Key Points
EVI1-r AMLs have recurrent mutations in RAS and other signaling genes, splicing factors, and at a lower frequency, IKZF1 and TP53.
EVI1-r AMLs show a characteristic transcriptome profile marked by high expression of MECOM, PREX2, MYCT1, PAWR, and VIP.
Abstract
The genetic and transcriptional signature of EVI1 (ecotropic viral integration site 1)-rearranged (EVI1-r) acute myeloid leukemias (AMLs) remains poorly defined. We performed RNA sequencing of 12 EVI1-r AMLs and compared the results with those of other AML subtypes (n = 139) and normal CD34+ cells (n = 17). Results confirm high frequencies of RAS and other activated signaling mutations (10/12 AMLs) and identify new recurrent mutations in splicing factors (5/12 AMLs in SF3B1 and 2/12 AMLs in U2AF1), IKZF1 (3/12 AMLs), and TP53 (3/12 AMLs). Mutations in IKZF1, a gene located on chromosome 7, and monosomy 7 are mutually exclusive in this disease. Moreover IKZF1 expression is halved in monosomy 7 leukemias. EVI-r AMLs are also characterized by a unique transcriptional signature with high expression levels of MECOM, PREX2, VIP, MYCT1, and PAWR. Our results suggest that EVI1-r AMLs could be molecularly defined by specific transcriptomic anomalies and a hitherto unseen mutational pattern. Larger patient cohorts will better determine the frequency of these events.
Introduction
The EVI1 (ecotropic viral integration site 1) gene, also termed MECOM (MDS1 and EVI1 complex locus), is located on chromosomal band 3q26.2. It is expressed at high levels in normal CD34+ cells1 and in 5% to 10% of human acute myeloid leukemias (AMLs).2,3 In a subset of these patients, EVI1 is rearranged. The typical cytogenetic anomaly inv(3)(q21q26.2)/t(3;3)(q21;q26.2), included in 2008 in the World Health Organization classification under the category “AML with recurrent genetic abnormalities,” represents one such rearrangement. This entity is characterized by very poor overall survival and by distinct morphologic features including atypical megakaryocytes, multilineage dysplasia, and normal or elevated blood platelet counts.4-6
The most frequent cytogenetic anomaly associated with EVI1-rearranged (EVI1-r) AMLs is monosomy 7, found in 33% to 66% of cases of inv(3)/t(3;3)5-9 and in one third of AMLs with other EVI1 rearrangements.6 Targeted analyses have revealed mutations in RUNX1, NRAS, KRAS, and NF1 in 20% to 33% of cases.8 However, the full mutation spectrum and expression profile remain unknown for this rare disease.
Material and methods
Material and methods are detailed in the supplemental Methods, available on the the Blood Web site. Samples were collected between 2001 and 2014, according to Banque de Cellules Leucémiques du Québec procedures, in accordance with the Declaration of Helsinki. Libraries were constructed with the TruSeq Protocols, and sequencing was performed using an Illumina HiSequation 2000. Variants were identified using Casava 1.8.2 and splice isoforms with Tophat 2.0.7 and Cufflinks 2.1.1. All variants in cancer-related genes (listed in supplemental Table 1) are reported. In other genes, only recurring variants (≥2 EVI1-r samples) were analyzed. Somatic or germline origin of new variants (not in the COSMIC database) were confirmed by Sanger DNA sequencing. Fisher’s exact test was used in the analysis of contingency tables. Analysis of differential gene expression was performed with the Wilcoxon rank-sum test (Mann-Whitney), and the false discovery rate method was applied for global gene analysis.
Results and discussion
Analysis of acquired mutations in EVI1 AMLs
Twelve EVI1-r AMLs were analyzed, including 7 with inv(3)/t(3;3) and 5 with other EVI1 rearrangements, as well as a cohort of 139 AMLs from other cytogenetic groups and 17 control cord blood–derived normal CD34+ cells (clinical and laboratory characteristics and sequencing statistics are detailed in supplemental Tables 2-4). The most frequent mutations in EVI1-r AMLs are presented in Figure 1A-B and in supplemental Table 5, and statistical analyses are shown in supplemental Table 6.
