT3 (internal tandem duplication [ITD] or tyrosine kinase domain), NPM1, NRAS, KRAS, IDH1-2, KIT, WT1, and CEBPA and EVI1 overexpression were determined as described previously17-21
DNMT3A mutation analysis
It was recently reported that most mutations in DNMT3A occurred in the second part of the gene.14-16 Therefore, DNMT3A mutations were determined in the present study by cDNA amplifications with the use of FWA, 5′-ACGACAGCGATGAGAGTGAC-3′, and REVA, 5′-CCCAATCACCAGATCGAATG-3′, or FWC, 5′-TGAGGACTCCATCACGGTG-3′, and REVC, 5′-CGGTATTTCCGCCTCTGTG-3′. All PCRs were performed in the presence of 25mM deoxynucleoside triphosphate, 20 pmol primers, 1mM MgCl2, Taq polymerase, DMSO, and 10 times buffer (Invitrogen). Cycling conditions were as follows: 1 cycle for 5 minutes at 94°C, 35 cycles for 1 minute at 94°C, 1 minute at 56°C, 1 minute at 72°C, and 1 cycle for 10 minutes at 72°C. All PCR products were subsequently sequenced with the appropriate primers (FWA, 5′-ACGACAGCGATGAGAGTGAC-3′, and FWB, 5′-GCTTTCTGGAGTGTGCGTAC-3′, or FWC, 5′-TGAGGACTCCATCACGGTG-3′, and FWD, 5′-TGATTGATGCCAAAGAAGTGTC-3′) applying an ABI-PRISM3100 genetic analyzer (Applied Biosystems). DNMT3A mutations are considered new somatic mutations in cases in which they were not reported as single nucleotide polymorphism (SNP) according to dbSNP version 132. DNMT3A mutations in AML around the hotspot site (R882) were confirmed by cDNA amplifications with FWD, 5′-TGATTGATGCCAAAGAAGTGTC-3′, and REVE, 5′-GACTGGCACGCTCCATGAC-3′, followed by denaturing HPLC analyses with the use of a Transgenomic WAVE system. Products were run at 60.4°C.
Gene expression profiling and methylation profiling
All AML cases were previously profiled for gene expression22 (Gene Expression Omnibus; National Center for Biotechnology Information; www.ncbi.nlm.nih.gov/geo; accession no. GSE6891) and profiled for methylation1 (accession no. GSE18700). For gene expression–based or gene methylation–based classification of DNMT3A in different defined intermediate risk subtypes, gene expression profiling and DNA methylation profiling (HpaII tiny fragment enrichment by ligation-mediated PCR) data of the HOVON cohort were used to derive predictive signatures with the use of a logistic regression model with Lasso regularization as reported previously.23
Statistics and survival analyses
The relation between DNMT3A mutations and various patient characteristics was determined by the chi square test or Fisher exact test for the categorical variables and by the Mann-Whitney U test for continuous variables. We distinguished the following cytogenetic risk categories: (1) favorable: t(8;21) or inv(16); (2) adverse: inv(3)/t(3,3), t(6;9), 11q23 abnormalities other than t(9;11), del5, del5(q), del7, del7(q), or t(9;22) and monosomal karyotypes24 ; and (3) intermediate risk: the remaining AML cases. Overall survival (OS) end points were death (failure) and alive at last follow-up (censored), as measured from entry onto trial. Relapse-free survival (RFS) end points were disease relapse or death from any cause, measured from complete remission (CR) onto trial. Estimates of the survival distribution for OS and RFS were calculated by the Kaplan-Meier method. Cox proportional hazards model was used in both univariate and multivariate analyses, in which the reported P values correspond to the Wald test. Andersen-Gill model was used to analyze survival outcome in relation to a time-dependent covariate.
