Key Points
3D bone marrow niche model recapitulates in vivo interactions of tumor and bone cells in a more biologically relevant system than in 2D.
Differential expression levels of miRs in MSCs provide novel insights into mechanisms of regulation of osteoblasts in multiple myeloma.
Abstract
Clonal proliferation of plasma cells within the bone marrow (BM) affects local cells, such as mesenchymal stromal cells (MSCs), leading to osteolysis and fatality in multiple myeloma (MM). Consequently, there is an urgent need to find better mechanisms of inhibiting myeloma growth and osteolytic lesion development. To meet this need and accelerate clinical translation, better models of myeloma within the BM are required. Herein we have developed a clinically relevant, three-dimensional (3D) myeloma BM coculture model that mimics bone cell/cancer cell interactions within the bone microenvironment. The coculture model and clinical samples were used to investigate myeloma growth, osteogenesis inhibition, and myeloma-induced abnormalities in MM-MSCs. This platform demonstrated myeloma support of capillarylike assembly of endothelial cells and cell adhesion–mediated drug resistance (CAM-DR). Also, distinct normal donor (ND)- and MM-MSC miRNA (miR) signatures were identified and used to uncover osteogenic miRs of interest for osteoblast differentiation. More broadly, our 3D platform provides a simple, clinically relevant tool to model cancer growth within the bone—useful for investigating skeletal cancer biology, screening compounds, and exploring osteogenesis. Our identification and efficacy validation of novel bone anabolic miRs in MM opens more opportunities for novel approaches to cancer therapy via stromal miR modulation.
Introduction
Increasing evidence demonstrates that matrix stiffness, geometry, chemistry, and spatial dimensionality, along with neighboring cells and soluble factors, regulate cellular behavior and tissue formation.1 However, current in vitro multiple myeloma (MM) research is conducted on 2D in vitro culture plates, highlighting the need for more realistic 3D in vitro models of myeloma growth.2 Many 3-dimensional (3D) culture and coculture systems have been described for MM and have validated the importance and relevancy of using 3D rather than 2D culture systems to more accurately model myeloma growth. Some of these models have used hydrogels (made from permutations of collagen, fibronectin, and Matrigel3,4 ), which are, as with our model, advantageous as simple, controllable, and reproducible 3D culture microenvironments useful for studying pharmaceuticals or biological pathways. However, our system transcends these properties to comprise a model representative of a mineralized bone microenvironment using bone marrow (BM)-derived mesenchymal stromal cells (MSCs) that are stimulated to undergo osteogenic differentiation on the strong, porous silk scaffolds, which does not occur on softer substrates. This is a critical component to a 3D model of myeloma and bone, because myeloma cells respond differently to undifferentiated MSCs compared with MSCs differentiated into osteoblasts and osteocytes.5 On the other end of the spectrum are the models that use 100% biologically relevant patient-derived, whole-bone cores,6 taken directly from patients, which have the advantage of providing a hard, mineralized, bony matrix but that lack the reproducibility, adaptability, scalability, controllability, and simplicity that characterize our tissue-engineered bone (TE-bone) model. Although this is beneficial for small-scale, individualized patient analyses, patient samples vary widely in results and responses in terms of myeloma growth and drug response, making large drug screens or biological pathway analyses impossible. Moreover, the 3D bioreactor system necessary for patient-bone core culture makes the system much more time- and cost-consuming than 3D TE-bone, which can be completely user-defined in terms of size, shape, porosity, and other parameters, and can be produced as hundreds of identical samples. Silk scaffolds, the platform of our TE-bone, can also be modified in terms of pore size, dimensions, Young's modulus, degradation speed, and seeded cellular components. Finally, our TE-bone can be used in vitro or in vivo, monitored using live, nondestructive optical imaging, and processed using flow cytometric techniques for analysis of cellular populations. Herein we use this novel disease model to demonstrate real-time inhibition of osteogenic differentiation in response to myeloma cells.
Osteolytic cancers such as MM develop via forward-feedback mechanisms with local MSCs in the BM, leading to devastating skeletal consequences (ie, pain, hypercalcemia, osteolysis, and fracture) and accelerated tumor growth.7 MM cells insidiously overtake normal bone homeostasis to decrease osteoblastic activity and increase osteoclastic activity by altering local microenvironment cells.8 MM patient–derived MSCs (MM-MSCs) exhibit decreased proliferation and osteogenesis and an inability to repair osteolytic damage, and they display great patient-to-patient heterogeneity in their ability to undergo differentiation and induce changes in MM cells.8-10 The tumor BM microenvironment also supports tumor growth,11 induces chemoresistance, and selects for tumor-initiating clones.12 Therefore, a realistic model of the abnormal BM seen in MM patients would greatly benefit translational research scientists.
In myeloma patients, bone lesions with concomitant bone fractures and osteoporosis often persist despite bisphosphonate or bortezomib administration, tumor cell ablation, or disease remission.13,14 This is partially explained by functional and gene expression differences between MM-MSCs and normal donor (ND)-MSCs.8,15-18 However, mechanisms governing ineffectual MM-MSC osteogenesis remain unclear, and the roles of microRNAs (miRs) in this process are unknown. This highlights our need for stroma-specific targets and therapies, which can be identified only with more realistic 3D bone cancer models.
Our 3D in vitro BM model recapitulates interactions among tumor cells, stroma cells (MSCs), and endothelial cells, and the osteogenic process in normal and myeloma conditions. Our purpose was to examine dynamic cell-to-cell interactions between tumor cells and supportive cells, to determine the inhibitory effects of MM cells on osteogenesis and to develop a robust preclinical model to accelerate the rate of discovery and development of efficacious cancer treatments.
Methods
Study approval
Approval for these studies was obtained from the Dana-Farber Cancer Institute or Brigham and Women's Institutional Review Boards. Informed consent was obtained from all patients and healthy volunteers in accordance with the Declaration of Helsinki.
