Key Points
A genome-wide study of the association of over 5 million SNPs with methotrexate clearance in 1279 patients treated with HDMTX in multicenter COG trials 9904 and 9905.
We replicated the finding that inherited variations in SLCO1B1 are the most important genetic variations influencing methotrexate clearance.
Abstract
Methotrexate clearance can influence the cure of and toxicity in children with acute lymphoblastic leukemia (ALL). We estimated methotrexate plasma clearance for 1279 patients with ALL treated with methotrexate (24-hour infusion of a 1 g/m2 dose or 4-hour infusion of a 2 g/m2 dose) on the Children's Oncology Group P9904 and P9905 protocols. Methotrexate clearance was lower in older children (P = 7 × 10−7), girls (P = 2.7 × 10−4), and those who received a delayed-intensification phase (P = .0022). A genome-wide analysis showed that methotrexate clearance was associated with polymorphisms in the organic anion transporter gene SLCO1B1 (P = 2.1 × 10−11). This replicates findings using different schedules of high-dose methotrexate in St Jude ALL treatment protocols; a combined meta-analysis yields a P value of 5.7 × 10−19 for the association of methotrexate clearance with SLCO1B1 SNP rs4149056. Validation of this variant with 5 different treatment regimens of methotrexate solidifies the robustness of this pharmacogenomic determinant of methotrexate clearance. This study is registered at http://www.clinicaltrials.gov as NCT00005585 and NCT00005596.
Introduction
High-dose methotrexate (HDMTX) is an important element of chemotherapy in the treatment of acute lymphoblastic leukemia (ALL) and other malignancies.1-4 Mikkelsen et al5 showed that a 1 g/m2 dose given over 24 hours resulted in significantly greater active methotrexate polyglutamates in leukemic cells than the same dose given over 4 hours. In a randomized clinical trial, investigators from the Children's Oncology Group (COG) tested the efficacy and toxicity of a 2 g/m2 dose over 4 hours versus a 1 g/m2 dose over 24 hours.6,7 The systemic exposure to methotrexate (ie, plasma concentration over time) is related to cure and toxicity in children with ALL.8,9 In a group of children with ALL who were treated and monitored at a single institution (St Jude Children's Research Hospital), with extensive prospective therapeutic drug monitoring, even including pharmacokinetically guided dosage adjustments, we identified variants in the SLCO1B1 gene that were associated with methotrexate clearance in a genome-wide association study (GWAS).10,11 Herein, we sought to test whether we could replicate these GWAS findings in a large cohort of patients treated with alternative HDMTX schedules on the COG multi-institutional trials P9904 and P9905.
Methods
This study was approved by the institutional review boards of all participating institutions, and informed consent was obtained in accordance with the Declaration of Helsinki. Patients in P9904 included National Cancer Institute (NCI) standard-risk (age 1.00-9.99 years and initial white blood cell count [WBC]< 50 000/μL) patients with an ETV6-RUNX1 translocation or simultaneous trisomies of chromosomes 4 and 10, whereas patients in P9905 included a mixture of NCI standard-risk patients without favorable genetic lesions, NCI high-risk (age ≥ 10 years and/or initial WBC ≥ 50 000/μL) patients with favorable genetic changes, and other NCI high-risk patients who did not meet age-, WBC-, and sex-specific criteria for especially high-risk disease originally described by Borowitz et al6 and Shuster et al.12 Patients were randomized in a 2 × 2 manner to 1 of 4 arms for consolidation: (A) 24-hour methotrexate infusion (1 g/m2 given as a 200 mg/m2 bolus over 20 minutes, followed by 800 mg/m2 over 23.6 hours) and no delayed intensification (DI) phase; (B) 4-hour methotrexate infusion (2 g/m2) with no DI; (C) 24-hour methotrexate infusion with DI; and (D) 4-hour methotrexate infusion with DI. Leucovorin at 10 mg/m2 was given every 6 hours (3 doses), beginning at 42 hours after the start of the infusion and continued until the plasma methotrexate level was less than 0.2μM. Plasma methotrexate concentrations were requested to be drawn at the end of infusion (4 or 24 hours) and 24 hours later (postinfusion), and leucovorin was increased based on plasma methotrexate concentration (details in supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). All patients with t(1;19)(q23;p13) were in P9905 as a separate stratum—all were assigned to receive a DI phase and randomized to arms C or D for the methotrexate infusion. Patients with simultaneous trisomies of chromosomes 4 and 10 all were assigned to receive no DI and randomized to arms A or B for the methotrexate infusion. Patients who did not respond to induction chemotherapy were not eligible.