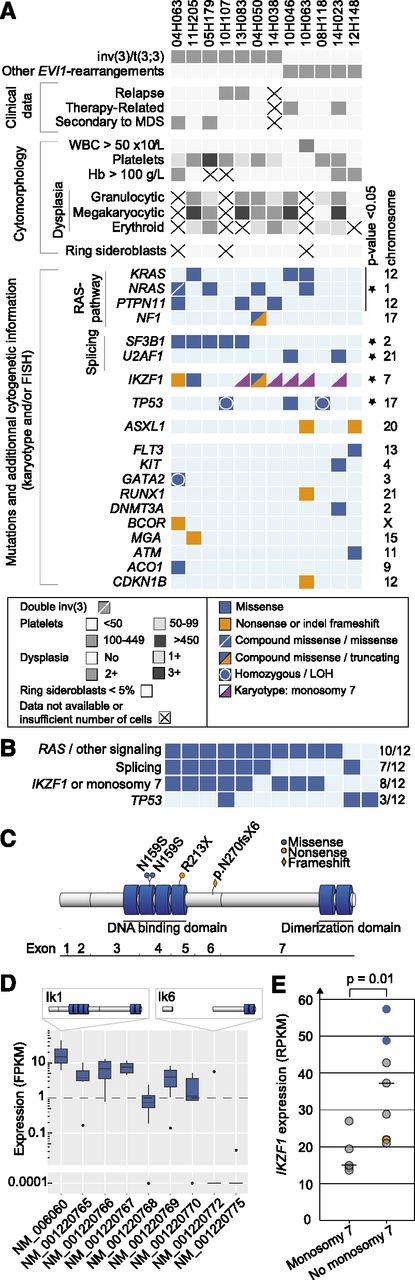
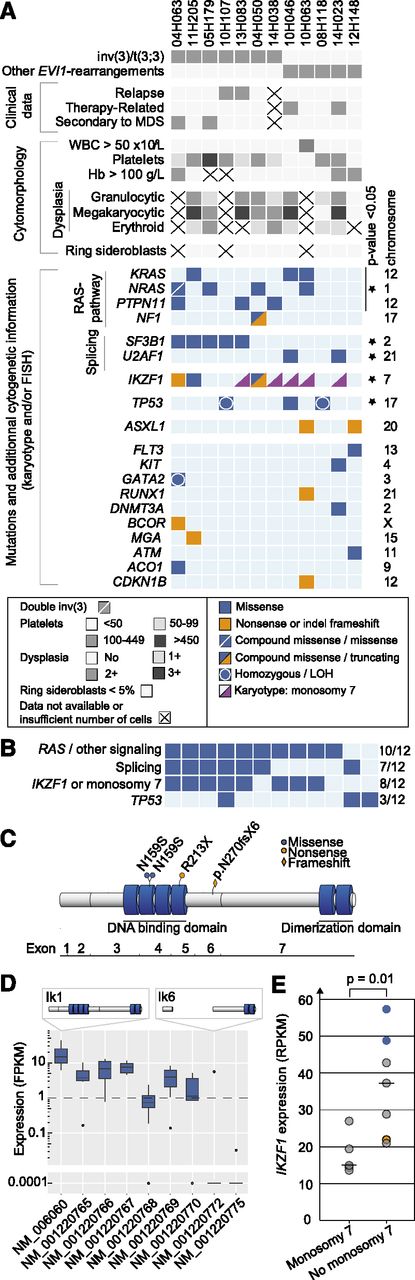
Mutations in EVI-r AMLs. (A) Clinical, morphologic, cytogenetic, and mutational data in EVI1-r AML samples. LOH, loss of heterozygosity. (B) Summary of commonly mutated pathways. (C) IKZF1 protein and mutations. Four N-terminal (DNA-binding domain) and 2 C-terminal (dimerization domain) zinc fingers are represented by blue cylinders. (D) Distribution of IKZF1 isoforms in EVI1-r samples. The only sample expressing isoform NM_001220772 (Ik6) harbors compound heterozygous mutations of IKZF1. (E) IKZF1 expression levels in EVI1-r AMLs with and without monosomy 7. Blue dots represent samples with IKZF1 N159S mutation; orange dot indicates IKZF1 frameshift mutation. Analysis is performed with all 12 specimens except for D, where n = 9.
Mutations in EVI-r AMLs. (A) Clinical, morphologic, cytogenetic, and mutational data in EVI1-r AML samples. LOH, loss of heterozygosity. (B) Summary of commonly mutated pathways. (C) IKZF1 protein and mutations. Four N-terminal (DNA-binding domain) and 2 C-terminal (dimerization domain) zinc fingers are represented by blue cylinders. (D) Distribution of IKZF1 isoforms in EVI1-r samples. The only sample expressing isoform NM_001220772 (Ik6) harbors compound heterozygous mutations of IKZF1. (E) IKZF1 expression levels in EVI1-r AMLs with and without monosomy 7. Blue dots represent samples with IKZF1 N159S mutation; orange dot indicates IKZF1 frameshift mutation. Analysis is performed with all 12 specimens except for D, where n = 9.
