Key Points
A mutation preventing interaction between c-Myb and p300 prevents transformation and leukemia induction by MLL-AF9 and AML1-ETO9a oncogenes.
Identifying agents that block the c-Myb-p300 interaction may be a valuable approach to developing a therapy for acute myeloid leukemia.
Abstract
The MYB oncogene is widely expressed in acute leukemias and is important for the continued proliferation of leukemia cells, suggesting that MYB may be a therapeutic target in these diseases. However, realization of this potential requires a significant therapeutic window for MYB inhibition, given its essential role in normal hematopoiesis, and an approach for developing an effective therapeutic. We previously showed that the interaction of c-Myb with the coactivator CBP/p300 is essential for its transforming activity. Here, by using cells from Booreana mice which carry a mutant allele of c-Myb, we show that this interaction is essential for in vitro transformation by the myeloid leukemia oncogenes AML1-ETO, AML1-ETO9a, MLL-ENL, and MLL-AF9. We further show that unlike cells from wild-type mice, Booreana cells transduced with AML1-ETO9a or MLL-AF9 retroviruses fail to generate leukemia upon transplantation into irradiated recipients. Finally, we have begun to explore the molecular mechanisms underlying these observations by gene expression profiling. This identified several genes previously implicated in myeloid leukemogenesis and HSC function as being regulated in a c-Myb-p300-dependent manner. These data highlight the importance of the c-Myb-p300 interaction in myeloid leukemogenesis and suggest disruption of this interaction as a potential therapeutic strategy for acute myeloid leukemia.
Introduction
Chromosomal translocations involving transcription factors are commonly associated with the pathogenesis of acute myeloid leukemia (AML).1-3 Two of the most common are the t(8;21)(q22;q22) translocation involving RUNX1(AML1) and RUNX1T1 (ETO) and that involving the KMT2A (MLL) gene on 11q23. The latter can be translocated to many other loci to generate fusions with different partners, although a small subset represents the majority of cases.4 AMLs with t(8;21)(q22;q22) are generally regarded as having a relatively favorable prognosis, although at least 30% of patients do not survive beyond 5 years postdiagnosis1 ; in contrast MLL translocations confer a very poor prognosis.1,2
The precise mechanism by which the AML1-ETO fusion protein leads to leukemia development is still not fully understood,5 although recent studies have highlighted the importance of oligomerization to form an AML1-ETO–containing transcription factor complex.6 Recently reviewed by Hatlen et al,7 several studies suggest that the protein functions as both a transcriptional repressor and an activator, and it appears to block differentiation and confer self-renewal ability upon hematopoietic progenitors during the process of leukemic transformation. Indeed, the identification and characterization of downstream targets and pathways involved in transformation by AML1-ETO has been reported.8-14
Transformation and leukemogenesis by MLL fusion oncogenes such as MLL-AF9 and MLL-ENL have also been studied intensively.15 Two key partners recruited by these fusions have been identified as the histone methyltransferase DOT1L16 and the transcriptional elongation factor P-TEFb,17 consistent with widespread transcriptional dysregulation as a major activity. Correspondingly, a number of studies18-21 have identified downstream targets of these oncoproteins.
The c-Myb protein is a sequence-specific DNA-binding transcription factor that plays an essential role in hematopoiesis. c-Myb null mice present with defects in hematopoiesis, including a lack of functional progenitors, and die before birth.22,23 Subsequently, several studies have reinforced the requirement for at least one wild-type (WT) allele of c-Myb for normal hematopoietic differentiation.24-27
Although it is required for the proliferation of human AML cells,28,29 only recently has a causal role been attributed to MYB in some human leukemias. Recurrent duplications, genomic gain, and chromosomal translocations have been reported in the MYB locus in T-cell acute lymphoblastic leukemia and subsets of AML.30-34 c-Myb has also been shown to coordinate a self-renewal program that is essential for the maintenance of MLL-AF9–induced AML in a mouse model.21 Thus, despite being unaltered in most AML cases, MYB expression appears to be a general requirement for the proliferation of leukemic cells.
Several coregulators have been shown to interact with c-Myb through various domains, including CBP/p300, Mybbp1a, TIP60, Menin, and several corepressors.35-39 We have previously shown that CBP/p300 is an essential coactivator for the intrinsic transforming capacity of c-Myb in vitro.40 We have also recently demonstrated that CBP/p300 is required for the ability of c-Myb to repress several key target genes involved in myeloid differentiation and that p300 is recruited to Myb-binding sites near c-Myb-activated and c-Myb-repressed genes.41
Here, we address the role of the c-Myb-CBP/p300 interaction in the development of AML. We used two mutant mouse strains, Booreana42 and Plt6,26 that carry mutations in c-Myb and Ep300, respectively, that interfere with the interaction of c-Myb with CBP/p300. By using these models, we show that abrogation of the c-Myb-CBP/p300 interaction blocks transformation and leukemogenesis by both AML1-ETO and MLL fusions, suggesting that the c-Myb-CBP/p300 interaction could be targeted as a therapeutic strategy for AML. We also carried out expression profiling to identify CBP/p300-binding–dependent c-Myb targets that could contribute to the c-Myb dependence of transformation.
Materials and methods
Retroviral vectors
AML1-ETO, MLL fusion, and Myb oncogenes were expressed by using MSCV-IRES-GFP or pMYs-IRES-Venus vectors. For in vitro studies, virus was generated by cotransfection of 293 T cells with the corresponding plasmids and the pEQEco packaging construct.
Gene expression profiling
RNA isolated from vector– and AML-ETO9a–transduced Kit+GFP+ bone marrow cells was analyzed 48 hours posttransduction on Illumina MouseRef-8 v2.0 arrays.