TE-bone
Porous, aqueous 8% (wt/wt) silk fibroin scaffolds were made following the silk processing steps previously described19 but were specifically designed with pores of 500 to 600 μm and cut into cylinders (5-mm × 3-mm height). Scaffolds were autoclaved for sterilization and soaked in media containing 10% fetal bovine serum 1 day before seeding. 1 × 106 MSCs were seeded onto scaffolds in regular MSC culture media and grown for 1 day, and then changed to osteogenic media. Osteogenic media consisted of α modified Eagle medium (αMEM) supplemented with 10% FBS, 100 U/mL penicillin, 10 μg/mL streptomycin (Invitrogen), 2 mmol/L l-glutamine (Invitrogen), 0.05 mM ascorbic acid, 100 nM dexamethasone, and 10 mM β-glycerophosphate. When cultured with MM1S (or without but used as controls for coculture studies), dexamethasone was excluded from the media.
Cell culture
The human multiple myeloma cell line MM1S was purchased from ATCC (American Type Culture Collection), engineered to express green fluorescent protein (GFP) and firefly luciferase (Luc+/GFP+ MM1S cells) as previously described,20 and was cultured in 500 µg/mL geneticin (Invitrogen) for selection. OPM2 MM cells were labeled with red fluorescent protein (RFP) and firefly luciferase and provided by Dr Andrew Kung, Columbia University. Primary human BM–derived MSCs obtained from normal healthy subjects (ND-MSCs) or MM patients (MM-MSCs) were isolated and cultured as previously described21 in expansion media of Dulbecco’s modified Eagle medium (DMEM)+20% FBS and used at passages 2 to 4. Clinical samples were collected from patients or healthy donors from the Dana-Farber Cancer Institute and Brigham and Women’s Hospital. Primary patient samples were isolated from MM patient BM aspirates using MACS technology (Miltenyi Biotec) with beads for CD138 as recommended by the manufacturer, and the negative fraction was seeded to flasks and grown as previously described to isolate BM-MSCs.21 For imaging assays, MSCs were labeled with either the Celltracker dye DiD (Invitrogen) or calcein for live-cell imaging (Invitrogen), or they were stably transfected with the TurboRFP gene (Thermo Scientific) subcloned into pCW307 lentivirus vector (Addgene). Primary patient myeloma cells and MSCs were cocultured in “50-50” medium: a base of 50-50 F12-DMEM (Invitrogen) supplemented with 10% FBS, 100 U/mL penicillin, 10 μg/mL streptomycin (Invitrogen), and 2 mmol/L l-glutamine (Invitrogen). RFP-labeled HUVECS (RFP-HUVECS) were purchased from Angioproteomie and expanded in Endothelial Medium (EGM-2 BulletKit media; Lonza). All experiments were performed at 37°C, 5% CO2 in normoxia.
3D scaffold coculture
Fluorescent (TurboRFP or DiD+) MSCs were seeded onto scaffolds as described.19 1 × 106 MSCs were seeded in MSC growth media 1 day before seeding with 1 × 106 GFP+MM1S cells. GFP+MM1S cells alone, MSCs alone, or cocultures of MSCs+ GFP+MM1S cells were cultured on 3D silk scaffolds in dexamethasone-free osteogenic media for the duration of the coculture experiments. Cells were monitored using confocal microscopy and isolated using fluorescent-activated cell sorting (FACS) or were fixed for histology. Primary patient MM cells were labeled with the lipophilic CellTracker dye (DiI, Invitrogen) and seeded onto scaffolds (0.5 × 106/scaffold) that had been preseeded (1 day prior) with MSCs (1 × 106) labeled with a different cell-tracker dye (DiD; Invitrogen), and cultured at 37°C, 5% CO2. For endothelial cocultures, RFP-HUVECs were cultured with or without GFP+MM1S cells in HUVEC media and imaged using confocal microscopy over 1 month.
Drug resistance
For 3D assays, scaffolds with MSCs, GFP+MM1S cells, or cocultures were cultured on scaffolds in 50-50 media with or without 5 nM bortezomib (Selleck). Bortezomib was diluted in dimethylsulfoxide and stored at −20°C until use, and was then diluted in culture medium immediately before use. Scaffolds were seeded with 0.5 × 106 MSCs per scaffold and 0.5 × 106 GFP+Luc+MM1S cells per scaffold the following day, and 50-50 media with or without bortezomib was added immediately before use and changed twice per week. For 2D assays, 0.5 × 104 GFP+Luc+ MM1S cells were seeded into 96-well plates with or without a confluent layer of ND-MSCs and cultured with or without media containing 5 nM bortezomib. Cells were quantified using bioluminescence imaging (BLI) and imaged with confocal microscopy.
Fluorescent microscopy
For imaging of MSCs on scaffolds, cells were labeled for live-dead staining with calcein or the LIVE/DEAD Fixable Red Dead Cell Stain Kit (Invitrogen) following the manufacturer’s instructions. For cocultures of GFP+MM1S and MSCs, scaffolds were nondestructively imaged weekly, using 24-well glass-bottomed dishes (1.5 mm; MatTek) with a Leica SP5X Laser Scanning Confocal Microscope using Leica LAS acquisition software. Scaffolds were imaged with 10× dry, 20× water immersion, or 63X Plan Apo objectives using 488 nm Argon, 405 nm UV diode, or white light lasers (470-670 nm). Photomultiplier tubes collected fluorescence signal from autofluorescent scaffolds (405 nm/420-440 nm), GFP+MM1S (488 nm/500-520 nm), calcein (493/509-525), DiI (552/563-573 nm), DiD (647/660-685nm), DiR (750/775-825), TurboRFP-MSCs (553/564-616 nm), and RFP-HUVECs (555/576-619 nm), which were given pseudocolors as described. Z-stack images were acquired and processed using LeicaLite or LeicaLAS software to create single maximum projection 3D-like images or videos. Nonconfocal fluorescent microscopy was performed using an Olympus CKX41 microscope with appropriate filter cubes and an Olympus DP72 Camera and dry ×10 or ×20 objectives.
Bioluminescence imaging quantification
GFP+Luc+MM1S cells or scaffolds seeded with GFP+Luc+MM1S cells were measured for bioluminescent signal after placement into opaque, white 96-well plates with 100 µL of media and 5 µL sterile firefly d-luciferin (7.5 mg/mL) (Caliper). After incubation for 5 minutes at 37°C, the signal from MM1S cells was measured on a FLUOstar Optima plate reader.