Results
DNMT3A is frequently mutated in AML
We performed mutation analysis of the second part of DNMT3A, encompassing the PHD and methyltransferase domains, in 415 patients with newly diagnosed AML. This part of the gene, which has been reported to harbor > 95% of the mutations found in DNMT3A, represents exons 11 until the last exon, that is, exon 23. We found mutations in 96 of the 415 patients (23.1%), that is, 58 missense mutations at position R882 and 38 mutations at other positions (supplemental Figure 1 and supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). In 2 AML cases 2 DNMT3A mutations were found simultaneously (supplemental Table 1). All DNMT3A mutations were considered somatic according to dbSNP (version 132). Patients with AML with DNMT3A mutations presented with significantly higher age, higher white blood cell (WBC) counts, and higher platelet counts compared with DNMT3Awild-type AMLs (Table 1). In line with previous reports, 37.5% of FAB M5 AMLs (36 of 100) carried mutations in DNMT3A. Seventy-two of 96 DNMT3Amutant AMLs (75%) carried a normal karyotype (NK-AML). AMLs with t(8;21), inv(16), inv(3;3)/t(3,3), and trisomy 21 never carried DNMT3A mutations. Furthermore, DNMT3A mutations were rare in complex karyotype AMLs (Table 1). We also separately analyzed 25 patients with acute promyelocytic leukemia (APL), that is, with a t(15;17), and found only 1 patient with a DNMT3A mutation (R882; supplemental Table 1).
DNMT3A mutations significantly associated with ITDs in FLT3 (FLT3ITD; P = .002) as well as with mutations in NPM1 (P < .001) or IDH1 (P < .001). In contrast, DNMT3A mutations were significantly underrepresented in EVI1-overexpressing AMLs. The presence of DNMT3A mutations did not correlate with recurrent mutations in CEBPA, NRAS, KRAS, WT1, FLT3 (TKD), or KIT (Table 1).
The frequency of mutations in DNMT3A in patients with AML > 60 years, which were analyzed separately, was 10 of 63 (16%; supplemental Table 1). Four of 16 AMLs with myelodysplastic syndrome–related changes carried DNMT3A mutations. Mutations in DNMT3A were also found in 2 of 11 therapy-related AML cases (supplemental Table 1).
DNMT3Amutant is an independent prognostic marker in AML
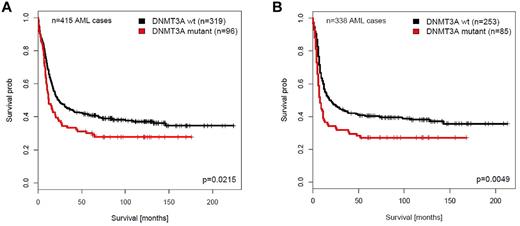
The median follow-up of the 415 patients with AML that remained alive was 115.7 months (range, 7.2-224.1 months). The median OS of patients with DNMT3Amutant AML was shorter than that of patients with DNMT3Awild-type AML (11.9 months vs 24.0 months). DNMT3Amutant AMLs did not show a significantly different CR rate compared with patients with DNMT3Awild-type AML (ie, 88.54% for DNMT3Amutant vs 79.3% for DNMT3Awild-type AML; P = .058). The univariate Cox regression analysis showed that the presence of DNMT3A mutation is an unfavorable prognostic factor for OS (P = .022; hazard ratio [HR], 1.38; 95% confidence interval [CI],1.04-1.81) and RFS (P = .005; HR, 1.52; 95% CI, 1.13-2.05) among all patients with AML (Figure 1A-B).
Survival analyses of patients with AML with or without DNMT3A mutations. Survival analyses were performed on the AML patient cohort < 60 years of age and from which the APLs were excluded. (A) Overall survival (OS). (B) Relapse-free survival (RFS).
Survival analyses of patients with AML with or without DNMT3A mutations. Survival analyses were performed on the AML patient cohort < 60 years of age and from which the APLs were excluded. (A) Overall survival (OS). (B) Relapse-free survival (RFS).
Multivariate analysis showed that DNMT3Amutant is an independent prognostic indicator for unfavorable OS (P = .003; HR, 1.82; 95% CI, 1.2-2.7) and RFS (P < .001; HR, 2.2; 94% CI, 1.4-3.3) in the entire patient group when we consider age (continuous variable), WBC count, cytogenetic risk, NPM1mutant, FLT3ITD, CEBPA double mutation (CEBPADM), NRASmutant, IDH1mutant, IDH2mutant, WT1mutant, cKITmutant, and EVI1 overexpression (Table 1). The variables that independently associated with OS were DNMT3Amutant, NPM1mutant, FLT3ITD, CEBPADM, cytogenetic risk, and WBC count. Of these factors, FLT3ITD and WBC count were not independently associated with RFS (Table 2). DNMT3Amutant showed no independent prognostic value for CR in multivariate analyses.