Serial replating assays
Bone marrow cells were isolated from WT C57BL/6 and Booreana mice and enriched for c-Kit–expressing cells by using MACS CD117 microbeads (Miltenyi). Cells were transduced with retroviral supernatants by spinoculation using fibronectin (Sigma; F-1141) or Retronectin (Takara) according to the manufacturer’s guidelines. Cells were cultured in Iscove's modified Dulbecco medium with 20% fetal calf serum and a cocktail of interleukin-3 (IL-3), IL-6, and stem cell factor cytokines produced in-house. Serial replating assays were carried out as described previously.43
Leukemia induction
Single-cell suspensions from day 14.5 fetal livers were incubated overnight with GP+E86 packaging cells producing empty vector, AML1-ETO9a, and MLL-AF9 retroviruses in the presence of recombinant IL-11, IL-6, and stem cell factor. Then, 1.5 × 106 of these fetal liver cells were retro-orbitally injected into irradiated B6.SJL-Ptprca (CD45.1) recipients (11 Gy split dose). Mice were monitored for signs of illness and for GFP positivity (GFP+) of and surface marker expression by blood cells obtained by tail vein sampling over the course of the study. Sick mice were euthanized by asphyxiation, femurs were flushed, and spleen and liver were crushed to obtain cells for fluorescence-activated cell sorting. Acquisition was carried out on a Cyan ADP cytometer (Beckman Coulter, Brea, CA) and analysis was performed by using FlowJo software (Tree Star Inc., Ashland, OR). All animal studies were approved by the University of Queensland Animal Ethics Committee. Additional detail regarding materials and methods can be found in the supplemental Data (available on the Blood Web site).
Results
AML1-ETO and MLL fusions are unable to transform hematopoietic cells from Booreana mice
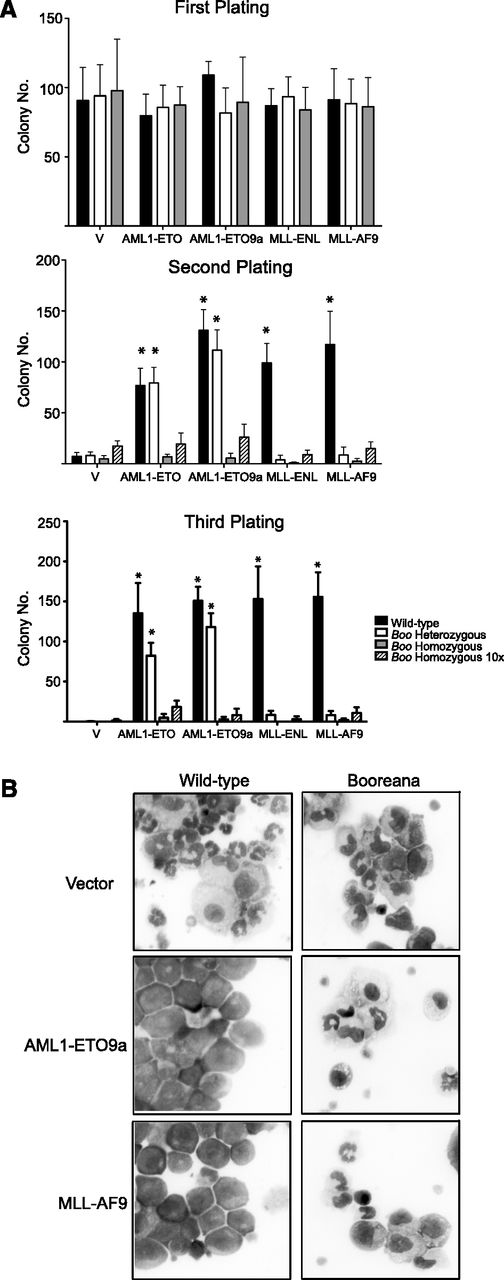
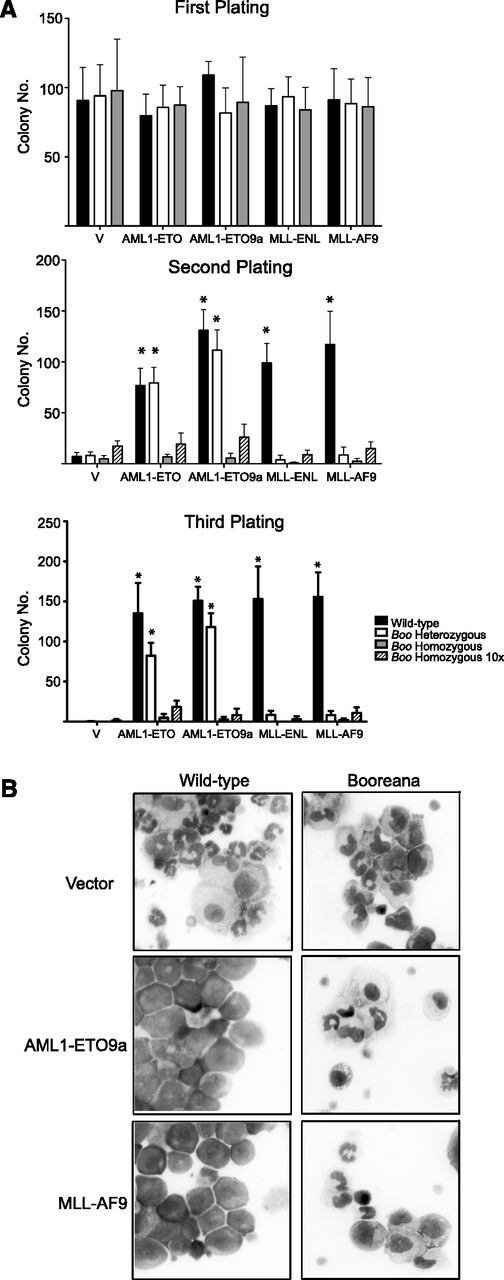
To study the role of the c-Myb-CBP/p300 interaction in the development of AML, we used the Booreana (Boo) mouse strain.42 In this strain, c-Myb has a substitution, E308G, that disrupts its interaction with CBP/p300 (supplemental Figure 1A) in several assays,42 including as we show here, coimmunoprecipitation in c-Kit–enriched bone marrow progenitors (supplemental Figure 1B). The transforming ability of AML1-ETO and AML1-ETO9a44 oncogenes was assayed by their ability to induce colony formation in methylcellulose over serial rounds of replating43 as a measure of their ability to induce sustained self-renewal. In this assay, both oncogenes were able to transform Kit-enriched cells from WT mice; however, there was a striking loss of transformation when using cells from Boo homozygous mice (Figure 1A). Previous studies have shown a stringent requirement for c-Myb in MLL-ENL–mediated transformation,20,45 and indeed we found that neither MLL-ENL nor MLL-AF9 could induce serial colony formation in cells from Boo heterozygous mice. This lack of colony-forming ability is not an inherent property of hematopoietic cells from Boo mice because similar numbers of colonies from WT, Boo/+, and Boo/Boo cells were seen in the first round of plating. The inability of these AML oncogenes to induce serial colony formation is unlikely to be the result of decreased numbers of any particular compartment of the myeloid lineage in Boo mice, because even a 10-fold increase in the number of cells did not lead to a significant increase in colony numbers in replating rounds 2 and 3.