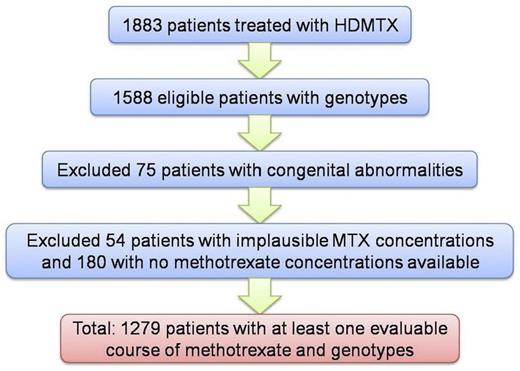
Data were retrieved from the COG Statistical and Data Center research databases. Of the 1883 patients treated with HDMTX on P9904 and P9905, 1588 were eligible and had germline DNA evaluable.6 Patients who had congenital abnormalities (usually Down syndrome, which is known to affect methotrexate clearance)13-15 or who did not have any courses that passed methotrexate quality control were excluded (Figure 1). Demographic information on the patients included can be found in supplemental Table 1. Although participating institutions in COG P9904/9905 were asked to submit data on plasma methotrexate concentrations, the data had not previously undergone any analysis or quality control. Therefore, it was necessary to establish a quality control approach to exclude courses with implausible data; for example, courses with undetectable plasma concentrations of methotrexate at the end of infusion were considered implausible and likely due to errors in drawing samples or in data entry. Courses with only 1 sample were excluded, as were courses for which the concentration of the postinfusion sample (28 or 48 hours) was greater than the end of infusion sample (4 or 24 hours). In addition, samples were removed that fell outside the expected 1-99th percentile range of methotrexate concentrations predicted with the use of the methotrexate pharmacokinetic parameters observed in nearly 500 prospectively monitored patients from St Jude's Total XV study (supplemental Table 2).10,16
Flow chart of patients included in this analysis. A total of 1883 patients were treated with HDMTX on P9904 and P9905, and 1588 were eligible and had germline DNA evaluable. Patients who had congenital abnormalities (usually Down syndrome) were excluded because they may not have received the full methotrexate dose. Patients with implausible or no evaluable methotrexate plasma concentrations were excluded (see “Methods” for details).
Flow chart of patients included in this analysis. A total of 1883 patients were treated with HDMTX on P9904 and P9905, and 1588 were eligible and had germline DNA evaluable. Patients who had congenital abnormalities (usually Down syndrome) were excluded because they may not have received the full methotrexate dose. Patients with implausible or no evaluable methotrexate plasma concentrations were excluded (see “Methods” for details).
In practice, this step resulted in the following: for the 24-hour infusion arms, end of infusion samples less than 2μM or greater than 38.5μM were excluded, and 48-hour samples greater than 7.5μM were excluded; for the 4-hour infusion arms, end of infusion samples less than 25μM or greater than 310μM were excluded, and 28-hour samples greater than 25μM were excluded. Low postinfusion samples were not excluded because they were considered plausible, whereas undetectable concentrations at the end of infusion (Figure 2) must be related to sampling or data entry errors. Thus, 252 of 4205 24-hour infusion courses (15 of 730 patients) and 653 of 4277 4-hour infusion courses (39 of 749 patients) were excluded on the basis of implausible pharmacokinetic data (likely due to data entry or sample acquisition error; Figure 2). Overall, the median clearance for all courses after excluding the outliers was similar to the median clearance prior to excluding the outliers (144.6 vs 141.2 mL/min/m2 for the 4-hour and 162.5 vs 162.3 mL/min/m2 for the 24-hour infusions).
