Abstract
The importance of human leukocyte antigen (HLA) matching in unrelated donor transplantation for nonmalignant diseases (NMD) has yet to be defined. We analyzed data from 663 unrelated marrow and peripheral blood stem cell transplants performed from 1995 to 2007 for treatment of NMD. Transplantation from a donor mismatched at the HLA-A, -B, -C, or -DRB1, but not -DQB1 or -DPB1, loci was associated with higher mortality in multivariate analyses (P = .002). The hazard ratio for mortality for single (7/8) and double mismatched (6/8) transplants was 1.29 (0.97-1.72; P = .079) and 1.82 (1.30-2.55; P = .0004), respectively, compared with 8/8 matched transplants. HLA mismatches were not associated with acute or chronic GVHD, but were strongly associated with graft failure. After adjustment for other factors, the odds ratio for graft failure for 7/8 and 6/8 (allele and/or antigen) matched pairs compared with 8/8 matched transplants was 2.81 (1.74-4.54; P < .0001) and 2.22 (1.26-3.97; P = .006), respectively. Patients with NMD should receive transplants from allele matched (8/8) donors if possible. Unlike the case with malignancies, HLA mismatching in NMD is associated with graft failure rather than GVHD.
Introduction
Current standards for human leukocyte antigen (HLA) matching in unrelated marrow and peripheral blood stem cell (PBSC) transplantation have been defined by studies largely or exclusively involving patients with hematologic malignancies.1-5 These studies have provided the basis for the US National Marrow Donor Program's (NMDP) current recommendation of allele level matching donors with recipients at the HLA-A, -B, -C, and -DRB1 loci.6 The effect of HLA mismatching, however, has not been well characterized in transplantation for nonmalignant diseases (NMD). Although its influence in severe aplastic anemia (SAA) has been reported in smaller studies,7-9 little is known in other NMD.
It cannot be assumed that the HLA-matching criteria derived from studies of patients with hematologic malignancies are appropriate for patients with NMD. In transplantation for malignancies,10 the harmful effects of acute graft-versus-host disease (GVHD), a complication that has been strongly associated with HLA mismatching,1-5 are partly negated by its beneficial antitumor effect.11 In NMD, however, there is no established benefit to GVHD.11 In addition, although there are notable exceptions, graft failure, a complication associated with histoincompatibility, appears to be more common in transplantation for many nonmalignant diseases.7,8,12,13 To better define the effect of HLA matching on the outcomes of unrelated hematopoietic stem cell transplantation (HSCT) for patients with NMD, we analyzed donor-recipient pairs in the NMDP/Center for International Blood and Marrow Transplant Research (CIBMTR) registry with high-resolution typing at 12 alleles of 6 loci—HLA-A, -B, -C, -DRBI, -DQBI, and -DPB1, given HSCT between 1995 and 2007.
Methods
Study sample
The study sample included 663 patients reported to the NMDP/CIBMTR registries who received an unrelated PBSC or marrow transplant between 1995 and 2007 for treatment of a NMD. Transplantations involving more than 2 mismatches were excluded, as were transplantations performed for patients who had myelodysplastic syndrome, acute leukemia, or other malignancy in addition to their NMD. All participants provided informed consent for inclusion of clinical data and biospecimens. Research was approved and conducted under supervision of the NMDP Institutional Review Board. Conditioning regimens were defined as myeloablative: (1) if the patient received total body irradiation (TBI) at a dose greater than 500 cGy if given as a single dose or greater than 800 cGy if given in fractions; (2) if the patient received busulfan dosed at or greater than 9 mg/kg; or (3) if the patient received melphalan at a dose greater than 150 mg/m2. All other regimens were considered either reduced-intensity conditioning (RIC) or nonmyeloablative, using established criteria.14 For the purposes of the multivariate analyses, diseases were categorized into 4 groups based on 2 factors influencing engraftment: marrow cellularity and cellular immunity. Patients with Fanconi anemia and other forms of SAA (acquired SAA and inherited forms of marrow failure other than Fanconi anemia) have reduced marrow cellularity, but normal to augmented immunity (eg, from transfusion-related alloimmunization). Because the transplantation approach used for Fanconi anemia diverges in important ways from that used for other forms of SAA, we separated it from the other forms of SAA. In a third group, we placed severe combined immune deficiencies and other severe T-cell