Cells from Booreana mice are resistant to transformation by AML oncogenes in vitro. (A) Bone marrow cells from WT and Booreana heterozygous but not Booreana homozygous mice form colonies in serial replating assays following transduction with AML oncogene-encoding retroviruses. Error bars represent standard deviation (SD) of four biological replicates; *P < .05 (Mann-Whitney U test). (B) AML oncogenes are unable to block differentiation of Booreana cells; these cells exhibit increased monocytic differentiation compared with transformed WT cells as seen on stained cytospin preparations.
Cells from Booreana mice are resistant to transformation by AML oncogenes in vitro. (A) Bone marrow cells from WT and Booreana heterozygous but not Booreana homozygous mice form colonies in serial replating assays following transduction with AML oncogene-encoding retroviruses. Error bars represent standard deviation (SD) of four biological replicates; *P < .05 (Mann-Whitney U test). (B) AML oncogenes are unable to block differentiation of Booreana cells; these cells exhibit increased monocytic differentiation compared with transformed WT cells as seen on stained cytospin preparations.
Furthermore, in suspension cultures, we saw a clear block of differentiation by AML1-ETO and AML1-ETO9a in cells from WT and Boo heterozygous mice as indicated by their immature morphology (Figure 1B) and low expression of the differentiation markers CD11b and Gr-1 (supplemental Figure 2). However, in cells from Boo homozygous mice, AML1-ETO was unable to induce a block of differentiation; these cells displayed predominantly monocytic differentiation as shown by morphology and high CD11b and low Gr-1 expression. Similarly, MLL-AF9–transduced WT but not Boo cells showed a clear morphologic differentiation block (but also expressed CD11b and Gr-1, as expected). These results suggest that the lack of c-Myb-CBP/p300 interaction renders Boo cells resistant to transformation by AML1-ETO, AML1-ETO9a, MLL-ENL, and MLL-AF9, in part by altering their ability to block myeloid differentiation.
AML1-ETO and MLL fusions are unable to transform hematopoietic cells from Plt6 mice
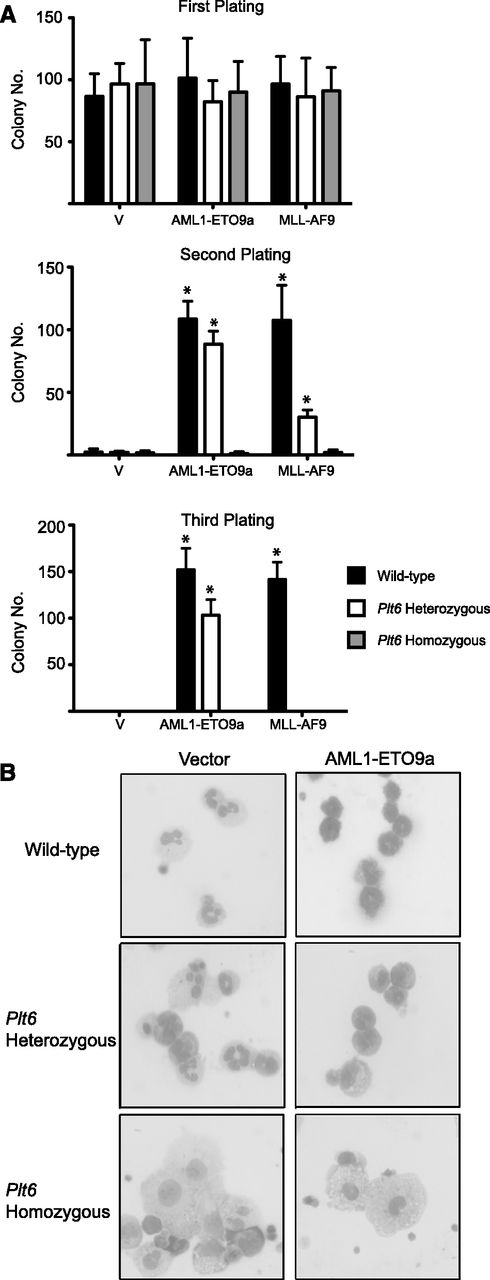
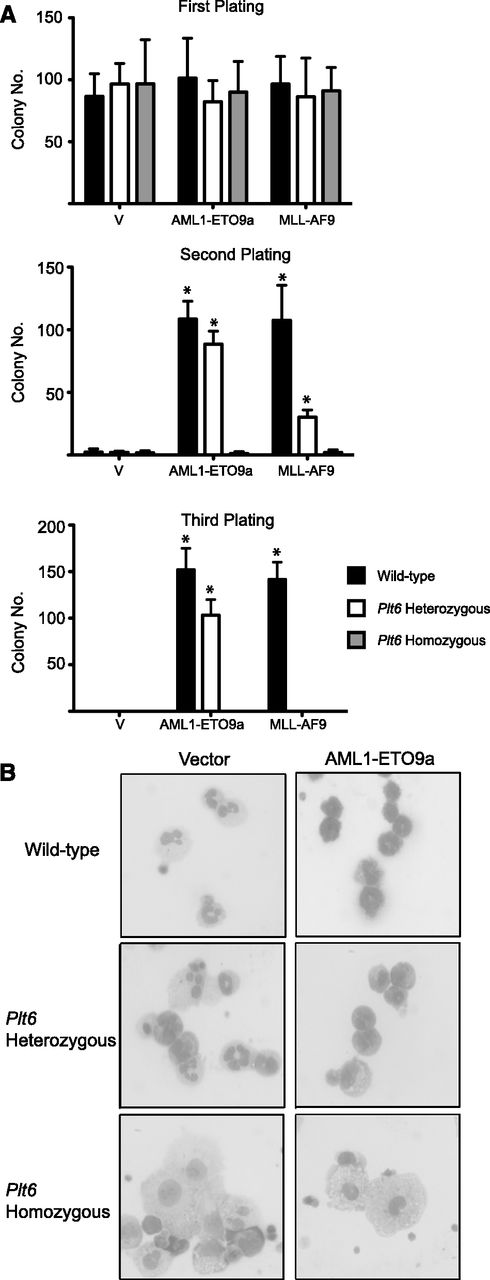
In addition to CBP/p300, at least two other factors, Menin and TIP60, are known to interact with c-Myb through its transactivation domain.36,39 To ensure that the effects we observed with the Boo mutation were solely due to disruption of interaction with CBP/p300, we carried out transformation experiments using another mouse strain, Plt6, which harbors a mutation at tyrosine 630 of the KIX domain of p300 that occurs in the α2 helix of the interaction region (supplemental Figure 1A), which interferes with the ability of p300 to interact with c-Myb.26 These mice exhibit hematopoietic abnormalities that are somewhat similar to those of the Boo mice, which is consistent with the phenotypic effects of both mutants being a result of a similar defect—interference with c-Myb-CBP/p300 binding.