Concentration versus time plots. (A) Twenty-four-hour methotrexate infusion of 1 g/m2. Concentrations shown were scheduled to be drawn at 24 hours (end of infusion) and at 48 hours (data are jittered [horizontal offset added] for better visualization). The pink lines represent the cut-offs (based on the range of the St Jude pharmacokinetic simulation) outside of which samples were excluded. There is no lower threshold for exclusion (pink line) for postinfusion samples. (B) Plasma concentrations were estimated for St Jude Total XV patients on the basis of a simulated dose of 1 g/m2 methotrexate over 24 hours (solid black line) and COG patients (dotted black line) receiving the same dose. The 1-99th percentile range is shown with shading for the St Jude Total XV patients (blue) and COG patients (red). (C) 4-hour methotrexate infusion of 2 g/m2. Concentrations shown were scheduled to be drawn at 4 hours (end of infusion) and at 28 hours (data are jittered for better visualization). (D) Plasma concentrations were estimated for St Jude Total XV patients based on a simulated dose of 2 g/m2 methotrexate over 4 hours (solid black line) and for COG patients (dotted black line) receiving the same dose. The 1-99th percentile range is shown with shading for the St Jude Total XV patients (blue) and COG patients (red).
Concentration versus time plots. (A) Twenty-four-hour methotrexate infusion of 1 g/m2. Concentrations shown were scheduled to be drawn at 24 hours (end of infusion) and at 48 hours (data are jittered [horizontal offset added] for better visualization). The pink lines represent the cut-offs (based on the range of the St Jude pharmacokinetic simulation) outside of which samples were excluded. There is no lower threshold for exclusion (pink line) for postinfusion samples. (B) Plasma concentrations were estimated for St Jude Total XV patients on the basis of a simulated dose of 1 g/m2 methotrexate over 24 hours (solid black line) and COG patients (dotted black line) receiving the same dose. The 1-99th percentile range is shown with shading for the St Jude Total XV patients (blue) and COG patients (red). (C) 4-hour methotrexate infusion of 2 g/m2. Concentrations shown were scheduled to be drawn at 4 hours (end of infusion) and at 28 hours (data are jittered for better visualization). (D) Plasma concentrations were estimated for St Jude Total XV patients based on a simulated dose of 2 g/m2 methotrexate over 4 hours (solid black line) and for COG patients (dotted black line) receiving the same dose. The 1-99th percentile range is shown with shading for the St Jude Total XV patients (blue) and COG patients (red).
Nonlinear mixed-effects modeling (Monolix 3.1; Lixoft) was used to determine population and individual posthoc methotrexate pharmacokinetic parameters. Specifically, a 2-compartment pharmacokinetic model with first-order elimination was fit to the data. The distribution of the parameters (both interoccasion and interindividual) was assumed log-normal. A proportional residual error model was used assuming normal distribution of the residuals. Furthermore, because of the sparse nature of the sampling in the current study, we included as a prior distribution the methotrexate pharmacokinetics from the St Jude Total XV study (supplemental Table 2). For the genome-wide analysis, we used the average methotrexate clearance value per patient.10
Germline DNA was isolated at the time of remission from peripheral blood. DNA was extracted and genotyped using the Genome-Wide Human SNP Array 6.0 as described (Affymetrix, Santa Clara, CA).17 DNA also was genotyped by the Illumina GoldenGate platform for 7 single-nucleotide polymorphisms (SNP; SNP Center, Johns Hopkins University). We did not perform sequencing or genotype for any rare variants. Genotype calls were coded as 0, 1, or 2 for the number of B alleles (AA, AB, BB). SNPs with a minor allele frequency less than 1% or a call rate less than 95% were excluded. Imputation of SNPs in the 1000 Genomes dataset not represented on the Affymetrix SNP 6.0 array was performed on each ancestral group independently using MACH1.0 (see supplemental Methods).18
A GWAS between germline SNP genotypes and log2-transformed methotrexate clearance was performed with the use of a general linear model assuming an additive effect of allelic dosage, with all models including age, genetic ancestry, sex, and treatment regimen as covariates, as we previously described.10,11 Genetic ancestry was estimated with STRUCTURE as previously described (see supplemental Methods).17 Age was coded in years and treated as a continuous variable. Treatment regimen was coded as a categorical variable. Adjusted methotrexate clearance refers to the residual from a linear regression including the clinical covariates. A meta-analysis was performed by combining the statistics from the current GWAS with those obtained from the GWAS of methotrexate clearance in St Jude patients.10 We calculated the combined P values from the meta-analysis using the Stouffer method.19,20
Results
We initially tested covariates for their association with methotrexate clearance in a multivariate general linear model (Table 1). Receipt of the 4-hour (rather than the 24-hour infusion; P < 2 × 10−16; Figure 3A) treatment arm, including a DI phase (P = .0010; supplemental Figure 1), older age (P = 9 × 10−7; supplemental Figure 2, consistent with previous reports),10 and female sex (P = .000 28, supplemental Figure 3) were all associated with lower clearance. Methotrexate clearance was not related to genetically defined patient ancestry in this study (supplemental Figure 4). In sum, we were able to explain 42.1% of the interindividual variability in methotrexate clearance with the clinical covariates and 1 SNP in SLCO1B1, rs4149056 (Table 1), in the same range as in our previous study.11
Histogram of each patient's average methotrexate clearance. (A) Unadjusted methotrexate clearance for the 655 COG patients receiving the 4-hour infusion (blue), and the 624 receiving the 24-hour infusion (red). (B) Methotrexate clearance in all COG patients (n = 1279), adjusted for age, sex, race, and treatment arm (residual from a linear regression model).