To investigate whether allogeneic transplantation modifies the prognostic effect of DNMT3Amutant, we applied Cox proportional hazards model with DNMT3A and allogeneic transplantation as a time-dependent covariate, including also an interaction of the 2. For overall survival, we observed a HR of 1.3507 (P = .066; 95% CI, 0.98-1.86) for DNMT3Amutant. The interaction term with allogeneic transplantation was not significant (P = .615; HR, 1.173; 95% CI, 0.623-2.189).
When we analyzed DNMT3A mutations at position R882 only, we observed an association with inferior outcome as well, that is, OS (P = .018; HR, 1.49; 95% CI, 1.07-2.07) and RFS (P = .029; HR, 1.50; 95% CI, 1.04-2.15) in this series of patients with AML (supplemental Figure 2A-B).
DNMT3Amutant predicts poor treatment response in NPM1wild-type/FLT3wild-type/CEBPAwild-type AML
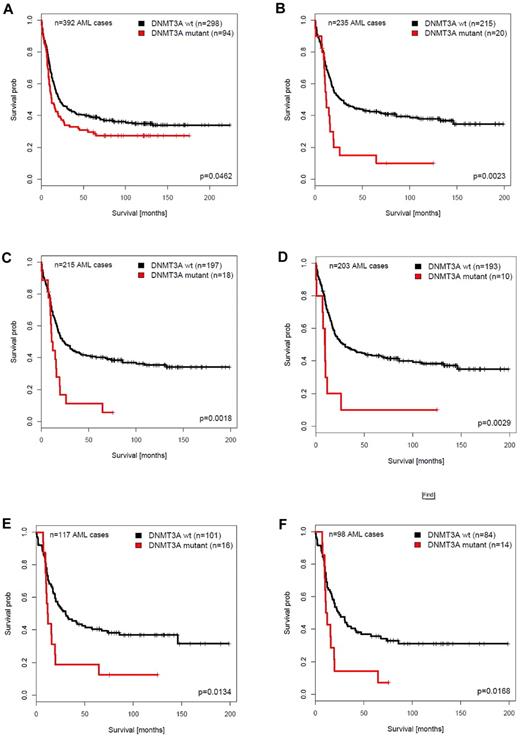
Mutations in DNMT3A particularly were associated with molecular abnormalities, for example, mutations NPM1, FLT3 (FLT3ITD), IDH1, or IDH2 (Table 1). DNMT3A mutations were not preferentially found in CEBPADM AMLs (Table 1). We studied the predictive value of DNMT3A mutations in the distinct molecularly defined AML subcategories. Subgroup analysis found that in the AML cohort from which CEBPADM cases were excluded, DNMT3A mutations still predicted inferior OS (P = .046; HR, 1.325; 95% CI, 1.01-1.75) or RFS (P = .023; HR, 1.414; 95% CI, 1.05-1.91; Table 3; Figure 2A; supplemental Figure 3A). We next assessed the prognostic value of DNMT3A mutations in each of the following composite subgroups: FLT3ITD/NPM1mutant, FLT3wild-type/NPM1mutant, FLT3ITD/NPM1wild-type, or FLT3wild-type/NPM1wild-type. A significant inferior OS (P = .002; HR, 2.167; 95% CI, 1.32-3.56) and RFS (P < .001; HR, 2.781; 95% CI, 1.63-4.75) for DNMT3Amutant AMLs was observed in the NPM1wild-type/FLT3wild-type (no FLT3ITD) subset (Figure 2B; supplemental Figure 3B), whereas DNMT3A mutations had no predictive value in the NPM1mutant/FLT3wild-type and the NPM1mutant/FLTITD subgroups (Table 3). The predictive value for inferior OS (P = .002; HR, 2.268; 95% CI, 1.36-3.79) and RFS (P = .002; HR, 2.503; 95% CI, 1.42-4.42) was also evident in the NPM1wild-type/FLT3wild-type/CEBPAwild-type (no CEBPADM) subset (Figure 2C; supplemental Figure 3C).