We observed strikingly similar results when comparing transformation of hematopoietic progenitors from Plt6 and Boo mice by AML1-ETO9a and MLL-AF9. Neither of these oncoproteins was able to induce serial colony formation in cells from Plt6 homozygous mice (Figure 2A). The ability of MLL-AF9 to transform cells from Plt6 heterozygous mice was also decreased threefold on the first replating and abolished in subsequent rounds. As assessed by cell morphology (Figure 2B), it was evident that the inability of these oncogenes to transform Plt6 progenitors could again be attributed to their inability to induce a block of differentiation.
Cells from Plt6 mice are resistant to transformation by AML oncogenes in vitro. (A) Fetal liver cells from WT mice but not Plt6 mice serially form colonies in replating assays following transduction with AML oncogene–encoding retroviruses. Error bars represent SD of three biological replicates; *P < .05 (Mann-Whitney U test). (B) These exhibit a more differentiated morphology with increased numbers of monocytes.
Cells from Plt6 mice are resistant to transformation by AML oncogenes in vitro. (A) Fetal liver cells from WT mice but not Plt6 mice serially form colonies in replating assays following transduction with AML oncogene–encoding retroviruses. Error bars represent SD of three biological replicates; *P < .05 (Mann-Whitney U test). (B) These exhibit a more differentiated morphology with increased numbers of monocytes.
Ectopic expression of c-Myb rescues the sensitivity of Booreana cells to transformation
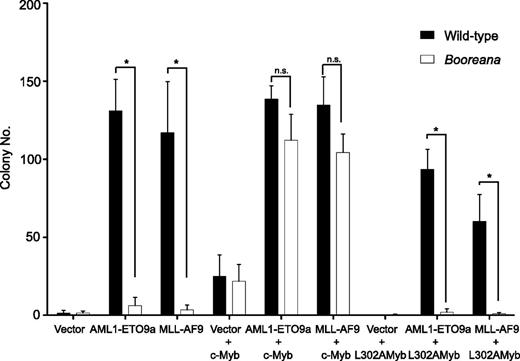
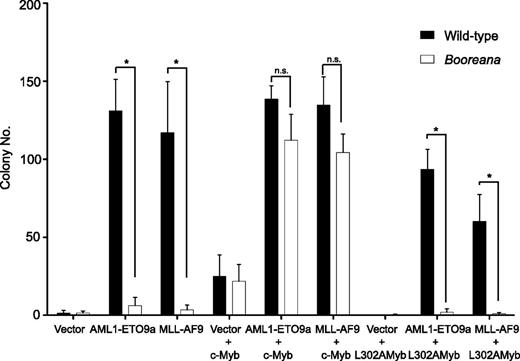
We next addressed whether ectopic expression of wild-type c-Myb would rescue the induction of serial replating capacity by the oncogenic fusion proteins AML1-ETO9a and MLL-AF9 in cells from Boo mice. To do this, we first inserted c-Myb and the transactivation domain mutant, c-Myb L302A, into the pMYs-IRES-Venus retroviral vector. The L302A mutation completely abrogates the ability of c-Myb to interact with CBP/p300.40 Hematopoietic cells were transduced with c-Myb or c-Myb L302A as well as AML1-ETO9a or MLL-AF9, and then sorted for expression of both GFP and Venus fluorescent protein. Figure 3 shows that expression of c-Myb, but not c-Myb L302A, was able to restore the serial replating ability of the AML oncogene-transduced cells. Note that c-Myb itself can induce a relatively weak differentiation block, leading to the formation of a small numbers of colonies46 (Figure 3); however, the number of colonies formed by cells expressing both GFP (AML1-ETO9a) and Venus (Myb) was at least 3-fold higher than that observed with c-Myb alone. These results reinforce the importance of the interaction of c-Myb with p300 in transformation by oncogenic fusions and exclude the possibility that the resistance of Boo cells to transformation is due to a mutation other than that in c-Myb.