Histology, immunohistochemistry, and alizarin red staining
Scaffolds were fixed in 4% paraformaldehyde (PFA) overnight at 4°C, paraffin-embedded, sectioned onto glass slides, and stained with alizarin red, hematoxylin and eosin (H&E), or anti-human CD138 immunohistochemistry (primary antibody,#M7228; Dako) by the Dana-Farber Cancer Institute Specialized Histopathology Core. Immunohistochemistry was run at a 1:50 dilution and stained on Leica’s Bond-III autostainer using a Leica Bond Polymer Refine Detection kit. Slides were antigen-retrieved using Epitope Retrieval I (Leica) for 30 minutes. For alizarin red staining, MSCs differentiated in osteogenic media (or osteogenic dexamethasone-free media in studies with GFP+MM1S) were fixed for 15 minutes in 1% formaldehyde, rinsed with water, stained with Alizarin Red Solution (Sigma-Aldrich) (2% wt/vol, 4.2 pH) for 10 minutes, rinsed 3 times with water, and then imaged. Staining was quantified by dissolving Alizarin Red Solution stain from wells in 6-well plates in 1 mL of decalcification solution (Cal-EX Decalcifier; Fisher Scientific) and reading absorbance of the solution at 405 nm (200 µL per well, 96-well plates in a FLUOstar Optima plate reader).
Scanning electron microscopy and micro–computed tomography
Scanning electron microscopy images of scaffolds were taken on a Nikon Eclipse 80i microscope with a DSFi1 Nikon Color Camera with NIS Elements AR Software. Scanning electron microscopy was done using a JEOL scanning electron microscope with gold sputter coating on scaffolds after fixation in 4% PFA. Microcomputed tomography (μCT) imaging was performed on scaffolds fixed overnight in 4% PFA and transferred to 70% ethanol in 1.5 mL Eppendorf tubes on a Siemens Inveon multimodality machine (positron emission tomography–single-photon emission tomography–μCT) at the Dana-Farber Cancer Institute Lurie Imaging Facility Core.
Cell counting
MM-MSCs (n = 4 donors) and ND-MSCs (n = 4 donors) were seeded to 12-well plates (5000 stromal cells/cm2) with or without GFP+MM1S (1250 MM1S cells/cm2) and cultured for 9 days in 50-50 culture media. Cells were then fixed and stained with a 10% neutral-buffered formalin, 1 µg/mL Hoechst (Invitrogen) solution for 10 minutes, and photographed (at least 3 representative fields of view/well) using brightfield and fluorescent (UV filter) microscopy with a Nikon Eclipse 80i microscope (20×), a DSFi1 Nikon Color Camera, and an NIS Elements AR Software, and then counted by a blinded investigator using the ImageJ Cell Counter plugin (v1.47). The mean number of cells/cm2 ± standard error of the mean (SEM) was calculated and graphed.
Matrigel and fibrin-hydrogel culture
Fibrinogen (Sigma-Aldrich) and thrombin (human BioUltra recombinant, Sigma-Aldrich) were mixed to create 4-mg/mL fibrin hydrogels. Matrigel (BD Biosciences) was diluted 1:3 in phosphate-buffered saline. Both hydrogels were immediately mixed with cells (cell tracker dye DiR+MSCs, RFP+HUVECs, GFP+MM1S cells, or a combination) before seeding into 96-well plates (20 000 cells/well in 100 µL) and were cultured and imaged with fluorescent confocal microscopy over 12 days.
mRNA and miR isolation and qRT-PCR
miRNAs and mRNAs were isolated from cells using the miRNeasy isolation mini-kit (Qiagen), quantified, and tested for quality and contamination using a Nanodrop machine (ThermoScientific),21 and then subjected to quality control minimum standards of 260/230 > 2 and 260/280 > 1.8 before further use for quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR) or nanoString analysis. For qRT-PCR, sample mRNA was reverse-transcribed into cDNA for either miR using the High Capacity cDNA Reverse Transcription Kit (Invitrogen) or for mRNA using SuperScript III First-String SuperMix (Invitrogen), according to the manufacturer’s instructions. qRT-PCR was performed using SYBR Master Mix (SA Bioscience). Analysis was done using the 2–ΔΔCt method, normalized to RNU6B (miR) or glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (mRNA). Primers were designed using the method at http://primerdepot.nci.nih.gov (2013), shown in supplemental Table 1 available on the Blood Web site. miR stem-loop sequences were defined using miRBase Sequence Database Release 20 (http://www.mirbase.org). Experiments were performed in a StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA) and consisted of an initial denaturation step of 10 minutes at 95°C, followed by 50 cycles of 95°C for 15 seconds and 60°C for 60 seconds. Data were analyzed with StepOne Software v2.1 (Applied Biosystems), which provides a threshold cycle value that was considered as the cycle in which the fluorescence begins to be distinguished from the background. All PCR reactions were run on an Applied Biosystems AB7500 Real Time PCR system using technical triplicates and were plotted as means of at least 3 different donors ± SEM.
mRNA and miR nanoString profiling
For miR and mRNA profiling, MSC mRNA (from normal or myeloma donors) or miRNA (from normal or myeloma donors or from MSCs after 2 weeks of culture in the 3D model alone or with GFP+MM1S) was analyzed using the nanoString platform. Expression levels from the nanoString cancer gene reference code set (nCounter GX Human Cancer Reference Kit) containing 230 cancer-associated mRNAs were compared between ND-MSCs and MM-MSCs (5 donors each) normalized to the average of 6 housekeeping genes (PGK1, TUBB, CLTC, GAPDH, GUSB, and HPRT1), graphed as heat maps, and analyzed using dChip software (DNA-Chip Analyzer; Cheng Li and Wing Wong Labs, http://www.hsph.harvard.edu/cli/complab/dchip/ [2013]; >1.5-fold change (fc) and P < .05 required for significance). For miRNA expression analysis of the 3D model system, 5 normal stroma cultured alone or in coculture with MM1S myeloma cells, isolated by FACS after 2 weeks of coculture, were analyzed and compared with clinical samples, (3 ND-MSC and 7 MM-MSC samples analyzed at passage 2). Primary samples (normal donor or myeloma patient) and 3D model samples (coculture vs alone) were both analyzed for stromal cell expression of 800 miRNAs using the nanoString miR analysis platform (nCounter human miRNA Expression Assay) following the manufacturer’s protocol. Briefly, default settings for quality control on miR samples were used to assure high-quality miR and accuracy of the experimental process for 4 parameters (imaging, binding density, positive control linearity, and control limit of detection). A total of 100 ng of mRNA and miR was used as input into the sample preparation reaction for the nanoString nCounter assay. The miRNAs were all then normalized to the top 100 miRs per sample (by averaging the expression of the top 100 miR per sample, and then dividing all miR by this number), filtered for miRs with an average of >25 counts (cutoff in nanoString units of expression to establish real expression) and considered significant for P < .05 using dChip software analysis, following the manufacturer’s instructions and previously reported literature.22 An abundance of specific target molecules was quantified robotically on the nCounter Digital Analyzer by counting the individual fluorescent barcodes and assessing the target molecules on the sample cartridge with a charge-coupled device camera as reported previously.22 For each assay, a high-density scan setting encompassing 600 fields of view was used. miRs reaching a minimum threshold of 25 counts, fc of >1.5, and significance of P < .05 were identified as significant and were further investigated in miR mimic assays.