RAS mutations
RAS-pathway mutations (NRAS, KRAS, PTPN11, NF1) were significantly more frequent (8/12 AMLs) in EVI1-r AMLs than in other AMLs sequenced as part of this project (43/139 AMLs; P = .022). Allelic frequency determination revealed that at least 1 gene of this pathway was mutated in the dominant clone, contributing to a cumulative ratio of mutated/wild-type alleles of ∼50% (supplemental Table 5), possibly indicating a strong collaboration between RAS-pathway activation and EVI1. Two additional samples had mutations in KIT and FLT3; hence, “activated signaling” mutations were present in 10/12 samples (Figure 1B).
Splicing factors
Splicing was the second most commonly mutated pathway (7/12 AMLs). SF3B1 mutations were found in 5 samples, all within the inv(3)/t(3;3) subgroup, and at a higher frequency than in the control cohort (P < .0001). Mutations were all located in exons 14-15, corresponding to the same “hotspots” found in myelodysplastic syndromes (MDS): K700E (n = 3), G740E (n = 1), and R625C (n = 1). In MDS, SF3B1 mutations are associated with ring sideroblasts and normal or elevated platelet counts.10,11 In our cohort, ring sideroblasts were universally absent, and platelet counts were not different between SF3B1-mutated and wild-type samples (median, 109 vs 88 × 109/L, respectively; P = .34). Another splicing factor, U2AF1, was mutated in 2 additional samples at position Q157, as observed in MDS (comparative analysis with control AMLs, P = .03; Figure 1A).
IKZF1
Four IKZF1 mutations were found in 3/12 EVI1-r AMLs compared with none in the control cohort (Figure 1A-C; supplemental Figure 2; P = .0004). Although mutations of this gene were previously reported in BCR-ABL1-positive acute lymphoblastic leukemia (ALL)12 and in advanced myeloproliferative neoplasms,13 they were never found to be recurrent in any AML cohort. IKZF1-mutated samples displayed typical AML characteristics (supplemental Figure 3). In ALL, the frequent deletion of IKZF1 exons 3-6 (Δ3-6) compromises the DNA-binding domain of this protein. Such deletions were not found in our cohort (Figure 1D). Rather, nonsynonymous or truncating mutations were observed (eg, N159S [n = 2], R213X, N270fsx6; Figure 1C). Interestingly, the N159S mutation, occurring in a position critical for DNA binding,14 was observed in 2/10 samples.
The reported association between monosomy 7 and EVI1-r AMLs is most interesting, considering that IKZF1 is located on this chromosome and that IKZF1 expression levels were significantly lower in samples with monosomy 7 (Figure 1E). Monosomy 7 and IKZF1 mutations were never observed simultaneously (see pink triangles representing monosomy 7 specimens in Figure 1A), possibly indicating that IKZF1 is more important than anticipated in this disease. This is reminiscent of what has been found in ALL, where IKZF1 alterations are either limited to the gene itself (intragenic deletions or mutations) or involve larger chromosomal deletions, predicted to result in the expression of a dominant-negative isoform or in haploinsufficiency, respectively.12,15
Other mutations
Mutations were recurrently found in 2 additional genes; namely, TP53 (3/12 AMLs) and ASXL1 (2/12 AMLs) (Figure 1A-B). Additional nonrecurrent mutations are illustrated in Figure 1A. Among those, 1 sample had a GATA2 mutation with monoallelic expression (variant allele frequency, 91%; supplemental Table 5). This is most interesting, considering the recent data showing that MECOM overexpression in AML with 3q rearrangements is driven by an enhancer of the GATA2 gene.16
Transcriptome analyses
MECOM.
Many genes were differentially expressed in EVI1-r AMLs when compared with control leukemias (Figure 2A; supplemental Table 7A). As expected, MECOM was highly expressed in all EVI1-r samples, in 10.8% of non-EVI1-r leukemias (15/139 AMLs), and in all normal CD34+ cells (Figure 2B).