deficiencies, diseases where the host's capacity to react immunologically against the graft is severely limited, but marrow cellularity is normal. In a fourth group, we placed all other diseases (including hemogloblinopathies, metabolic storage disease, and other immune deficiencies), ones where host reactivity and marrow cellularity both range from normal to increased. The NMDP retrospectively obtained consent for study participation from 91.6% of surviving patients or parents/legal guardians in accordance with the Declaration of Helsinki; 8.4% did not consent and these patients were not included in the analysis. Consent was waived for deceased patients. To avoid bias from including all deceased patients but only a fraction of surviving patients, a weighted randomized model was applied to exclude a proportion of deceased patients similar to the fraction of nonconsenting surviving patients, based on patient characteristics associated with nonconsent. This approach is used with all NMDP studies to obtain an unbiased sample for analysis.15
HLA typing
High-resolution typing was performed for HLA-A, -B, -C, DRB1, -DQB1, and -DPB1, as described.1 Testing was done on banked samples as needed to complete the typing. Low-resolution (serologic or antigen level) disparities involved conversion of the DNA-based typing to its lower-level serologic equivalent, usually by collapsing the 4-digit typing result back to its first 2 digits, with the exception of a few HLA-B alleles that were mapped to their corresponding serologic specificities. Mismatches detectable only at the allele level were distinguished from antigen mismatches in the analyses. Directional mismatches (“graft vs host” or “host vs graft”) were considered as appropriate in the analysis of GVHD and graft failure, as described.5 Mismatches at homozygous alleles were considered single mismatches.
Biostatistical methods
Univariate probabilities for overall survival were calculated using the Kaplan-Meier estimator with variance estimated by the Greenwood formula. Comparison of survival curves was done using the log-rank test. The cumulative incidences of acute and chronic GVHD were calculated using a Taylor series linear approximation to estimate the variance.
Primary and secondary graft failure were considered together as a single outcome. Primary graft failure was defined as failure to achieve a postnadir absolute neutrophil count of 500 cells/mL or donor peripheral blood T-cell chimerism of at least 5% (in the absence of peripheral blood T-cell chimerism, unsorted blood, or marrow chimerism was used). Secondary graft failure was defined as initial donor engraftment followed by graft loss, evidenced by a persistent drop in the absolute neutrophil count to less than 500 cells/mL or loss of donor chimerism. Patients receiving second transplants were also considered to have graft failure. Only those cases of graft failure that occurred within a year of first transplantation were included in the graft failure analysis.
Multivariate analyses were performed for all outcomes to identify risk factors. For outcomes other than graft failure, we used proportional hazards models. All variables were tested for the affirmation of the proportional hazards assumption. Factors violating the proportional hazards assumption were adjusted via stratification. Then, a stepwise model building approach was used in developing models for the primary and secondary outcomes. For graft failure, a pseudo-value approach was applied with death being treated as a competing risk. Two-way interactions were checked between each selected variable and the main effect. Because of multiple testing, a threshold of 0.01 was used for statistical significance. SAS software (Version 9.2; SAS Institute) was used in all the analyses. Two secondary analyses were performed: in the first, because recent CIBMTR studies have indicated that C allele mismatching has little effect on outcome,2,3 we reanalyzed the data excluding C allele mismatched pairs; in the second, we split the single mismatched pairs into those with single allele mismatches and those with single antigen mismatches.
Results
Patient, disease, and transplantation characteristics
Of the 663 donor-recipient pairs, 372 were matched at 8 HLA alleles (HLA-A, -B, -C and -DRB1), 191 (28.8%) had 1 mismatch (allele or antigen), and 100 (15.1%) had 2 mismatches (antigen or allele; Table 1). In 12 (4.1%) of the mismatched pairs, the donor was homozygous at the mismatched locus (graft-vs-host direction only), while in 14 (4.8%) the recipient was homozygous at the mismatched locus (graft rejection direction). HLA-DQB1 matching was present in 86.4% of the donor-recipient pairs, while only 11.8% were matched at DPB1.