miR transfection
MM-MSCs were transfected with miR-199a-3p and miR-199a-5p miR mirVana mimics (Ambion) and a negative control (mirVana negative control mimic #1) or miRCURY (Exiqon) inhibitors for miR-181a-5p, miR181c-5p, miR-222-3p, miR-601, miR-146a-5p, and miRCURY negative control, following the manufacturer’s instructions. MSCs were cultured until they were 80% confluent and were then transfected with a final concentration of 30 nM of each miR mimic or 50 nM of each miRCURY Inhibitor for 24 hours using Lipofectamine 2000 (Invitrogen). Alizarin Red Solution staining and qRT-PCR were performed on samples after 10 days of culture in osteogenic (dexamethasone-free) media. Efficiency of transfection was validated by qRT-PCR for detection of miR levels at 24 hours and 10 days as previously described.21
Pathway enrichment analysis
The targets of miR-199a-5p were predicted by TargetScan23 and retrieved from online (http://www.targetscan.org/). We used pathways derived from 3 databases—BioCarta, KEGG, and Reactome—which were downloaded from MSigDB.24 Hypergeometric testing was used to assess the enrichment of pathways. The enrichment P values were adjusted to account for multiple testing, resulting in a false discovery rate for each pathway,25 and pathways were identified using a false discovery rate cutoff of 10%.
Statistics
Statistical analysis was performed using GraphPad/Prism Version 6.02 or Microsoft Excel. P values are based on Student's t tests (2-tailed) for 2-way comparisons, or analysis of variance (ANOVA) for multiple hypothesis testing using post hoc Dunnett (2-way ANOVA) or Fisher least significant difference (LSD) (1-way ANOVA) multiple comparison testing. Sample variance was determined using an F-Test, and normality was determined using a Normal Quantile Plot to test for non-normality (Q–Q probability plot). P < .05 was considered significant and P values are provided in the figures or their captions. Statistics for heat maps were done using dChip software using P < .05 as significant.
Results
We first developed our TE-bone model and characterized its unique ability to represent a mineralized 3D bone matrix, not afforded by any previously described model. Silk scaffolds, which recapitulate the high-compressive strength and porous nature of the BM trabecula,26 were seeded with ND-MSCs and differentiated into TE-bone using osteogenic media for 50 days. The TE-bone samples formed dense, calcified tissue, as demonstrated by μCT imaging (supplemental Figure 1A), scanning electron microscopy (supplemental Figure 1B), and nondestructive fluorescence confocal microscopy (Figure 1A). This 3D mineralized model served as a basis to begin studying osteogenesis in the context of myeloma.
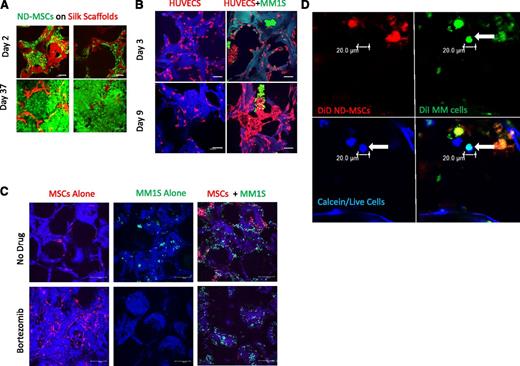
Development of an in vitro 3D BM niche model. (A) Confocal images of calcein-labeled ND-MSCs, passage 2, (calcein/live cells, green; silk scaffold, red) at 2 and 37 days of culture in osteogenic media. The scale bar represents 100 μm. (B) Confocal images of RFP+HUVECs (red) ± GFP+MM1S cells (green) on scaffolds (blue) (days 3 and 9; scale bar = 100 μm). Representative image of 3 experiments is shown here, cultured in endothelial growth media. (C) Confocal images at day 30 of culture of GFP+MM1S alone (left, green), DiD-labeled MSCs alone (middle, red), and cocultures (right) in 50-50 medium with bortezomib (top, 5 nM) or without bortezomib (bottom) on autofluorescent scaffolds (blue). (Scale bar = 100 μm.) (D) Confocal images of primary patient CD138+ MM cells (green with DiI) at day 7 seeded onto ND-MSCs (red with DiD). Channels show the myeloma cell (arrow) as alive (calcein+, blue), DiI+ (green), and DiD– (red). Overlay of green and blue appears cyan and demonstrates colocalization of calcein and DiI staining. Samples cultured in 50-50 media (n = 3); the scale bar represents 20 μm.
Development of an in vitro 3D BM niche model. (A) Confocal images of calcein-labeled ND-MSCs, passage 2, (calcein/live cells, green; silk scaffold, red) at 2 and 37 days of culture in osteogenic media. The scale bar represents 100 μm. (B) Confocal images of RFP+HUVECs (red) ± GFP+MM1S cells (green) on scaffolds (blue) (days 3 and 9; scale bar = 100 μm). Representative image of 3 experiments is shown here, cultured in endothelial growth media. (C) Confocal images at day 30 of culture of GFP+MM1S alone (left, green), DiD-labeled MSCs alone (middle, red), and cocultures (right) in 50-50 medium with bortezomib (top, 5 nM) or without bortezomib (bottom) on autofluorescent scaffolds (blue). (Scale bar = 100 μm.) (D) Confocal images of primary patient CD138+ MM cells (green with DiI) at day 7 seeded onto ND-MSCs (red with DiD). Channels show the myeloma cell (arrow) as alive (calcein+, blue), DiI+ (green), and DiD– (red). Overlay of green and blue appears cyan and demonstrates colocalization of calcein and DiI staining. Samples cultured in 50-50 media (n = 3); the scale bar represents 20 μm.