Transcriptomic profile of EVI1-r AMLs. (A) Comparative analysis of expressed genes in EVI-r AMLs compared with AMLs without EVI1 rearrangements. Among expressed genes (ie, those with an average log10 expression ≥3, corresponding approximately to a reads per kilobase of transcript per million reads mapped [RPKM] of 0.1), the 20 most differentially expressed in EVI1-r AMLs are highlighted (solid rhombuses, also detailed in supplemental Table 7A). Color of dots shows the false discovery rate q-values. (B) Comparative analysis of MECOM, PREX2, MYCT1, VIP, and PAWR expression between EVI1-r AML, AML without EVI1 rearrangements, and normal CD34+ cord blood-derived cells. *P < .05, **P < .005, and ***P < .0005.
Transcriptomic profile of EVI1-r AMLs. (A) Comparative analysis of expressed genes in EVI-r AMLs compared with AMLs without EVI1 rearrangements. Among expressed genes (ie, those with an average log10 expression ≥3, corresponding approximately to a reads per kilobase of transcript per million reads mapped [RPKM] of 0.1), the 20 most differentially expressed in EVI1-r AMLs are highlighted (solid rhombuses, also detailed in supplemental Table 7A). Color of dots shows the false discovery rate q-values. (B) Comparative analysis of MECOM, PREX2, MYCT1, VIP, and PAWR expression between EVI1-r AML, AML without EVI1 rearrangements, and normal CD34+ cord blood-derived cells. *P < .05, **P < .005, and ***P < .0005.
Other preferentially expressed genes.
PREX2, VIP, MYCT1, and PAWR are also overexpressed in EVI1-r AMLs compared with non-EVI1-r specimens (Figure 2A). High expression levels of these genes are very sensitive and specific to this disease (supplemental Table 7B). Most interestingly, as observed with MECOM, these genes are highly expressed in normal CD34+ cells (Figure 2B).
PREX2 encodes for a PIP3-dependent Rac exchanger,17 which amplifies PI3K signaling,18 possibly leading to RAC and AKT pathway activation. PAWR (PRKC, apoptosis, WT1, regulator, also termed Par-4) is a proapoptotic protein inactivated by PI3K/AKT signaling and participates in PTEN-mediated apoptosis.19 MYCT1 overexpression recapitulates many of the c-Myc phenotypes including tumorigenic conversion.20 EVI1 binding sites are found in the MYCT1 promoter (EpiTect ChIP qPCR Primers), which appears to be under the control of EVI1 in primary murine hematopoietic progenitor cells.21 VIP (vasointestinal peptide) is adjacent to MYCT1 on chromosomal band 6q25.2. It has been described as a potential growth-promoting factor in hematopoietic stem and progenitor cells22 and may have a role in megakaryocytic cell proliferation.23
In summary, this study provides in-depth transcriptome analyses of the rare EVI1-rearranged AMLs and shows that these leukemias are distinctly characterized by mutations in RAS/signaling, splicing factors, and IKZF1, a gene found on chromosome 7. Our work also documents an inverse correlation between IKZF1 mutations and monosomy 7, the nature of which will need larger patient cohorts for confirmation and functional characterization. A distinct expression signature characteristic of EVI1-rearranged AMLs was identified. These findings may provide new insights into the networks operating in this disease.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Isabel Boivin for data validation, Geneviève Boucher from the Institute for Research in Immunology and Cancer bioinformatics platform for data processing, and Pierre Chagnon and Marianne Arteau at the Institute for Research in Immunology and Cancer genomics platform for sequencing. The dedicated work of the Banque de Cellules Leucémiques du Québec staff, namely, Claude Rondeau and Sylvie Lavallée, is also acknowledged.
This work was supported by the Government of Canada through Genome Canada and the Ministère de l’Enseignement Supérieur, de la Recherche, de la Science et de la Technologie du Québec through Génome Québec to G.S., J.H., and S.L. G.S. and J.H. are recipients of a Canada Research Chair in Molecular Genetics of Stem Cells and of an Industrielle-Alliance leukemia chair. J.H. is supported by a grant from the Cancer Research Network of the Fonds de Recherche du Québec–Santé. RNA-Seq read mapping and transcript quantification were performed on the supercomputer Briaree from Université de Montréal, managed by Calcul Québec and Compute Canada. The operation of this supercomputer is funded by the Canada Foundation for Innovation, NanoQuébec, Réseau de Médecine Génétique Appliquée du Québec, and the Fonds de recherche du Québec–Nature et technologies. V.-P.L. is supported by a postdoctoral fellowship jointly supported by the Hôpital Maisonneuve-Rosemont’s Foundation and by the Cole Foundation.