Survival analysis of patients with AML with or without DNMT3A mutations in distinct AML subgroups. Survival analyses were performed on the AML patient cohort < 60 years of age and from which the APLs were excluded. (A) OS of patients with AML, excluding patients with CEBPADM (n = 392). (B) OS of patients with AML without FLT3ITD and without NPM1 mutations (FLT3wild-type/NPM1wild-type; n = 235). (C) OS of patients with AML without FLT3ITD, NPM1 mutations, and CEBPADM (FLT3wild-type/NPM1wild-type/CEBPAwild-type; n = 215). (D) OS of patients with AML without FLT3ITD, NPM1, IDH1, and IDH2 mutations (FLT3wild-type/NPM1wild-type/IDH1wild-type/IDH2wild-type; n = 203). (E) OS of patients with cytogenetically intermediate-risk AML without FLT3ITD and without NPM1 mutations (FLT3wild-type/NPM1wild-type; n = 117). (F) OS of patients with cytogenetically intermediate-risk AML without FLT3ITD, NPM1 mutations, and CEBPADM (FLT3wild-type/NPM1wild-type/CEBPAwild-type; n = 98).
Survival analysis of patients with AML with or without DNMT3A mutations in distinct AML subgroups. Survival analyses were performed on the AML patient cohort < 60 years of age and from which the APLs were excluded. (A) OS of patients with AML, excluding patients with CEBPADM (n = 392). (B) OS of patients with AML without FLT3ITD and without NPM1 mutations (FLT3wild-type/NPM1wild-type; n = 235). (C) OS of patients with AML without FLT3ITD, NPM1 mutations, and CEBPADM (FLT3wild-type/NPM1wild-type/CEBPAwild-type; n = 215). (D) OS of patients with AML without FLT3ITD, NPM1, IDH1, and IDH2 mutations (FLT3wild-type/NPM1wild-type/IDH1wild-type/IDH2wild-type; n = 203). (E) OS of patients with cytogenetically intermediate-risk AML without FLT3ITD and without NPM1 mutations (FLT3wild-type/NPM1wild-type; n = 117). (F) OS of patients with cytogenetically intermediate-risk AML without FLT3ITD, NPM1 mutations, and CEBPADM (FLT3wild-type/NPM1wild-type/CEBPAwild-type; n = 98).
Because DNMT3A mutations were most frequently found in intermediate-risk AMLs (Table 1), we performed the same survival analyses in subgroups within this particular subset, with comparable results (Table 3). In the FLT3wild-type/NPM1wild-type intermediate-risk AMLs, DNMT3A mutations were significantly associated with an inferior OS (P = .013; HR, 2.09; 95% CI, 1.16-3.77) and RFS (P = .001; HR, 2.70; 95% CI, 1.48-4.89; Figure 2E; supplemental Figure 3E). A significant predictive value of DNMT3A mutations for poor treatment response was also found for intermediate-risk NPM1wild-type/FLT3wild-type/CEBPAwild-type (no CEBPADM) AMLs (OS: P = .017; HR, 2.11; 95% CI, 1.145-3.907; RFS: P = .016; HR, 2.18; 95% CI, 1.153-4.125; Figure 2F; supplemental Figure 3F). We recently reported that IDH1 mutations have prognostic value in FLT3wild-type/NPM1wild-type as well.21 Although the numbers were too low for a reliable statistical analysis, the high hazard ratios (Table 3) favor the hypothesis that the predictive value of DNMT3A mutations is independent of the presence of IDH1 and IDH2 mutations as well.