Histogram of each patient's average methotrexate clearance. (A) Unadjusted methotrexate clearance for the 655 COG patients receiving the 4-hour infusion (blue), and the 624 receiving the 24-hour infusion (red). (B) Methotrexate clearance in all COG patients (n = 1279), adjusted for age, sex, race, and treatment arm (residual from a linear regression model).
When we adjusted for age, sex, and treatment arm, methotrexate clearance displayed a unimodal population distribution (Figure 3B). A GWAS was performed for SNP-genotype associations with clearance, including age, sex, genetic ancestry, and treatment arm as covariates. The GWAS showed a strong association between methotrexate clearance and SNPs in SLCO1B1 (Figure 4A; Table 2). There were 4 typed SNPs that reached a genome-wide significance threshold of P < 5 × 10−8 (rs4149081, rs4149056, rs11045879, and rs11045821). rs4149056 is a nonsynonymous coding SNP that has been shown to have reduced transport of methotrexate in vitro.11 Infusion length, treatment including a DI, age, and sex remained significantly associated with clearance in a multivariate model, including the rs4149056 genotype (Table 1); it is unclear why there was an association of DI with methotrexate clearance because it persisted after we adjusted for the sex, race, and age factors that might have confounded the association. The other 3 significant SNPs are in linkage disequilibrium (LD) with rs4149056. With each copy of the C allele at rs4149056, the adjusted methotrexate clearance was reduced by 12 mL/min/m2 (Table 2; supplemental Figure 5); thus, clearance is (on average) approximately13% lower in patients with CC versus TT genotypes at rs4149056, a magnitude of difference in exposure that could have significance for efficacy or adverse events.9
Manhattan plot of P values for a genome-wide association with methotrexate clearance. Each chromosome is plotted along the x-axis, whereas the y-axis plots the negative log10 of the P value for the association of each SNP with methotrexate clearance. Higher points on the graph indicate a stronger association with methotrexate clearance. Typed and imputed SNPs are shown in all panels (5.2 million SNPs). Methotrexate clearance is adjusted for age, sex, race, and treatment arm. (A) Association of each SNP with methotrexate clearance in COG patients (n = 1279). (B) Meta-analysis of St Jude (n = 699) and COG patients (n = 1279). (C) Meta-analysis of St Jude and COG patients, after we adjusted for rs4149056.
Manhattan plot of P values for a genome-wide association with methotrexate clearance. Each chromosome is plotted along the x-axis, whereas the y-axis plots the negative log10 of the P value for the association of each SNP with methotrexate clearance. Higher points on the graph indicate a stronger association with methotrexate clearance. Typed and imputed SNPs are shown in all panels (5.2 million SNPs). Methotrexate clearance is adjusted for age, sex, race, and treatment arm. (A) Association of each SNP with methotrexate clearance in COG patients (n = 1279). (B) Meta-analysis of St Jude (n = 699) and COG patients (n = 1279). (C) Meta-analysis of St Jude and COG patients, after we adjusted for rs4149056.