Next we tested the ability of myeloma cells to grow in osteogenic media, which would be required in coculture. However, we found dexamethasone to be toxic to both MM1S and OPM2 cells (data not shown) and hence adapted the osteogenic media for our purposes by removing dexamethasone, an alteration that has been previously described.27 We next tested the ability for OPM2 and MM1S cell lines to grow on the silk scaffolds and found that, although both cell types were able to adhere to ND-MSCs seeded on scaffolds, only MM1S could adhere to the silk scaffolds alone, as observed in confocal imaging (data not shown). Hence for subsequent studies we chose to use MM1S as the main myeloma cell line for this model.
We hypothesized that silk scaffolds would provide a more realistic platform to investigate endothelial cell–myeloma cell interactions in the bone marrow, so we compared coculture responses of these cells in 3D silk scaffolds and hydrogels. We cocultured fluorescent endothelial cells (RFP+HUVECs) with GFP+MM1S cells and observed cell-to-cell contact and interaction, as well as interesting assembly patterns unique to silk scaffold culture (Figure 1B, supplemental Figure 2A, and supplemental Video 1). Samples were imaged with confocal microscopy over 18 days and demonstrated GFP+MM1S cell adherence to RFP+HUVECs and incorporation into the capillarylike HUVEC structures. Myeloma cells also appeared to support the branching, tube-shaped formations of HUVECs (with observable lumens), which were not observed in HUVEC monocultures. GFP+MM1S cells clumped and colocalized at endothelial protrusions, perhaps mimicking some of the signaling and evolution of angiogenesis within bone tumors. Interestingly, GFP+MM1S cell association with endothelial cells in a tubelike formation may model the early stages of myeloma cell intravasation and extravasation, as well as contributions toward angiogenesis. None of these phenomena were observed in 3D fibrin hydrogel or Matrigel cultures (supplemental Figure 2B), supporting validation that the stiffer scaffold substrate more accurately recapitulates the in vivo conditions than do softer substrates.
The 3D silk scaffold model also recapitulated the ability for MSCs to protect GFP+MM1S cells from therapeutic agents such as bortezomib over a 30-day treatment period, as quantified with bioluminescent imaging (supplemental Figure 3A) and imaged with confocal microscopy (Figure 1C). This was not achieved in 2D culture (supplemental Figure 3B), defining the 3D system as a unique environment suitable for long-term drug studies. Similarly, in vitro growth of primary MM tumor cells in 2D lacks the realistic complexity of a 3D milieu, explaining why primary patient MM cell growth was observed on MSC-seeded scaffolds over 11 days but was not possible under the same conditions in 2D culture.28 Primary MM cells were labeled with cell-tracker dyes and imaged with confocal microscopy using calcein to assess cell viability (Figure 1D and supplemental Figures 4 and 5). They were further identified with H&E and human-CD138 stains on fixed scaffold samples to ensure plasma cell identity (supplemental Figure 6). Together, these findings indicate that our 3D BM model allows for cancer-bone modeling in a more biologically relevant system than does 2D culture or soft 3D culture.
To address the study of myeloma-induced osteogenesis inhibition, we first confirmed prior reports8 that proliferation and osteogenesis are significantly inhibited by myeloma in clinical samples (MM-MSCs vs ND-MSCs) and in vitro 2D cocultures of ND-MSCs and myeloma cells (supplemental Figures 7 and 8). We also assessed the mRNA profile of clinical samples (ND-MSCs and MM-MSCs) by analyzing 230 mRNAs involved in cell proliferation, differentiation, migration, and other vital signaling. Unsupervised analysis demonstrated distinct clustering between ND-MSCs and MM-MSCs, confirming inherent differences between normal and myeloma stroma (supplemental Figure 9A). Forty-nine mRNAs were found to have significantly different expression between ND-MSCs and MM-MSCs (P < .05, 1.5-fc; supplemental Figure 9B), including the cell-cycle regulators CDKN2A (p16, previously reported29 ) and CDKN1A (p21, not previously reported), which may contribute to the decreased MM-MSC proliferation, and Collage1A1, likely contributing to decreased bone matrix formation.
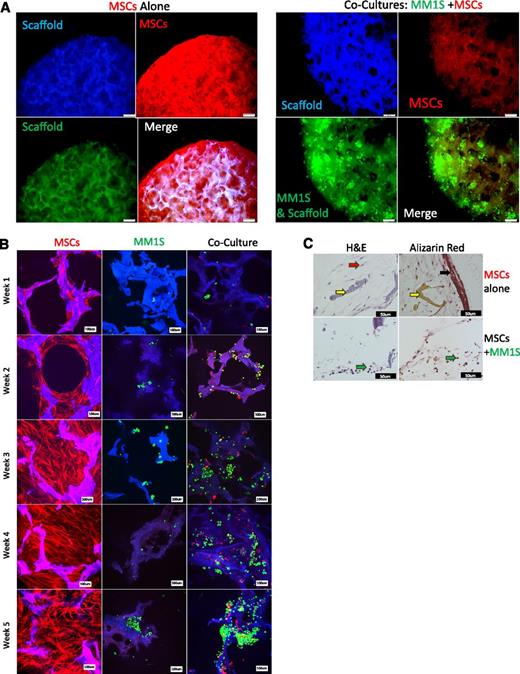
We then attempted to model inhibited osteogenesis of MSCs in our 3D BM model. ND-MSCs and GFP+MM1S were cultured alone or together on scaffolds in osteogenic media over 5 weeks. Confocal and fluorescent microscopy showed that GFP+MM1S cells inhibited ND-MSC proliferation, migration, and tissue production in scaffolds (Figure 2A-B and supplemental Videos 2 and 3). Alizarin Red Solution and H&E staining histology of scaffolds after 5 weeks demonstrated cellular tissue formation and mineralization in ND-MSC samples grown alone. Conversely, a lack of mineralization, as well as poor tissue formation and decreased cell numbers, were observed in samples of ND-MSCs cocultured with MM1S cells (Figure 2C). Interestingly, by week 2 the inhibition of MSC growth was evident in confocal imaging, and this trend continued over the full 5-week period, whereas MSCs alone proliferated, filled in scaffold pores, and formed mineralized, TE-bone. In sum, this system is useful for investigating myeloma effects on 3D osteogenesis in a more realistic setting than in 2D.