DNMT3A mutations are enriched in a methylation and gene expression cluster of NPM1mutant/FLT3ITD AMLs
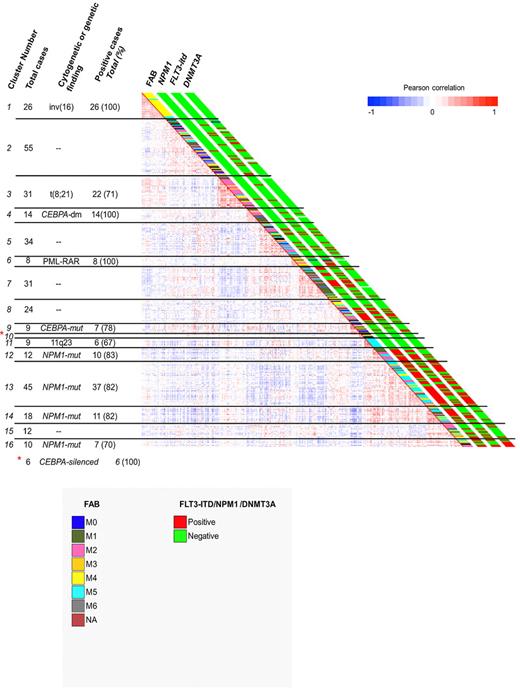
We investigated whether DNMT3Amutant AMLs were associated with unique DNA methylation25 or gene expression signatures.22 Although DNMT3Amutant AMLs did not express a exclusive methylation signature, as determined with the HELP assay25 (Figure 3), methylation cluster 13 was enriched for DNMT3Amutant AMLs (71.1% [n = 32 of 45] DNMT3Amutant AML; Figure 3). Importantly, this cluster mainly contains FAB M4/M5 NPM1mutant/FLT3ITD AMLs.25 This group of AMLs also showed a unique gene expression signature (supplemental Figure 4). In fact, patient samples within this cluster overexpress multiple HOX genes, a signature that is particularly associated with NPM1mutant AMLs.20,26,27 It has been proposed that the increased levels of HOX genes in these leukemias are caused by loss of methylation as the result of mutations in DNMT3A16. However, we observed no difference in HOX gene expression, for example, HOXB5 expression levels between NPM1mutant/DNMT3Amutant and NPM1mutant/DNMT3Awild-type AMLs (supplemental Figure 5). Thus, HOXB5 overexpression, representative for the expression of multiple HOX genes in human AML, does not associate specifically with the presence of DNMT3A mutations.
Clustering of AML patient samples according to methylation profiling with HELP. Distribution of DNMT3Amutant and DNMT3Awild-type AMLs. Pearson correlation view based on methylation profiling of 334 AML cases (see previous figure from Figueroa et al25 ). The presence (red) or absence (green) of NPM1 mutation, FLT3-ITD, or DNMT3A mutation is indicated for the individual AML cases adjacent to the correlation matrix. The distinct FAB types are indicated in different colors.
Clustering of AML patient samples according to methylation profiling with HELP. Distribution of DNMT3Amutant and DNMT3Awild-type AMLs. Pearson correlation view based on methylation profiling of 334 AML cases (see previous figure from Figueroa et al25 ). The presence (red) or absence (green) of NPM1 mutation, FLT3-ITD, or DNMT3A mutation is indicated for the individual AML cases adjacent to the correlation matrix. The distinct FAB types are indicated in different colors.
Supervised analysis with the use of a logistic regression model with Lasso regularization was performed to compare the gene expression as well as the gene methylation signatures of DNMT3Amutant versus DNMT3Awild-type AMLs in distinct subgroups, that is, in NPM1mutant, FLT3ITD, NPM1mutant/FLT3ITD, NPM1wild-type/FLT3ITD, NPM1mutant/FLT3wild-type, and NPM1wild-type/FLT3wild-type AMLs. We did not find a strong predictive signature for each dataset in any of the aforementioned AML subtypes within the intermediate risk group. Therefore, the role of mutated DNMT3A on DNA methylation and gene expression in human AMLs remains elusive.
Discussion
In this study, we identified mutations in DNMT3A in 96 of 415 newly diagnosed AMLs (23.1%). Of the 96 DNMT3A mutant AMLs, 85 belonged to the group of AMLs with an intermediate-risk cytogenetic profile. In fact, mutations in DNMT3A are as frequent in this group of AMLs as other common molecular abnormalities, such as mutations in FLT3 (FLT3ITD), NPM1, IDH1, or IDH2. As a matter of fact, DNMT3A mutations frequently are associated with these particular mutations, which is in strong concordance with data reported previously.14-16
Note that, with the use of th approach we chose to determine mutations in DNMT3A, a small number of DNMT3Amutant AMLs may have been missed. We performed mutation analysis on cDNA in the second half of the DNMT3A gene only, that is, the region containing the PDH and methyltransferase domains and which, according to previous studies, harbors > 95% of the DNMT3A mutations.14-16 Missense or frameshift mutations, leading to nonsense-mediated RNA decay, which represented only a minority of the mutations in previous studies, may have been missed as well because we used cDNA for nucleotide sequencing. However, the mutation rate we found is comparable with frequencies reported by others, suggesting that we identified most DNMT3A mutations in our cohort of AMLs and that the conclusions drawn in our study are valid.