In a meta-analysis in which we combined patients treated with the COG and St Jude protocols, SLCO1B1 remained significantly related to methotrexate clearance (Figure 4B). Of the 103 typed or imputed SNPs that reached a Bonferroni-corrected genome-wide significance threshold of P < 9.62 × 10−9 (supplemental Table 3), 101 were in SLCO1B1. The meta-analysis P value for the top SNP in SLCO1B1, rs4149080, is 5.61 × 10−21 (supplemental Table 3). This SNP is in LD (r2 = 0.897) with rs4149056 in European patients and was imputed in both cohorts. The SNP typed in both the COG and St Jude cohorts that had the smallest P value was rs11045879, which is also in LD with rs4149056 (r2 = 0.865), and had a P value of 3.13 × 10−19 in the meta-analysis. Two SNPs in SLCO1A2 also were related to methotrexate clearance at the genome-wide level (7.6 × 10−10 and 1.9 × 10−9); however, this is most likely because they are very near SLCO1B1 on chromosome 12 and in LD with the rs4149056 SNP in SLCO1B1 (Figure 5).21,22 They are not in LD with the rs11045891 and rs4148981 SNPs that are SLCO1A2 enhancers.23 SLCO1B3 is a closely related family member that also transports methotrexate.24,25 Although they did not reach genome-wide significance, 2 SNPs in SLCO1B3 were related to methotrexate clearance (P < 3 × 10−6), and the association remained after we adjusted for SNPs in SLCO1B1. Four SNPs in SLC19A1 (the reduced folate carrier) were interrogated in this study on the Affymetrix array (2 are in LD with functional polymorphisms rs1051266 and rs1131596, r2 > 0.9), but none reached genome-wide significance for an association with methotrexate clearance.
LD of SNPs with P < 10−5 in meta-analysis spanning SLCO1B3, LST-3TM12, SLCO1B1, and SLCO1A2 generated with the use of Haploview software21 in European-ancestry patients only (COG n = 806 and SJ n = 459). The darkness of each box represents the association between 2 SNPs (R2), with white being R2 = 0, black being R2 = 1, and 0 < R2 < 1 being shades of gray. LD blocks, highlighted with the yellow lines, were defined by confidence intervals.22 Assembly hg18 from http://genome.ucsc.edu used for display.
LD of SNPs with P < 10−5 in meta-analysis spanning SLCO1B3, LST-3TM12, SLCO1B1, and SLCO1A2 generated with the use of Haploview software21 in European-ancestry patients only (COG n = 806 and SJ n = 459). The darkness of each box represents the association between 2 SNPs (R2), with white being R2 = 0, black being R2 = 1, and 0 < R2 < 1 being shades of gray. LD blocks, highlighted with the yellow lines, were defined by confidence intervals.22 Assembly hg18 from http://genome.ucsc.edu used for display.
The nonsynonymous rs4149056 (T521C) SNP has been extensively characterized in relation to other phenotypes26-28 and shown to reduce methotrexate transport in vitro.11 After we adjusted for the rs4149056 genotype, a significant association remained in SLCO1B1 at the genome-wide significance level (Figure 4C). The SNP with the lowest P value after adjustment for rs4149056 was an intronic SNP, rs4149040 (P = 1.2 × 10−12, supplemental Table 4), which is in LD with the nonsynonymous SNP rs2306283. Together, rs2306283 and rs4149056 define the most common haplotypes of SLCO1B1 (*1a, *1b, *5, and *15)29 and are in partial LD (D′ = 0.91, r2 = 0.25). As we previously demonstrated, the relationship between methotrexate clearance and rs2306283 genotype is evident only in an analysis stratified for rs4149056 genotype (supplemental Figure 6, P = 4 × 10−11),11 indicating a highly reproducible SNP-by-SNP interaction. Patients with at least 1 C allele at rs4149056 have lower clearance than those with 2 T alleles (supplemental Figure 5); however, within each rs4149056 genotypic group, the A allele at rs2306283 was associated with even lower clearance, whereas the variant G allele was associated with increased clearance (supplemental Figure 6). We also replicated that the rs11045872 variant G allele was associated with greater methotrexate clearance (P = 3.8 × 10−7 in COG patients, supplemental Table 3; supplemental Figure 7).