Inhibited osteogenesis induced by myeloma in a 3D bone model. (A) Fluorescent imaging at week 5 (TurboRFP+MSCs, red; GFP+MM1S, green; scaffold, blue). Overlaid channels (merge) shows increased pore infiltration, elongation, and proliferation by MSCs when grown in the absence of myeloma cells (left) compared with when grown with MM1S (right). The scale bar represents 200 μm. (B) Confocal images of TurboRFP+MSCs (red) and GFP+MM1S (green), alone or in coculture, on silk scaffolds (blue) from 1 to 5 weeks of culture in osteogenic media. The scale bar represents 100 μm. (C) Histologic analysis of scaffolds after 5 weeks of osteogenesis for MSCs alone (top) or in coculture with GFP+MM1S (bottom) stained for mineralization (Alizarin Red, right) or hematoxylin and eosin (H&E) (left). Black arrow indicates mineralization found only in MSCs cultured alone. Yellow arrows indicate silk scaffold. Red arrow indicates stromal cells, which are found throughout the MSC alone samples and sparsely through coculture samples. Green arrows indicate MM1S plasma cells found only in coculture samples. The scale bar represents 50 μm.
Inhibited osteogenesis induced by myeloma in a 3D bone model. (A) Fluorescent imaging at week 5 (TurboRFP+MSCs, red; GFP+MM1S, green; scaffold, blue). Overlaid channels (merge) shows increased pore infiltration, elongation, and proliferation by MSCs when grown in the absence of myeloma cells (left) compared with when grown with MM1S (right). The scale bar represents 200 μm. (B) Confocal images of TurboRFP+MSCs (red) and GFP+MM1S (green), alone or in coculture, on silk scaffolds (blue) from 1 to 5 weeks of culture in osteogenic media. The scale bar represents 100 μm. (C) Histologic analysis of scaffolds after 5 weeks of osteogenesis for MSCs alone (top) or in coculture with GFP+MM1S (bottom) stained for mineralization (Alizarin Red, right) or hematoxylin and eosin (H&E) (left). Black arrow indicates mineralization found only in MSCs cultured alone. Yellow arrows indicate silk scaffold. Red arrow indicates stromal cells, which are found throughout the MSC alone samples and sparsely through coculture samples. Green arrows indicate MM1S plasma cells found only in coculture samples. The scale bar represents 50 μm.
We then investigated the role of miRs in the dysfunctional osteogenesis of MSCs cultured with myeloma cells. TurboRFP+ ND-MSCs were cultured alone or with GFP+MM1S cells in the 3D model in osteogenic media for 2 weeks, sorted and collected using FACS, and analyzed for miR changes (coculture vs monoculture) using nanoString analysis of 800 miRs. Fifty-three miRs (28 up- and 25 downregulated) showed significantly altered expression in MSCs during coculture with MM1S (Figure 3A and supplemental Table 2). To compare with clinical samples, miR profiling was also performed on BM stroma samples from normal, healthy donor or myeloma patients. These samples demonstrated 41 miRs (34 up- and 7 downregulated) with significantly altered expression in MSCs from myeloma patient donors (MM) vs normal donor MSCs (ND) (Figure 3B and supplemental Table 2). Of these, six were found to be similarly downregulated (1 miR) or upregulated (5 miRs) in the 3D coculture system in the 3D model compared with the clinical samples (Table 1). The correlation between the patient and normal samples, and the BM niche 3D model contributes additional evidence that our system can reliably recapitulate many of the in vivo effects of MM cells on MSCs and suggests miRs that may govern the inhibited osteogenesis seen in patient MSCs. All miR data can be found in the GEO database under accession number GSE60423.
Alterations in MSC miRs in 3D modeling and patient vs normal data, and the resulting changes in MSCs after mimic-induced increased expression of miR-199a. (A) Heat map of the 53 mRNAs identified from nanoString analysis from the 3D model samples that are significantly different between cocultured (Co-culture) and monocultured ND-MSCs (MSCs). Filtering was done on original 800 miRs based on high expression (>25 average counts), significance between myeloma vs normal donor groups (P < .05), and high fc threshold (fc >1.5). (B) Heat map of 41 miRs identified from nanoString analysis that are significantly different between patient samples (myeloma [MM] and normal donor [ND] sample MSCs). Filtering was done on original 800 miRs based on high expression (>25 average counts), with significance between myeloma vs normal donor groups (P < .05), and high fc threshold (fc >1.5). (C) Alizarin Red staining quantification of mineralization produced by MSCs transfected with negative control mimic or miR-199a-5p mimic after 10 days in osteogenic no-dexamethasone media. Data plotted as mean ±SEM, n ≥3 different donors. (D) Alizarin Red staining representative images showing mineralization of MM-MSCs in 6-well plates transfected with negative control mimics or miR-199a-5p mimics to increase miR-199a-5p expression after 10 days in osteogenic no-dexamethasone media. Images are representative of n ≥3 different donors. The scale bars represent 200µm (original magnification ×4), 100 µm (×10), 50 µm (×20), and 20 µm (×40). (E) MM-MSCs transfected to increase expression of miR-199a-3p (E) or 199a-5p (F) demonstrate increased expression of osteogenic markers after 10 days of culture, measured by q-RT-PCR, gene expression normalized to negative control (Control) for each gene. ALPL, alkaline phosphatase; BSP, integrin-binding sialoprotein; Col1a1, collagen type I α 1; OP, osteopontin; OC, osteocalcin; RUNX2, runt-related transcription factor 2. Data plotted as mean ± SEM and analyzed with 1-way ANOVA and a post hoc Fisher least significance difference test for multiple comparisons (each gene vs negative control). Day 10 after transfection with miR mimics or controls and grown in osteogenic-no dexamethasone medium. n ≥ 3 different donors, **P < .05, **P < .01.