There were 39 diseases represented in the sample. Six diseases comprised 77% of the sample: acquired SAA (38.8%), Fanconi anemia (13.1%), histiocytic disorders (9.7%), severe combined immune deficiencies, and other severe T-cell deficiencies (5.3%), Wiskott-Aldrich syndrome (5.1%), and Hurler syndrome (5.0%). Most diseases, like sickle cell anemia (0.5%), thalassemia major (1.1%), Chediak-Higashi syndrome (0.3%), leukocyte adhesion deficiencies (0.6%), and osteoporosis (1.4%), though, were rare. The distribution of diseases did not differ significantly across the 3 HLA-matching groups. The median age was 9 years (range, < 1-71) and also did not differ between groups.
There were significant differences in other baseline characteristics. Transplantations involving 8/8 matched pairs were more likely to have been performed after 2004 (P < .0001) and to have shorter follow-up (P < .0001), but less likely to have been performed using an ex vivo T cell–depleted graft (P = .0009). Patients with 8/8 matched donors were more likely (P < .05) to be white, have a CMV-seronegative donor if they were CMV seronegative, receive less intensive conditioning, and receive a higher PBSC CD34+ cell count.
Impact of DQ and DP mismatches on outcomes
Fully HLA-A, -B, -C, -DRB1 matched pairs were less likely to be mismatched at HLA-DQB1 than HLA-mismatched pairs (P = .040). The proportion of DPB1 mismatching also varied by group (P = .034; Table 1). In the univariate analysis, there was no association between overall survival and DQB1 mismatching (P = .64) or DPB1 (P = .58) mismatching. Similarly, there was no association between HLA mismatching at these loci and other outcomes. Matching at these loci, therefore, was not considered in subsequent analyses.
Mismatching at the HLA-A, -B, -C, and -DRB1 loci and outcome
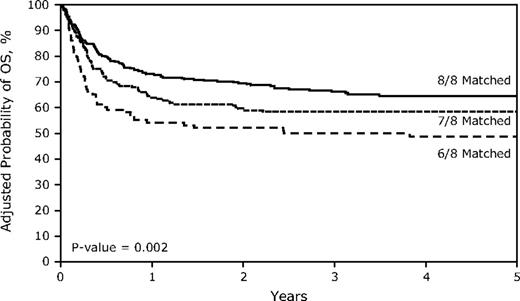
Unadjusted overall survival at 5 years was negatively associated with HLA mismatching (P = .003; Table 2). Estimated survival was 65% (95% CI, 60%-70%) in 8/8 matched transplants, 57% (50%-64%; vs 8/8, P = .07) in 7/8 matched transplants, and 46% (37%-56%; P = .001) in 6/8 matched transplants. After adjustment for graft cell dose, performance score, year of transplantation, and type of disease in the multivariate analysis, mismatching remained associated with mortality (P = .005; Table 3 and Figure 1). The hazard ratios for mortality for patients with single mismatch and double mismatched grafts were 1.29 (95% CI, 0.97-1.72; P = .079) and 1.82 (1.30-2.55; P = .0004). Other factors significantly associated with mortality included a diagnosis of Fanconi anemia, a pretransplantation Karnofsky performance score of less than 90, a PBSC graft mononuclear cell dose < 2 × 108/kg, a bone marrow graft nucleated cell dose < 5 × 108/kg, and a year of transplantation between 1995 and 1999 (data not shown).
Because the association of a single mismatch with mortality trended toward significance, we estimated the sample size that would be needed to achieve significance, using a 2-sided log-rank test at the 1% significance level. The calculation indicated that a sample size of 2352 would be needed to achieve statistical significance.
HLA mismatching was not associated with the unadjusted cumulative incidence of grades II-IV (P = .82) or grades III-IV (P = .59) acute GVHD (Table 2). Similarly, there was no association with chronic GVHD (P = .62). In the multivariate analyses, HLA mismatch was not associated with risk for either acute or chronic GVHD (Table 3).