Re-expression of c-Myb, but not c-Myb L302A, in cells from Booreana mice rescues their transforming ability, inducing colony formation in the presence of AML1-ETO9a and MLL-AF9 oncogenes. Data shown are for the second serial replating. Error bars represent SD of three replicates. *P < .05 (Mann-Whitney U test).
Re-expression of c-Myb, but not c-Myb L302A, in cells from Booreana mice rescues their transforming ability, inducing colony formation in the presence of AML1-ETO9a and MLL-AF9 oncogenes. Data shown are for the second serial replating. Error bars represent SD of three replicates. *P < .05 (Mann-Whitney U test).
AML1-ETO9a– and MLL-AF9–transduced Booreana cells are unable to generate leukemias
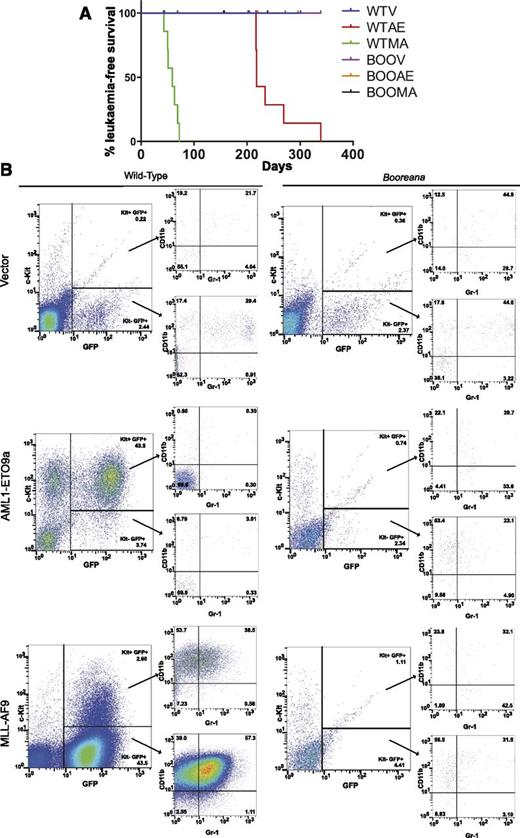
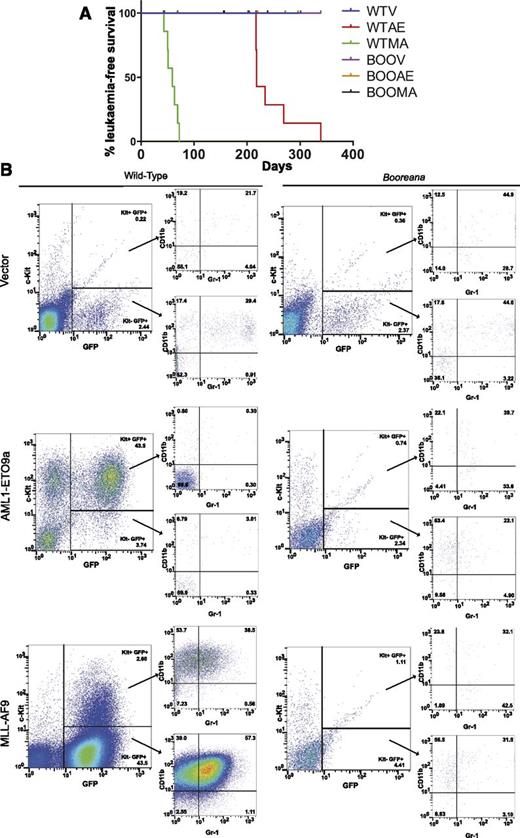
We next wanted to determine whether the effects of the Boo mutation on in vitro transformation were also reflected in vivo. Fetal liver cells (e14.5) were obtained from donor WT and Booreana mice, transduced with empty vector, AML1-ETO9a, or MLL-AF9 retroviruses, and transplanted into irradiated CD45-congenic recipients. By monitoring these mice over nearly 11 months (Figure 4A), we found that all of the assessable mice transplanted with WT oncogene-transduced cells (but none of the vector controls) developed AML. As expected,11 the latency of leukemia development was much longer for AML-ETO9a recipients than for MLL-AF9 recipients. The development of leukemia was indicated by a rapid rise in the proportion of GFP+ myeloid cells in peripheral blood compared with healthy and control mice (as illustrated in Figure 4B and supplemental Figure 3). In the case of AML-ETO9a, these were predominantly Kit+Gr-1+CD11b+, while the leukemic cells in MLL-AF9 recipients were predominantly CD11bhi with a lower proportion of Kit+ cells (Figure 4B). Analyses of bone marrow (supplemental Figure 4A), spleen (supplemental Figure 4B), and liver (data not shown) confirmed the presence of GFP+ myeloid cells in these organs also. In striking contrast, none of the assessable animals transplanted with Booreana cells developed AML, as indicated by illness or death and analysis of blood and other organs (Figure 4 and supplemental Figures 3 and 4), strongly suggesting that these cells were refractory to leukemic transformation.
Cells from Booreana mice are resistant to transformation by AML oncogenes in vivo. (A) Percentage of leukemia-free surviving irradiated recipient mice (n = 8) receiving AML1-ETO9a– and MLL-AF9–expressing WT and Booreana donor cells. Only the former succumb to a myeloid leukemia with (B) increased numbers of GFP+ cells in their peripheral blood. Cells in the peripheral blood of AML1-ETO9a–expressing WT donor mice exhibit a Kit+ phenotype, whereas those of MLL-AF9–expressing WT donor mice exhibit a Kit+CD11b+Gr-1+ phenotype, reminiscent of a monocytic leukemia.