Discussion
We have replicated the association between SLCO1B1 and methotrexate clearance10,11 with 2 additional schedules of intravenous methotrexate administration in the setting of a cooperative group trial. We estimated methotrexate clearance using 2 plasma methotrexate concentrations per course via a Bayesian pharmacokinetic modeling approach. The mean and range of clearances that we estimated from only 2 plasma concentrations was similar to that estimated from an entirely different group of patients with more samples per course (St Jude Total XV participants).10,11 Further external validity of methotrexate clearance estimates from these large multicenter trials is provided by the fact that we replicated the association of methotrexate clearance with age, sex, and treatment regimen. We previously found that black patients had the greatest methotrexate clearance10 ; although this association was not replicated here, there were relatively few black patients (5%) in this COG cohort.
SLCO1B1 is an organic anion transporter that is known to transport methotrexate; minor alleles for SNPs in the SLCO1B1 gene are associated with both increased and decreased transporter function and span multiple functional domains in the gene.11 We found SNPs associated with both increased and decreased methotrexate clearance in the COG cohort, replicating our previous finding in the St Jude cohort (supplemental Figures 5-7).11 We improved our power to detect significant associations by performing a meta-analysis combining the COG and St Jude cohorts. The meta-analysis showed that only SNPs on chromosome 12 near SLCO1B1 were related to methotrexate clearance in these patients, many of which were not significant at the genome-wide threshold in either cohort alone. The nonsynonymous rs4149056 (T521C) SNP has been the subject of multiple clinical studies of statins and other agents.26,30-34 However, even after adjustment for the rs4149056 SNP, other SNPs in SLCO1B1 remained significantly related to methotrexate clearance, indicating that there are multiple variants in SLCO1B1 that can influence methotrexate clearance. In this independent cohort, we replicated our previous finding, that is, that the rs2306283 G allele was associated with greater clearance after we adjusted for the rs4149056 genotype.11
Variants in SLCO1B1 could be clinically useful in identifying patients at risk of low methotrexate clearance, particularly in settings in which plasma methotrexate concentrations are not readily available. Toxicity associated with 1 g/m2 and 2 g/m2 doses has been reported.35,36 One possible intervention would be to increase IV hydration and/or alkalinization in the subset of patients with a “low-clearance” SLCO1B1 genotype to force greater clearance of methotrexate, but the effects of this intervention on toxicity are not known. Tools for aggregation of SLCO1B1 variants into clinically important haplotypes are available (http://www.pharmgkb.org/gene/PA134865839), and such haplotypes are already used for dosing decisions for other drugs, such as statins.37 In addition, it is likely that the coadministration of HDMTX with other SLCO1B1 substrates or inhibitors will result in increased plasma methotrexate concentrations.38-40
In summary, we have replicated the finding that SLCO1B1 is related to methotrexate clearance. This validates our previous GWAS10 in a separate group of patients who received different dosages of HDMTX in a multi-institutional clinical trial, solidifying the robustness of this pharmacogenomic determinant of methotrexate clearance.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to physicians and clinical research associates at the COG institutions participating in this study for cooperation with the sample submission requirements. The authors thank Shaherah Rankins for assistance in preparation of the manuscript.
This work was supported by grants CA 36401, GM 92666, CA 98453, CA98413, CA114766, and CA 21765 from the National Institutes of Health and the American Lebanese Syrian Associated Charities. The authors thank the Jeffrey Pride Foundation and the National Childhood Cancer Foundation for their financial support for genome-wide genotyping of COG specimens. S.P.H. is the Ergen Family Chair in Pediatric Cancer.
National Institutes of Health
Authorship
Contribution: L.B.R., C-H.P., W.E.E., and M.V.R. designed research, analyzed and interpreted data, and drafted the manuscript; J.C.P., C.S., W.Y., Y.F., C.C., and M.D. analyzed and interpreted data and performed statistical analysis; and N.J.W., P.L.M., S.P.H., and M.L. collected, analyzed and interpreted data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mary V. Relling, PharmD, St Jude Children's Research Hospital, 262 Danny Thomas Place, MS 313, Memphis, TN 38105; e-mail: mary.relling@stjude.org.