Patients with DNMT3Amutant have significantly worse OS than patients with DNMT3Awild-type, independent of a number of NPM1, FLT3, or CEBPA mutations, and regardless of age, cytogenetic risk, and WBC count. Interestingly, by univariate analyses we show that in a composite genotypic subset of cytogenetic intermediate-risk AML without FLT3ITDs and NPM1 mutations this association was particularly evident (OS, P = .013; RFS, P = .001). A negative prognostic effect of DNMT3A mutations on AML with a NK and with wild-type NPM1 and wild-type FLT3 was previously reported by Thol et al,15 emphasizing the relevance of this finding. Similar to the report by Thol et al,15 no effect of DNMT3A mutations on survival was found in the FLT3wild-type/NPM1mutant NK-AML subgroup. In contrast, the same investigators reported a significant worse OS for NK-AMLs with DNMT3Amutant/FLT3ITD/NPM1mutant compared with DNMT3Awidtype/FLT3ITD/NPM1mutant, which was not evident in our study. Even though we studied the effects DNMT3A mutations on intermediate-risk AMLs, the discrepancy is remarkable and emphasizes the requirement of studies in larger cohorts of AML.
Mutations in DNMT3A would predict that those AML cases show a distinct DNA methylation pattern than cases without mutations. Yan et al reported that DNMT3A mutations caused loss of methylase activity, and they proposed that these alterations resulted in hypomethylation and uncontrolled expression of multiple HOX genes.16 Although we showed that DNMT3Amutant AMLs did not show a strong methylation signature, one methylation cluster (cluster 13) showed enrichment of DNMT3Amutant cases (Figure 3).25 Importantly, this cluster had already been reported to mainly consist of FAB M4/M5 AMLs with NPM1mutant/FLT3ITD.1,20 Moreover, this group of patients also showed a unique gene expression signature, including overexpression of multiple HOX genes.20,26,27 Therefore, the question should be raised whether this HOX gene signature, that is, the strong increase of expression of multiple HOX genes, is indeed caused by hypomethylation of the HOX gene promoters as the result of mutations in the DNMT3A gene, as was reported by Yan et al.16 Multiple investigators have previously reported that the HOX gene signature is particularly associated with NPM1mutant AMLs.20,26,27 Our study found that DNMT3Amutant AMLs without NPM1 mutations did not present this HOX gene signature; that is, HOX genes were not overexpressed in those AMLs (supplemental Figure 5). Furthermore, HOX gene overexpression was also found in DNMT3Awild-type/NPM1mutant AMLs.20,26,27 Importantly, Vassiliou et al28 reported recently that expression of mutant NPM1 in the myeloid compartment, using a NPM1-mutant knock-in mouse model, was associated with a strong up-regulation of HOX genes. Therefore, the role for mutated DNMT3A in aberrant control of HOX genes remains unclear. DNMT3Amutant/NPM1mutant compound mice should further clarify what the effect of mutant DNMT3A is in myeloid progenitors and how it may affect HOX gene expression in the context of NPM1 mutations.
Currently, patient age, cytogenetics, and mutations in CEBPA, FLT3, and NPM1 at diagnosis are main prognostic factors in AML.29-31 In line with recently published data, our data suggest a prognostic effect of DNMT3Amutant in AML.14-16 We therefore propose to implement DNMT3A mutation analysis in the routine of the molecular diagnostics of human AML.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was performed within the framework of CTMM, the Center for Translational Molecular Medicine. This work was supported by Leukemia BioCHIP project (grant 03O-102), the Dutch Cancer Society (Koningin Wilhelmina Fonds), the Instituto de Salud Carlos III (ISCIII; grant CM08/00 027), the Spanish Ministry of Health, Spain, Capes/Brazil, and the Leukemia & Lymphoma Society Translational Research Program (grant 6196-09; A.M. and R.D.).
Authorship
Contribution: A.F.T.R. and M.P. performed experiments, analyzed data, and wrote the paper; C.E.-V., S.A., and A.Z. performed experiments; V.R. and M.S. analyzed data; M.E.F., A.M., and B.L. analyzed data and wrote the paper; and P.J.M.V. and R.D. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ruud Delwel, Erasmus University Medical Center Rotterdam, Department of Hematology, Ee1391a, Dr Molewaterplein 50, 3015 GE Rotterdam Z-H, The Netherlands; e-mail: h.delwel@erasmusmc.nl.
References
Author notes
A.F.T.R. and M.P. contributed equally to this study.
P.J.M.V. and R.D. contributed equally to this study.