Cells from Booreana mice are resistant to transformation by AML oncogenes in vivo. (A) Percentage of leukemia-free surviving irradiated recipient mice (n = 8) receiving AML1-ETO9a– and MLL-AF9–expressing WT and Booreana donor cells. Only the former succumb to a myeloid leukemia with (B) increased numbers of GFP+ cells in their peripheral blood. Cells in the peripheral blood of AML1-ETO9a–expressing WT donor mice exhibit a Kit+ phenotype, whereas those of MLL-AF9–expressing WT donor mice exhibit a Kit+CD11b+Gr-1+ phenotype, reminiscent of a monocytic leukemia.
However, it was important to rule out several alternative explanations for the results shown in Figure 4. First, the failure to generate leukemia was not the result of differences between WT and Booreana cells in either initial transduction frequency of or oncogene expression in donor fetal liver cells (supplemental Figure 5). Second, bleeds of most recipient mice that received transduced WT or Boo donor cells showed effective reconstitution by donor (CD45.2) cells at both early and later time points posttransplantation (supplemental Figure 6B). Third, the proportions of GFP+ (ie, transduced myeloid cells in recipient mice at these time points) were roughly similar in each case (supplemental Figure 6A). Taken together, these data show that the failure of Booreana recipients to develop AML did not reflect defects in or failure of oncogene expression, transduction, or long-term reconstitution. Furthermore, a similar result (ie, leukemia induction by WT but not Booreana cells) was obtained in an independent experiment using AML1-ETO9a–transduced bone marrow cells as donors (supplemental Figure 7). In this study, semiquantitative polymerase chain reaction analysis indicated ≥98% Boo allele contribution to the bone marrow of WT recipient mice (supplemental Table 1), reconfirming their capacity for long-term hematopoietic reconstitution.
Identification of key targets required for leukemic transformation
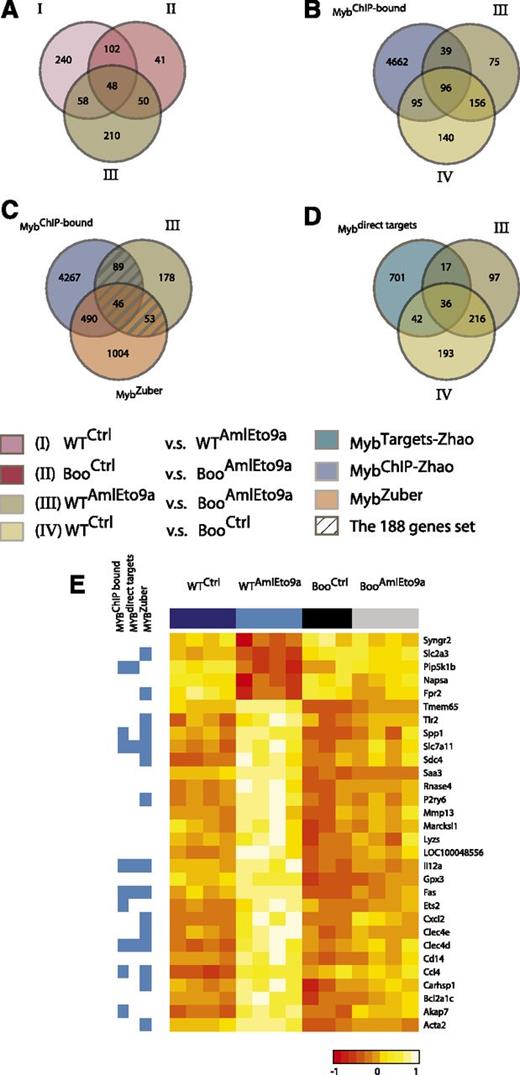
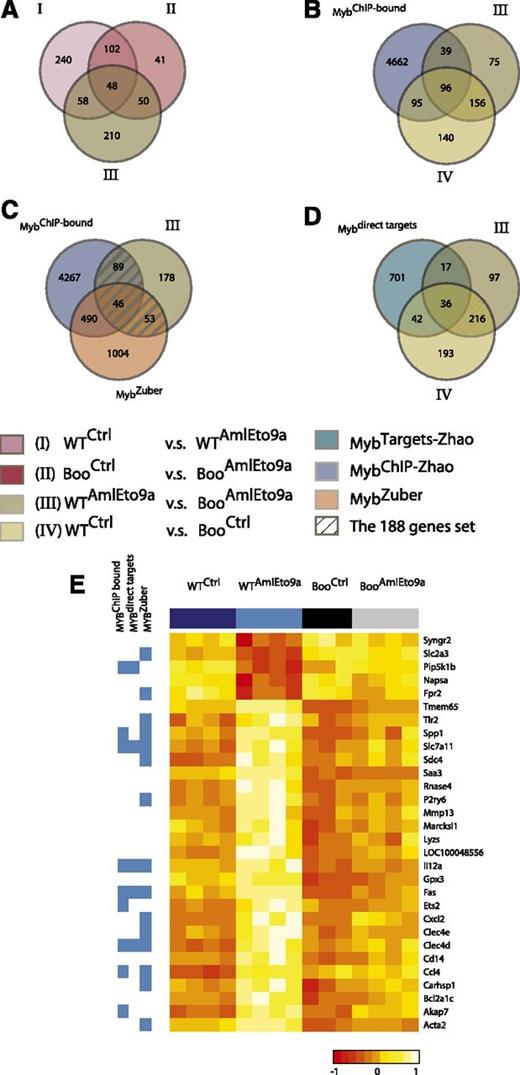
We showed above that the abrogation of c-Myb-CBP/p300 binding results in the inability of AML1-ETO and MLL fusion oncogenes to transform hematopoietic progenitors from Boo mice. This is likely to reflect the inability of mutant c-Myb to regulate target genes critical for transformation. To identify such genes, we carried out microarray expression profiling of hematopoietic progenitors from WT and Boo mice transduced with AML1-ETO9a retrovirus (Figure 5 and supplemental Table 2). These gene expression profiling data have been deposited in the GEO database under the accession number GSE34224. We did not analyze MLL fusion-transduced cells in this study because Zuber et al21 have recently reported a comprehensive study of c-Myb targets in MLL-AF9 leukemia cells, and because we found that the latter oncogene induces relatively few changes within in the short time frame used here, in agreement with previous studies.47 c-Kit–enriched progenitors were transduced with either the AML1-ETO9a retroviral vector or empty (control) vector and sorted for c-Kit+GFP+ cells after 48 hours. Expression profiles of these cells were corrected for batch effects (supplemental Figure 8), and comparisons were used to identify the following gene sets (Figure 5 and supplemental Table 2): I, an AML1-ETO9a–induced program in WT cells (WT AML1-ETO9a vs WT Control); II, an AML1-ETO9a–induced program in Boo cells (Boo AML1-ETO9a vs Boo Control); III, genes differentially expressed between WT and Boo cells transduced with AML1-ETO9a (WT AML1-ETO9a vs Boo AML1-ETO9a); and IV, genes differentially expressed between WT and Boo cells transduced with empty vector. Differentially expressed genes in each case were defined as those with fold change ≥1.5 and adjusted P value < .05.