![Figure 2. Concentration versus time plots. (A) Twenty-four-hour methotrexate infusion of 1 g/m2. Concentrations shown were scheduled to be drawn at 24 hours (end of infusion) and at 48 hours (data are jittered [horizontal offset added] for better visualization). The pink lines represent the cut-offs (based on the range of the St Jude pharmacokinetic simulation) outside of which samples were excluded. There is no lower threshold for exclusion (pink line) for postinfusion samples. (B) Plasma concentrations were estimated for St Jude Total XV patients on the basis of a simulated dose of 1 g/m2 methotrexate over 24 hours (solid black line) and COG patients (dotted black line) receiving the same dose. The 1-99th percentile range is shown with shading for the St Jude Total XV patients (blue) and COG patients (red). (C) 4-hour methotrexate infusion of 2 g/m2. Concentrations shown were scheduled to be drawn at 4 hours (end of infusion) and at 28 hours (data are jittered for better visualization). (D) Plasma concentrations were estimated for St Jude Total XV patients based on a simulated dose of 2 g/m2 methotrexate over 4 hours (solid black line) and for COG patients (dotted black line) receiving the same dose. The 1-99th percentile range is shown with shading for the St Jude Total XV patients (blue) and COG patients (red).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/121/6/10.1182_blood-2012-08-452839/4/m_zh89991301740002.jpeg?Expires=1765015118&Signature=IepmeLgfxeuW7pK7WHHbBNoqAAhOIWgmBRr3NTlBLr-pO1EeOlibzYFzfFHsPKSQz5sRjRpNn8YyVRTI-vm8C--6vGLDV2qnJRF8gKkJu0quoM~Ahr0hQwQika1C1p9nleks2KTVydmp-yG-d5dpw5wkwpR8fIbaG3vqeuHdZvas3qYe4xniWFwOqbXdanYsZDo-1DGSAkazsdSeDhmys28zu652D3-Qzb-ayIs95DX37ssfj7wEOC7eFs4PMiQ2CAhS-D0~5Z0zbMODFO7I0O-5cDXOeN74MSfHW7efZ9bCD6loet5tkTxy~hMa52D~UBVZG~-8HC0lY3oxoyit~g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)





![Figure 2. Concentration versus time plots. (A) Twenty-four-hour methotrexate infusion of 1 g/m2. Concentrations shown were scheduled to be drawn at 24 hours (end of infusion) and at 48 hours (data are jittered [horizontal offset added] for better visualization). The pink lines represent the cut-offs (based on the range of the St Jude pharmacokinetic simulation) outside of which samples were excluded. There is no lower threshold for exclusion (pink line) for postinfusion samples. (B) Plasma concentrations were estimated for St Jude Total XV patients on the basis of a simulated dose of 1 g/m2 methotrexate over 24 hours (solid black line) and COG patients (dotted black line) receiving the same dose. The 1-99th percentile range is shown with shading for the St Jude Total XV patients (blue) and COG patients (red). (C) 4-hour methotrexate infusion of 2 g/m2. Concentrations shown were scheduled to be drawn at 4 hours (end of infusion) and at 28 hours (data are jittered for better visualization). (D) Plasma concentrations were estimated for St Jude Total XV patients based on a simulated dose of 2 g/m2 methotrexate over 4 hours (solid black line) and for COG patients (dotted black line) receiving the same dose. The 1-99th percentile range is shown with shading for the St Jude Total XV patients (blue) and COG patients (red).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/121/6/10.1182_blood-2012-08-452839/4/m_zh89991301740002.jpeg?Expires=1765015119&Signature=mihFrdG1deqLIwQDGFtF6kSuA6WyrRm0ik7aLjY4bLfNVS9vTZOshfcjABZfiai83uDctDsxuhmIORNsoNA07tLhrCAFUAE7dzHjwohdIzGnn49PkLjNL343WJwRZCz~dthaQJE0M2EatEZlLkB8g3~olT89DE158OhrlUIYLzRXrIiZnuEFpe6k-QTvgjoXZn3XjkbBuwv4ElZR5tcoxeK9H7DCjnLpOfCYzrtwcLorYTFXNFhciBnqaNk9wlFlN3KJBh8MGgJwR45g23PSVvZFo51i1f0s1CsFSJ3BTcZLHBHNXTYGDOErQ9~G8WS9un7bqp3BkYqa5cELXI08EQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