The cumulative incidence of primary or secondary graft failure at 1 year was 17% (95% CI, 15%-20%). HLA mismatching was associated with incidence of graft failure (P < .0001); the cumulative incidence was 11% (95% CI, 8%-14%) for 8/8 matched transplants, 28% (95% CI, 21%-34%; P < .0001) for single mismatched transplants, and 24% (95% CI, 16%-33%; P = .008) for 2 mismatched transplants, respectively. Graft failure occurred in 4 of the 11 transplantations with a single mismatch in the host-versus-graft direction only and in the 1 transplantation with 2 mismatches in this direction. In multivariate analysis, the association between HLA mismatching and graft failure remained significant (P = .0001; Table 3). After adjustment for use of ex vivo T-cell depletion, disease type, and type of conditioning, the odds ratios for single and 2 mismatched transplants compared with 8/8 were 2.81 (95% CI, 1.74-4.54; P < .0001) and 2.24 (95% CI 1.26-3.97; P = .006), respectively. In this analysis, ex vivo T-cell depletion (2.38, 1.27-4.48; P = .007) and the use of RIC (2.09, 1.20-3.62; P = .009) were also associated with graft failure. Nonmyeloablative conditioning was associated with graft failure, but the result was not statistically significant (1.84, 0.82-4.12; P = .14). Disease type was not significantly associated with graft failure. Comparing single mismatches (allele or antigen) at the HLA-A, -B, -C, and -DRB1 loci, there were no significant differences in risk for graft failure. Using the HLA-A locus as the baseline, the odds ratios for the HLA-B, -C, and -DRB1 loci were 0.55 (95% CI, 0.21-1.42; P = .22), 0.75 (95% CI, 0.38-1.50; P = .41), and 0.25 (95% CI, 0.15-1.25; P = .12).
Of the 114 patients who suffered graft failure, 77 died (67.5%), accounting for 29.8% of all deaths. Of the patients who died after receiving 8/8 matched, single mismatched, and 2 mismatched transplantations, 21.3%, 41.5%, and 31.5% died after having graft failure.
Excluding donor-recipient pairs mismatched at the C allele did not materially alter the univariate or multivariate analyses (data not shown), suggesting that C allele mismatching was no better tolerated than other forms of mismatching. Splitting the single mismatch group into a single allele mismatch and a single antigen mismatch group also had little effect on the analyses; associations between allele mismatching and outcome and antigen mismatching and outcome differed little (data not shown).
Discussion
Since the late 1990s, there has been a succession of studies that have helped to define the current standards for HLA typing and matching in unrelated marrow and PBSC transplantation.1-5 The donor-recipient pair samples for these studies, however, were drawn either predominantly or exclusively from transplantations for patients with acute lymphocytic leukemia, acute myeloid leukemia, chronic myeloid leukemia, or myelodysplastic syndrome. Only 2 of these studies included patients with NMD, but in both studies such patients made up a small minority, comprising 9% of the sample in one and 10% in the other.1,4
Our findings indicate that in unrelated marrow and blood transplantation for NMD, HLA mismatching is associated with treatment failure just as it is in unrelated transplantation for malignancies.1-5 However, the primary immunologic consequence of HLA mismatching in transplantation for malignancies is GVHD,1-3 while in NMD it is graft failure. In addition, our results suggest that the association between HLA mismatching and mortality may not be as strong as it is in malignant diseases.1-3
Considering the influence of mismatching on mortality, our study, like recent CIBMTR studies of HLA matching and unrelated marrow and peripheral blood transplantation for malignancies, demonstrated that mismatching at the HLA-A, -B, -C, and -DRB1 loci, but not mismatching at the HLA-DQ or -DP loci, was associated with mortality.2,3 In our study, double, but not single, mismatches at the HLA-A, -B, -C, or -DRB1 loci were significantly associated with mortality. The recent CIBMTR studies of HLA matching in transplantation for malignant diseases, on the other hand, did demonstrate significant associations between mortality and single mismatches.2,3 In both, with their larger sample sizes, single allele and single antigen mismatches were considered separately. In the study of marrow transplantation, both allele and antigen mismatches were associated with mortality, while in the study of PBSC transplantation, only antigen mismatches were.2,3 Although our study suggests that a single mismatch may not increase the risk of mortality, this finding should be interpreted cautiously; the hazard ratio for a single mismatch (HR = 1.29) in our study was similar to ratios for single allele and antigen mismatches in the most recent CIBMTR study of marrow transplantation for malignant diseases, and our power calculations indicated that if our sample had been as large as that study's (n = 3857), a significant association may have been detected.2