Expression profiling reveals Myb-p300–regulated genes important for AML1-ETO–mediated transformation. (A-D). Venn diagrams and key below showing gene numbers and overlaps between gene sets I, II, III, and IV as defined in the text and supplemental Table 2, genes identified as binding c-Myb by chromatin immunoprecipitation and as Myb targets in Zhao et al,41 and genes affected by c-Myb short hairpin RNA knockdown in an MLL-AF9 leukemia model21 (supplemental Table 3). (E) Expression levels (heatmap) of a subset of the 58 genes in (A) regulated by AML1-ETO9a in WT but not in Boo cells, and their membership in c-Myb-related gene sets (blue side bar).
Expression profiling reveals Myb-p300–regulated genes important for AML1-ETO–mediated transformation. (A-D). Venn diagrams and key below showing gene numbers and overlaps between gene sets I, II, III, and IV as defined in the text and supplemental Table 2, genes identified as binding c-Myb by chromatin immunoprecipitation and as Myb targets in Zhao et al,41 and genes affected by c-Myb short hairpin RNA knockdown in an MLL-AF9 leukemia model21 (supplemental Table 3). (E) Expression levels (heatmap) of a subset of the 58 genes in (A) regulated by AML1-ETO9a in WT but not in Boo cells, and their membership in c-Myb-related gene sets (blue side bar).
Gene set enrichment analysis of set I showed, as expected, enrichment in AML1-ETO–related gene sets from other studies (supplemental Figure 9A). Indeed much, but not all, of this AML1-ETO–induced program was also represented in AML1-ETO9a–transduced Boo cells (set II above) as shown in Figure 5A and by rank-rank hypergeometric overlap analysis (supplemental Figure 9B and supplemental Methods). We also note that 191 genes (39% of set IV) differentially expressed between control WT and Boo cells (set IV) have c-Myb binding sites that we identified by ChIP-Seq in the ERMYB cell line41 (Figure 5B), and of these, 78 genes (16% of set IV) were also regulated by c-Myb in those cells (Figure 5D). This shows that, as expected, many of the c-Myb-p300–dependent genes are indeed direct c-Myb targets. Conversely, while differences in cell type and indirect regulation undoubtedly contribute to the lack of complete correspondence between set IV and direct c-Myb targets, such discordance also supports the notion that many other c-Myb targets may not require p300 for regulation.
Comparison of sets I, II, and III identified 58 genes that are potentially coregulated by AML1-ETO9a and c-Myb (genes belonging to sets I and III but not II; Figure 5A and supplemental Figure 10). About half these genes, as shown in Figure 5E, were regulated by AML1-ETO9a in WT cells but showed similar expression levels in control WT, control Boo, and AML1-ETO9a–transduced Boo cells. Moreover, 188 genes (hatched in Figure 5C) were identified by comparison of the potentially c-Myb-dependent genes that are differentially expressed between AML-ETO9a–transduced WT and Boo cells (set III) with genes previously identified as c-Myb targets by ChIP-Seq41 or short hairpin RNA knockdown in MLL-AF9–transformed cells.21 These genes, listed in supplemental Table 3, are likely to represent p300-dependent (and mostly) direct c-Myb target genes that are required for the process of AML1-ETO9a–induced transformation.
Discussion
That c-Myb expression is required for the maintenance of leukemic cell proliferation has been known for two decades28,29 ; however, the mechanisms underlying this requirement are yet to be fully elucidated. Studies with MLL fusion proteins have previously uncovered the requirement for c-Myb expression for the transforming ability of these oncogenes20,45 ; more recently, this has been attributed to a self-renewal program controlled by c-Myb in MLL-AF9 leukemias.21 We previously reported comprehensive analyses of the transcriptional program controlled by c-Myb, uncovering novel target genes that could play a role in maintaining a block of differentiation and conferring self-renewal properties in hematopoietic cells.41 From this and other work,40 we established p300 to be an essential coactivator and context-dependent corepressor in the intrinsic transforming ability of c-Myb. In this study, we show that the interaction between c-Myb and p300 is essential for the transforming and leukemogenic capacities of AML1-ETO and MLL fusion oncoproteins, which are products of two of the most frequently occurring chromosomal translocations in human AML.
Our results indicate that the Myb-p300 interaction is essential for the ability of AML1-ETO and MLL fusions to confer self-renewal properties on myeloid progenitor cells, which subsequently progress in vivo to induce AML. In the absence of this interaction, the fusion oncogenes are unable to impose a block of differentiation, thus leading instead to terminal differentiation. To ensure that our results are not the consequence of disruption of other interacting partners of c-Myb, we show that myeloid progenitors from Plt6 mice, which have a mutation in p300, are also refractory to transformation by the two oncogenic fusions. Importantly, re-expression of WT c-Myb is able to restore the transformation of cells from Boo mice by AML1-ETO9a and MLL-AF9 oncoproteins. Taken together, these results imply that the specific interaction between c-Myb and p300 is required to control a transcriptional program, which is essential for the acquisition of self-renewal and possibly other leukemogenic properties upon expression of fusion oncoproteins.