Alterations in MSC miRs in 3D modeling and patient vs normal data, and the resulting changes in MSCs after mimic-induced increased expression of miR-199a. (A) Heat map of the 53 mRNAs identified from nanoString analysis from the 3D model samples that are significantly different between cocultured (Co-culture) and monocultured ND-MSCs (MSCs). Filtering was done on original 800 miRs based on high expression (>25 average counts), significance between myeloma vs normal donor groups (P < .05), and high fc threshold (fc >1.5). (B) Heat map of 41 miRs identified from nanoString analysis that are significantly different between patient samples (myeloma [MM] and normal donor [ND] sample MSCs). Filtering was done on original 800 miRs based on high expression (>25 average counts), with significance between myeloma vs normal donor groups (P < .05), and high fc threshold (fc >1.5). (C) Alizarin Red staining quantification of mineralization produced by MSCs transfected with negative control mimic or miR-199a-5p mimic after 10 days in osteogenic no-dexamethasone media. Data plotted as mean ±SEM, n ≥3 different donors. (D) Alizarin Red staining representative images showing mineralization of MM-MSCs in 6-well plates transfected with negative control mimics or miR-199a-5p mimics to increase miR-199a-5p expression after 10 days in osteogenic no-dexamethasone media. Images are representative of n ≥3 different donors. The scale bars represent 200µm (original magnification ×4), 100 µm (×10), 50 µm (×20), and 20 µm (×40). (E) MM-MSCs transfected to increase expression of miR-199a-3p (E) or 199a-5p (F) demonstrate increased expression of osteogenic markers after 10 days of culture, measured by q-RT-PCR, gene expression normalized to negative control (Control) for each gene. ALPL, alkaline phosphatase; BSP, integrin-binding sialoprotein; Col1a1, collagen type I α 1; OP, osteopontin; OC, osteocalcin; RUNX2, runt-related transcription factor 2. Data plotted as mean ± SEM and analyzed with 1-way ANOVA and a post hoc Fisher least significance difference test for multiple comparisons (each gene vs negative control). Day 10 after transfection with miR mimics or controls and grown in osteogenic-no dexamethasone medium. n ≥ 3 different donors, **P < .05, **P < .01.
Finally, we examined whether any of the 6 miRs identified in the 3D model and in the clinical samples could serve as targets for inducing osteogenesis. Inhibition of miRs overexpressed in MM-MSCs (miR-181a-5p, miR181c-5p, miR-222-3p, miR-601, miR-146a-5p) using miRCURY inhibitors did not alter the mineralization potential of MM-MSCs as assessed by Alizarin Red Solution (data not shown) and hence were not further pursued. However, increasing the expression of miR-199a-5p significantly increased mineralized matrix production, indicative of osteogenic potential, and further supported the function of miR-199a as an osteogenic-promoting miR (Figure 3C-D). Using miR mimic transfection in ND-MSCs and MM-MSCs, we increased expression of miR-199a and observed that increasing the expression of both miR-199a-5p and miR-199a-3p in MM-MSCs significantly increased expression of several common osteogenic markers previously described,30 namely, alkaline phosphatase, integrin-binding sialoprotein, collagen type I α 1, osteopontin, osteocalcin, and runt-related transcription factor 2 (Figure 3E-F and supplemental Figure 10). Similar results were found with transfection of ND-MSCs with miR-199a-3p and 199a-5p mimics (data not shown).
To explore the potential pathways regulated by hsa-miR-199a-5p, we performed a pathway enrichment analysis of its predicted target genes. Analysis of miR-199a-5p targets revealed 19 pathways that were significantly enriched (supplemental Table 3). Interestingly, among them the ErbB signaling pathway was identified and is also reported to be involved in osteogenesis.31,32 The MAPK signaling pathway was also identified by us, as well as by others,33 as an miR199a-5p target pathway, and has been shown to play a role in osteogenic differentiation via the ErbB1 and ErbB2 pathways.34 Moreover, 3 pathways centered on semaphorins were also identified and may play a role in osteogenesis formation,35,36 although this is currently not well-defined.37 Hence, the pathways identified here may explain the mechanisms by which miR199a-5p regulates mRNAs that have anti-osteogenic effects (such as semaphorin4D) and may suggest novel pathways that could be targeted to normalize the osteogenic differentiation of MM-MSCs.
Discussion
3D culture models with material properties similar to those found in vivo are materializing as essential tools in cancer biology, owing to their ability to replicate tissue- or organ-specific structural features, biomechanical properties, and cell-cell or cell–extracellular matrix interactions more accurately than conventional 2D culture. Our new preclinical bone cancer model has the capacity to support long-term culture and imaging for expansion of primary myeloma cells, high-throughput drug screening, vessel formation, and osteogenesis in the presence of cancer. Prior published models have used soft, hydrogel matrices that cannot be mineralized and therefore cannot mimic the bone microenvironment.3,4 Our 3D model uses silk protein–based scaffolds that allow for active cell attachment and adherence to scaffolds rather than passive encapsulation in 3D hydrogel cultures. In addition, the tissue-engineering approach represents a more controllable model compared with culturing whole-patient bone biopsies,6 because it allows for user-designed introduction of cells of interest, increasing the reproducibility, adaptability, and scalability of the model. Therefore, the silk-based 3D TE-bone model presented herein represents a unique model to examine the interactions of bone and cancer cells in a 3D microenvironment, with mechanical properties similar to bone.
It remains to be determined why a decrease in certain miRs may lead to inhibited osteogenesis in myeloma and what mRNA targets drive this, but it is evident that overexpressing certain miRs within MSCs can increase their osteogenic potential, and our 3D model helped to identify 199a-5p as one such miR. MiR-199a has been described as “flexible and versatile as a chameleon,”33 because it has a wide variety of important functions across many cell types and systems. In terms of osteogenesis, miR-199a-5p specifically has been shown to have a pro–stem cell differentiation effect in BM-derived human MSCs both in vitro and in vivo, whereas inhibition with siRNAs blocking miR-199a-5p reduced osteogenesis of hMSCs.38,39 Pathways implicated in this are still uncertain but include HIF1a, TWIST, NADPH-oxidase, PI-3 kinase, mitogen-activated protein kinase, and NF-κB pathways, which are being investigated for their roles in osteogenesis.38,40 miR-199a is also a BMP2-responsive miR,41 suggesting that altered BMP2 signaling may be involved in the observed effects of these miRs. Decreased 199a-5p may also increase fibronectin in MM BM, which is elevated in MM patient serum42,43 and has been shown to cause increased MM tumor accumulation within the BM and dictate CAM-DR.44 Although the exact composition and interaction of mRNAs inhibited by miR-199a appear to be complex, it is clear that miR-199a represents the first miR identified as abnormally downregulated, and one of the first abnormally expressed45 in bone cancer patients, that may be a therapeutic strategy for enhancing bone formation.