Authorship
Contribution: V.-P.L. analyzed the next generation sequencing data of all samples, generated the figures and tables, and cowrote the paper; G.S. contributed to analyses and cowrote the paper; J.H. analyzed and provided all the AML samples, analyzed the data, and edited the manuscript; P.G. processed the raw NGS data; G.D. performed microscopic analyses (morphology, immuno-cytochemistry) for all specimens in this study; and S.L. was responsible for supervision of the bioinformatics team.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Guy Sauvageau, Institute for Research in Immunology and Cancer, PO Box 6128, Station Centre-Ville, Montreal, QC, Canada, H3C 3J7; e-mail: guy.sauvageau@umontreal.ca; or Josée Hébert, Banque de Cellules Leucémiques du Québec, 5415 L'Assomption Blvd, Montreal, QC, Canada, H1T 2M4; e-mail: josee.hebert@umontreal.ca.

![Figure 2. Transcriptomic profile of EVI1-r AMLs. (A) Comparative analysis of expressed genes in EVI-r AMLs compared with AMLs without EVI1 rearrangements. Among expressed genes (ie, those with an average log10 expression ≥3, corresponding approximately to a reads per kilobase of transcript per million reads mapped [RPKM] of 0.1), the 20 most differentially expressed in EVI1-r AMLs are highlighted (solid rhombuses, also detailed in supplemental Table 7A). Color of dots shows the false discovery rate q-values. (B) Comparative analysis of MECOM, PREX2, MYCT1, VIP, and PAWR expression between EVI1-r AML, AML without EVI1 rearrangements, and normal CD34+ cord blood-derived cells. *P < .05, **P < .005, and ***P < .0005.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/1/10.1182_blood-2014-07-591529/4/m_140f2.jpeg?Expires=1770406614&Signature=rk7G7KqH0zFOnxqbUZDuCJ9NfRWoNVjI55Cb1PHON8iTPdNrEAu1SDkYCPJ6U3XRcTrr8m3DIIEUELdhiEqgFyZ0i80MrunQ9sKPFdKvksLvASHiW96vRgfA9E~JOd6xyZfPEnXV0UIHQHDt9BL977x3SGfXmgAUhXNyLkNMNIBqt5-8vMkMgmLBxe4Z1kuda9FvYNuusHuYFMBh5GpDypxBFmhL6sZvIfLIPYzzAGmzhVgSXPFXuopBMrpfK89zf01TsNvr7sy7VM3dJ0GrESudllCKPVwunlLV4NF0P-dyIQJXJXE4rTbvCRq5CEQQWsELnMP1vHxHa749Dl8~Fg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 2. Transcriptomic profile of EVI1-r AMLs. (A) Comparative analysis of expressed genes in EVI-r AMLs compared with AMLs without EVI1 rearrangements. Among expressed genes (ie, those with an average log10 expression ≥3, corresponding approximately to a reads per kilobase of transcript per million reads mapped [RPKM] of 0.1), the 20 most differentially expressed in EVI1-r AMLs are highlighted (solid rhombuses, also detailed in supplemental Table 7A). Color of dots shows the false discovery rate q-values. (B) Comparative analysis of MECOM, PREX2, MYCT1, VIP, and PAWR expression between EVI1-r AML, AML without EVI1 rearrangements, and normal CD34+ cord blood-derived cells. *P < .05, **P < .005, and ***P < .0005.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/1/10.1182_blood-2014-07-591529/4/m_140f2.jpeg?Expires=1770406615&Signature=Y1eGmh-XMyZvlxYhaeDxMzpZ9W9aquNqRA4W5KG5Of7C1Lc6rXaHA9D6AIbp-qKgf-tMOWOqtjThCnmCjVhLUoOI5Xm-4OueLTR0oSF1dARyqh-6bvtu3lDxvlAvlYYBfqzdHe9e-WESTGN-v29DaVdORuZtX7GkhpyVqbLi2DSaOifBewGbm0dIVkwNp8vm3bAm958FJUuiuyWP-QyEpd03930oqf1YCjSuMQBZeeEKjemHofaNRVwLweGeTp3MDtnfLxsfKVM1DRyWzxW1~oopiv-YAi1f8butDT~t28C6D-Cz5mP7ZW4dK3qc3l0Z25A4ga2T3UW5rsUOpan-xQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)