The major effects of the Boo mutation on transformation in vitro appear to be suppression of AML1-ETO– and MLL fusion–induced self-renewal and differentiation blockage. Such effects are likely to be associated with dysregulation of hematopoietic transcription factors, which is consistent with enrichment of processes such as immune and/or hematopoietic system development and proliferation in gene set III (supplemental Figure 11). Inspection of genes differentially expressed between AML-ETO9a–transduced WT and Boo cells (ie, set III) that have associated c-Myb binding sites (Figure 5C) revealed downregulation of Gata2 and Gfi1b along with upregulation of Gfi1 and Ets2 (supplemental Table 3 and supplemental Figure 10). Interestingly, this same pattern was seen in two other mouse models of AML driven by MLL-ENL and MOZ-TIF2, respectively.48 Importantly, that report also showed that ectopic expression of Gata2 suppressed proliferation, cell cycle progression, and colony formation by MLL-ENL–transformed cells. Moreover, apparent loss-of-function mutations in GATA2 have recently been reported in familial myelodysplastic syndrome and AML,49,50 and GATA2 expression was reported to be lower in poor-prognosis NPM1-WT human AMLs compared with NPM1-mutant AMLs.51 Furthermore, we have recently shown that GFI1 is necessary and sufficient for MYB’s ability to block monocytic differentiation of a human AML cell line.52 We therefore suggest that c-Myb-p300–dependent regulation of some or all these genes is important for myeloid transformation by AML1-ETO and possibly other AML oncogenes.
A further implication of the requirement for c-Myb-p300 interaction in leukemic transformation by AML1-ETO and MLL fusion oncogenes is that inhibition of this interaction may represent an avenue for targeted therapeutic intervention. This assumes that the c-Myb-p300 interaction is required for leukemia maintenance as well as initiation, an issue not directly addressed by this study. Testing this assumption will require development of an inhibitor of the interaction that can be applied to established leukemias. However, knockdown studies have repeatedly shown the essential role of MYB in the proliferation of leukemic cells, suggesting that the c-Myb-p300 interaction also may well be essential for maintenance.
Because of the essential role of c-Myb in establishing and maintaining normal hematopoiesis,22,53 complete or near-complete inhibition of c-Myb activity may not be a desirable therapeutic option. However, targeting the c-Myb-p300 interaction could provide a more specific approach, given that Booreana homozygous mice are viable, fertile, and have a near-normal lifespan in contrast with the embryonic lethality of c-Myb null mice; furthermore, we show that cells from Booreana mice are capable of long-term hematopoietic reconstitution. Moreover, given the widespread requirement for MYB in AML, therapeutics that target the c-Myb-p300 interaction may be applicable in many other subtypes of AML.
Our finding that the c-Myb-p300 interaction is required for the transforming ability of MLL-ENL and MLL-AF9 is consistent with a particularly stringent requirement for c-Myb expression in leukemias harboring these two fusion proteins, as previously reported.20,21,45 In fact, Booreana heterozygous mice are phenotypically indistinguishable from WT mice, but hematopoietic cells from the former are refractory to transformation by MLL fusions. This suggests that there could be an even greater therapeutic window in the treatment of this subtype of AML for agents developed to disrupt the c-Myb-p300 interaction.
Finally we note that inhibition of interactions between oncogenic transcription factors (Notch, Bcl6) and their respective coregulators has been achieved by using cell-permeable peptides and/or small molecules.54-56 Such agents were able to suppress leukemia/lymphoma cell proliferation in vitro and in vivo, suggesting that similar approaches could be used to inhibit interaction between c-Myb and p300 in the treatment of leukemias induced by AML1-ETO, MLL fusions, and probably other AML oncogenes.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Liang Zhao and members of the Gonda Laboratory for helpful discussions, Dr Michael Rist and Dr Robert Wadley for valuable assistance with cell sorting, and Prat Thiru of the Whitehead Institute Bioinformatics and Research Computing facility for microarray analysis.
This work was supported by a Leukemia Foundation (Australia) Scholarship (D.R.P.), a Fellowship under Program 1016647 (W.S.A.), Infrastructure Support Grants (Independent Research Institutes Infrastructure Support Scheme 361646) from the Australian National Health and Medical Research Council (NHMRC), the Australian Cancer Research Fund, a Victorian Government Operational Infrastructure Support grant, Career Development Fellowship (1033736) (I.G.W. and J.P.L.), a Research Fellowship (1044091) from the NHMRC, and by grants 1023175 and 1023179 from the NHMRC (T.J.G.).
Authorship
Contribution: D.R.P. designed and performed research, collected, analyzed, and interpreted data, and wrote the manuscript; K.S. performed research, contributed analytical tools, analyzed and interpreted data, and wrote the manuscript; C.M., V.B., K.K., P.H., and A.T. performed research and analyzed data; J.D. performed research; P.M. analyzed data; S.M.G., P.P., and W.S.A. contributed vital reagents and analytical tools; A.C.P., J.-P.L., and I.G.W. designed research, contributed new reagents, and analyzed and interpreted data; and T.J.G. designed research, analyzed and interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for D.R.P. is Whitehead Institute for Biomedical Research, Cambridge, MA.
The current affiliation for P.M. is QIMR Berghofer Medical Research Institute, Brisbane, Australia.
Correspondence: Thomas J. Gonda, School of Pharmacy, University of Queensland, Pharmacy Australia Centre of Excellence, 20 Cornwall St, Woolloongabba, QLD 4102, Australia; e-mail: t.gonda@uq.edu.au.
References
Author notes
C.M. and K.S. contributed equally to this study.
The online version of this article contains a data supplement.