The roles of specific miRs in osteogenesis and MSC-tumor feedback are currently enigmatic,39,46 but our results suggest that novel target miRs are useful for reactivating the osteogenic abilities of cancer-associated MSCs. Targeting these miRs may provide a new avenue for healing lesions and reversing the osteolytic cancer cycle in myeloma along with other osteotropic cancers. In conclusion, the novel 3D, in vitro bone cancer model developed provides a physiologically relevant platform to investigate osteogenesis, angiogenesis, and cancer growth, as well as drug response with primary samples and cell lines. It allows for nondestructive imaging over long periods and can be used for testing a multitude of other bone cancer hypotheses and modeling an array of biological processes involved in the inhibited osteogenesis of cancer-colonized bones. More broadly, many researchers would likely increase their in vivo success rates by first testing their hypotheses in our 3D model system. Our model allows biological questions to be investigated, compounds to be screened, and novel targets or therapeutics to be identified more quickly and cheaply and in a more realistic 3D BM niche setting than is currently available. Resulting research will be more clinically translatable and will advance more quickly and efficiently from the bench to the bedside of patients who have bone cancer.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Lisa Cameron, Grace O’Callaghan, Priya Dhir, Michele Moschetta, Susanna M. Santos, Wenjing Zhang, and Rucsanda Carmen Preda for their assistance.
This study was supported by the Department of Defense, Peer-review Cancer Research Program (W81XWH-13-1-0390), the National Institutes of Health P41 Center (EB002520) and National Cancer Institute grant R01CA154648.
Authorship
Contribution: M.R.R. designed the experiments, performed in vitro studies and data analysis, and wrote the manuscript; Y.M. created RFP-MSCs; YongZ., YuZ., and S.M. assisted with qRT PCR studies; J.E.R., Y.-T.T., A.S., and Y.A. provided primary MSCs or patient data; S.V.G., M.M., and Z.N.L. assisted with in vitro assays, cell culture, and qRT-PCR; J.S. performed microRNA pathway analysis; D.L.K. provided feedback and silk scaffolds; A.M.R. provided scientific assistance and advice; and I.M.G. supervised the project and edited the manuscript.
Conflict-of-interest disclosure: I.M.G. is on the advisory boards of Millennium/Takeda, Novartis, Onyx, and Celgene. All other authors declare no competing financial interests.
Correspondence: Irene M. Ghobrial, Medical Oncology, Dana-Farber Cancer Institute, 450 Brookline Ave, Boston, MA, 02115; e-mail: Irene_ghobrial@dfci.harvard.edu.

![Figure 3. Alterations in MSC miRs in 3D modeling and patient vs normal data, and the resulting changes in MSCs after mimic-induced increased expression of miR-199a. (A) Heat map of the 53 mRNAs identified from nanoString analysis from the 3D model samples that are significantly different between cocultured (Co-culture) and monocultured ND-MSCs (MSCs). Filtering was done on original 800 miRs based on high expression (>25 average counts), significance between myeloma vs normal donor groups (P < .05), and high fc threshold (fc >1.5). (B) Heat map of 41 miRs identified from nanoString analysis that are significantly different between patient samples (myeloma [MM] and normal donor [ND] sample MSCs). Filtering was done on original 800 miRs based on high expression (>25 average counts), with significance between myeloma vs normal donor groups (P < .05), and high fc threshold (fc >1.5). (C) Alizarin Red staining quantification of mineralization produced by MSCs transfected with negative control mimic or miR-199a-5p mimic after 10 days in osteogenic no-dexamethasone media. Data plotted as mean ±SEM, n ≥3 different donors. (D) Alizarin Red staining representative images showing mineralization of MM-MSCs in 6-well plates transfected with negative control mimics or miR-199a-5p mimics to increase miR-199a-5p expression after 10 days in osteogenic no-dexamethasone media. Images are representative of n ≥3 different donors. The scale bars represent 200µm (original magnification ×4), 100 µm (×10), 50 µm (×20), and 20 µm (×40). (E) MM-MSCs transfected to increase expression of miR-199a-3p (E) or 199a-5p (F) demonstrate increased expression of osteogenic markers after 10 days of culture, measured by q-RT-PCR, gene expression normalized to negative control (Control) for each gene. ALPL, alkaline phosphatase; BSP, integrin-binding sialoprotein; Col1a1, collagen type I α 1; OP, osteopontin; OC, osteocalcin; RUNX2, runt-related transcription factor 2. Data plotted as mean ± SEM and analyzed with 1-way ANOVA and a post hoc Fisher least significance difference test for multiple comparisons (each gene vs negative control). Day 10 after transfection with miR mimics or controls and grown in osteogenic-no dexamethasone medium. n ≥ 3 different donors, **P < .05, **P < .01.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/22/10.1182_blood-2014-02-558007/4/m_3250f3.jpeg?Expires=1769638141&Signature=ZXKcsBaID2eBQ6NbAIod4lvegBW3~B9LciaEbS4aV15Aif-pg8Zamx32t1EvorpXMPzLmKqszPDO6gMIQR-1CaveodAzANE8BHY9EkBWKr5l6MGnUW3l92aBwsFhlkxcJmxSiIWm0iy6GI~ri5ukfCdstVcDi39fWrerq17fzuBIqVtXHKkIEo44pOp4jzOQ6o9Vt~6Zmpq4thjJNbadTFR8ugc-Q36Tu99AOrxQ~Hn-LYo3VTgQNefHqOsJDo5jYVnDno4t9cp6O~Qtvs1gT8qd4KfByW0NKye9oS~RkrWFcnm0LrKHgbFjtzxNemwZ0bzCQanrwb8C3MiQMTouCA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