The Japanese Marrow Donor Program (JMDP) recently reported the results of a study in 301 patients with acquired SAA.9 The authors concluded from their results that certain single locus mismatches have little influence on survival and that even transplants with 2 mismatches that do not involve the HLA-A or -B loci may not diminish survival greatly. Their methodology differed from ours in important respects. For example, they considered the HLA-A and -B loci together and the HLA-C, -DRB1, and -DQB1 loci together. Because of these differences, caution should be used comparing our findings with theirs. One also should keep in mind that racial dissimilarities between our sample (predominantly white) and the JMDP sample may have influenced the relative effects of HLA mismatching in the 2 studies. Race-related differences in HLA haplotypes16 and matching17 could be important, as could differences in the impact of minor histocompatibility antigen mismatching.18,19
The high incidence of graft failure, 17%, is notable and appears to exceed its incidence in unrelated donor transplantation for malignant diseases. While graft failure has not been rigorously assessed in most studies of unrelated marrow and peripheral blood transplantation, one study in chronic myeloid leukemia reported an incidence of only 6%.20 There are several reasons that graft failure may have been more common in our study. Except for patients with severe T-cell immune-deficiency diseases (5.6% of our sample), the immune systems of patients with NMD are more able to respond against the graft than are the immune systems of patients with malignant diseases, particularly those who were previously treated with multiagent, highly immunosuppressive chemotherapy.21 This difference may be amplified in some types of NMD, like SAA, where the HSC-targeted isoimmunity (which is responsible for the disease) and transfusion-related alloimmunity both promote rejection.22 Treatment differences, including the more frequent use of RIC and of ex vivo and in vivo T-cell depletion in transplantation for NMD, may also contribute to the higher graft failure rate.23,24
Because of the small number of transplantations with recipient homozygosity in our cohort, we were unable to fully assess the impact of isolated host-versus-graft mismatching on the risk for graft failure. However, 5 of the 12 transplantations where there was isolated host-versus-graft mismatching were complicated by graft failure. A recent study of unrelated cord blood transplantation for malignant and nonmalignant diseases reported that the risk of graft failure was higher with isolated host-versus-graft mismatching.25 In contrast, unpublished data from a forthcoming CIBMTR study of unrelated donor marrow and peripheral blood transplantation for patients with malignancies receiving myeloablative conditioning suggest that isolated host-versus-graft mismatching is not more strongly associated with graft failure than bidirectional mismatching (A.W. and C.K.H., unpublished observation, January 2012). Until further research is done in this area, we believe it is prudent to avoid performing transplantations with isolated host-versus-graft mismatches whenever possible if a patient has a NMD.
It is plausible that alloimmunization, induced by the transfusion therapy that figures importantly in the care of many patients with nonmalignant diseases, could accentuate the effect of HLA mismatching on rejection.22,26 Recent studies of unrelated marrow, peripheral blood, and cord blood transplantation in patients with malignancies have examined the impact of alloimmunization on graft failure, using HLA antibodies as a marker. These studies indicate that recipients who have antibodies directed against donor-disparate HLA antigens are at greatly increased risk for graft failure.27-30 A limitation of our study is that we were unable to account for the influence of alloimmunization. Although further research is needed in this area in the NMD population, we recommend testing patients lacking 8/8 matched donors for HLA-directed antibodies to guide selection of a graft that lacks the stimulating HLA-mismatch.
Previous studies have indicated that DP mismatching is important in graft rejection. Donor-directed antibodies, which, as noted previously, are strongly associated with graft rejection, are often targeted at DP antigens.22,26 A study in unrelated donor transplantation for thalassemia suggested that DP mismatches can be categorized as permissive and nonpermissive DP mismatches and the latter heighten the risk for graft rejection.31 In our study, we did not observe an association between DP mismatching and graft rejection. Because the majority of the donor-recipient pairs in our sample were DP mismatched, however, our ability to assess the effects of mismatching at this locus likely had limited power. Moreover, we did not distinguish between permissive and nonpermissive mismatches. Because the majority of our donor-recipient pairs were DQ matched, our ability to assess the effects of mismatching at this locus was also limited.
Our findings are notable for the absence of an association between HLA mismatching and acute GVHD. This contrasts with the 2 most recent CIBMTR studies in malignancies2,3 and the JMDP SAA study,9 which all demonstrated an association between HLA mismatching and acute GVHD. Most patients in our series received a lymphocyte-depleting antibody such as antithymocyte globulin or alemtuzumab, and many also received an ex vivo T-depleted graft. It is possible that these measures reduced the influence of HLA disparity on acute GVHD.32,33 The very young age (median 9 years) of patients in our series may also have contributed to the lack of association between HLA mismatching and risk for GVHD. A recent CIBMTR study of unrelated donor transplantation for children with acute leukemia also found no association between HLA mismatching and acute GVHD.34
Our cohort included patients with a wide variety of diseases, reflecting the great diversity of severe nonmalignant diseases for which allogeneic transplantation is beneficial. While these diseases differ from one another in important ways, several of the more frequent diseases, like SAA (38.8% of the patients),8 Fanconi anemia (13.1%),13 and Hurler syndrome (5%),12 have previously been associated with an incidence of graft failure exceeding 15%. To account for differences between diseases, we adjusted for disease in our multivariate model of graft failure. Because the probability of graft failure does vary by disease, when clinicians attempt to estimate the extent to which the use of an HLA-mismatched graft will increase their patients' probability of rejection, it is important for providers to consider disease. For example, although our results suggest that mismatching nearly triples a patient's probability of rejection, if a patient's risk is otherwise low, a tripling of it may still yield a risk that is acceptable in absolute terms.
Unrelated cord blood units represent an alternative graft source for many patients with NMD. As with unrelated marrow and peripheral blood transplantation, the effects of HLA matching in unrelated cord blood transplantations have been assessed primarily for patients with malignant diseases.35 As the number of unrelated cord blood transplantations performed for NMD grows, it will be important to assess the impact of HLA mismatching in this setting.
In conclusion, our results suggest that to minimize the risk of graft failure, HLA-A, -B, -C, and -DRB1 allele matched (8/8) donors should be sought for patients with NMD. Our results also suggest that for patients lacking such a donor, a single allele or antigen-mismatched donor may be used without greatly increasing the risk of mortality, but that 6/8 matched donors should be avoided because of a pronounced increase in risk for mortality.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The CIBMTR is supported by Public Health Service Grant/Cooperative Agreement U24-CA76518 from the National Cancer Institute (NCI), the National Heart, Lung, and Blood Institute (NHLBI), and the National Institute of Allergy and Infectious Diseases (NIAID); Grant/Cooperative Agreement 5U01HL069294 from NHLBI and NCI; contract HHSH234200637015C with the Health Resources and Services Administration (HRSA/DHHS); grants N00014-06-1-0704 and N00014-08-1-0058 from the Office of Naval Research; and grants from Allos Inc; Amgen Inc; Angioblast; an anonymous donation to the Medical College of Wisconsin; Ariad; Be the Match Foundation; Blue Cross and Blue Shield Association; Buchanan Family Foundation; CaridianBCT; Celgene Corporation; CellGenix GmbH; Children's Leukemia Research Association; Fresenius-Biotech North America Inc; Gamida Cell Teva Joint Venture Ltd; Genentech Inc; Genzyme Corporation; GlaxoSmithKline; HistoGenetics Inc; Kiadis Pharma; The Leukemia & Lymphoma Society; The Medical College of Wisconsin; Merck & Co Inc; Millennium: The Takeda Oncology Co; Milliman USA Inc; Miltenyi Biotec Inc; National Marrow Donor Program; Optum Healthcare Solutions Inc; Osiris Therapeutics Inc; Otsuka America Pharmaceutical Inc; RemedyMD; Sanofi; Seattle Genetics; Sigma-Tau Pharmaceuticals; Soligenix Inc; StemCyte, A Global Cord Blood Therapeutics Co; Stemsoft Software Inc; Swedish Orphan Biovitrum; Tarix Pharmaceuticals; Teva Neuroscience Inc; THERAKOS Inc; and Wellpoint Inc.
The views expressed in this article do not reflect the official policy or position of the National Institutes of Health, the Department of the Navy, the Department of Defense, or any other agency of the US government.
National Institutes of Health
Authorship
Contribution: J.H., T.W., M.H., S.R.S., J.D., M.E., H.F., V.G., G.A.H., C.K.H., S.M., M.O., V.R., P.S., and S.J.L. critically revised the research plan; J.H., S.J.L., and A.W. drafted the manuscript; J.H., T.W., M.H., S.R.S., J.D., M.E., H.F., V.G., G.A.H., C.K.H., S.M., M.O., V.R., P.S., S.J.L., and A.W. analyzed and interpreted data, and critically revised the manuscript; T.W. and M.H. performed statistics; and A.W. drafted the research plan.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: John Horan, MD, Emory University, 1405 Clifton Rd, Atlanta, GA 30322; e-mail: john.horan@choa.org.