Key Points
Jarid1b is not required for steady-state hematopoiesis.
Jarid1b is required for HSC self-renewal.
Abstract
Jarid1b/KDM5b is a histone demethylase that regulates self-renewal and differentiation in stem cells and cancer; however, its function in hematopoiesis is unclear. Here, we find that Jarid1b is highly expressed in primitive hematopoietic compartments and is overexpressed in acute myeloid leukemias. Constitutive genetic deletion of Jarid1b did not impact steady-state hematopoiesis. In contrast, acute deletion of Jarid1b from bone marrow increased peripheral blood T cells and, following secondary transplantation, resulted in loss of bone marrow reconstitution. Our results reveal that deletion of Jarid1b compromises hematopoietic stem cell (HSC) self-renewal capacity and suggest that Jarid1b is a positive regulator of HSC potential.
Introduction
The capacity for hematopoietic stem cells (HSCs) to self-renew and regenerate the hematopoietic system throughout life is maintained through extrinsic and intrinsic mechanisms. Transcriptional regulation of HSC potential has been highly characterized, whereas more recently, epigenetic regulatory mechanisms, including DNA methylation and histone modifications, have been shown to be important in the maintenance and regulation HSC potential.1-4
Histone demethylases, including Lsd15 and Jarid1a,6 have been reported to be involved in the regulation of HSC and leukemic potential. Jarid1b (Kdm5b/Plu-1) is an H3K4me2/3 demethylase7,8 that regulates self-renewal and differentiation in a cell type–specific manner. Jarid1b is highly expressed in embryonic stem cells9-11 and in various primary cancers including melanoma,12 breast,13 and testicular,14 and is upregulated in leukemic cell lines.15 Recently, knockdown of Jarid1b in primitive hematopoietic cells has been reported to increase engraftment16 ; however, its functional role in HSCs is poorly defined.
Here, we characterize the role of Jarid1b within HSCs. We show that Jarid1b is highly expressed in human and murine primitive hematopoietic compartments and in acute myeloid leukemia (AML). Genetic deletion of Jarid1b does not disrupt steady-state hematopoiesis but does compromise HSC self-renewal potential after secondary transplant. Our results show that Jarid1b is required for HSC self-renewal.
Study design
Transplantation
Jarid1bfl/fl;RosaCreERT210 cryopreserved bone marrow (BM) was competitively transplanted into congenic recipients in a 1:1 ratio. Tamoxifen or vehicle control was administered intraperitoneally to induce deletion of Jarid1b.
HSC purification and analysis
For primary and secondary transplant analysis, BM cells were stained and sorted as previously described.17 For secondary transplants, donor cells isolated from primary transplant BM by fluorescence-activated cell sorting were competitively transplanted at a ratio of 1:1 into congenic recipients. See supplemental Methods, available on the Blood Web site.
Results and discussion
Jarid1b regulates self-renewal and differentiation in multiple cell types; however, its function in hematopoiesis is unclear. Interrogation of public expression data18 suggested that Jarid1b was highly expressed in primitive hematopoietic populations, including HSCs and multipotent progenitors (MPPs), but was downregulated in committed hematopoietic populations, including B lymphocytes, granulocytes, and T cells (Figure 1A). We validated these findings by qRT-PCR (Figure 1B). Expression profiling of human BM and hematopoietic stem/progenitor cells (HSPCs) revealed that JARID1B is more highly expressed in human HSPCs compared to unfractionated BM (supplemental Figure 1). Additionally, we found that JARID1B was significantly upregulated in AMLs compared to normal BM and that, in comparison with HSPCs, JARID1B was significantly upregulated in AMLs containing either the AML-ETO fusion t(8:21) or the t(15:17) translocation found in acute promyelocytic leukemia (supplemental Figure 1).19
Jarid1b is not required for steady-state hematopoiesis. (A) Expression of Jarid1b in hematopoietic populations.18 (B) qRT-PCR of Jarid1b expression in HSCs, MPPs (LSKCD34+), and MPs (Lineage–Sca1–ckit+). (C) Image of Jarid1b WT and KO spleen. Bar represents 1 cm. (D) Weight of Jarid1b control (WT or heterozygote) and KO spleens. FACS analysis of spleen (E) and thymus (F) from 8-week-old male Jarid1b control or KO mice. Blood count analysis of Jarid1b control and KO PB for leukocytes (G), lymphocytes (H), and erythrocytes (I). (J) FACS analysis of hematopoietic populations within the BM of Jarid1b KO mice. Error ± standard deviation, n = 5 mice per group; ***P < .001. (K) Frequency of HSCs, MPP1 (LSK34+Flk2−), and MPP2 (LSKCD34+Flk2+) within the BM LSK compartment. (L) Frequency of CMPs (LSKCD34+FcγRαmid), GMPs (LSKCD34+FcγRαhi), and MEPs (LSKCD34−FcγRα−) of the BM MP compartment. (M) Frequency of CLPs (Lineage−Flk2hiIL7Rα+) within live BM. Error ± standard deviation; n = 3. B cell, IgM+B220+; CLP, common lymphoid progenitor; CMP, common myeloid progenitor; Ctl, control; EP, erythroid progenitor (Ter119+CD71+); FACS, fluorescence-activated cell sorter; GMP, granulocyte-macrophage progenitor; Gran, granulocyte (Mac1+Gr1+); LSK, Lineage–Sca1+cKit+; MEP, megakaryocyte-erythroid progenitor; MP, myeloid progenitor; qRT-PCR, quantitative reverse-transcription polymerase chain reaction; SP, single positive; WT, wild-type.
Jarid1b is not required for steady-state hematopoiesis. (A) Expression of Jarid1b in hematopoietic populations.18 (B) qRT-PCR of Jarid1b expression in HSCs, MPPs (LSKCD34+), and MPs (Lineage–Sca1–ckit+). (C) Image of Jarid1b WT and KO spleen. Bar represents 1 cm. (D) Weight of Jarid1b control (WT or heterozygote) and KO spleens. FACS analysis of spleen (E) and thymus (F) from 8-week-old male Jarid1b control or KO mice. Blood count analysis of Jarid1b control and KO PB for leukocytes (G), lymphocytes (H), and erythrocytes (I). (J) FACS analysis of hematopoietic populations within the BM of Jarid1b KO mice. Error ± standard deviation, n = 5 mice per group; ***P < .001. (K) Frequency of HSCs, MPP1 (LSK34+Flk2−), and MPP2 (LSKCD34+Flk2+) within the BM LSK compartment. (L) Frequency of CMPs (LSKCD34+FcγRαmid), GMPs (LSKCD34+FcγRαhi), and MEPs (LSKCD34−FcγRα−) of the BM MP compartment. (M) Frequency of CLPs (Lineage−Flk2hiIL7Rα+) within live BM. Error ± standard deviation; n = 3. B cell, IgM+B220+; CLP, common lymphoid progenitor; CMP, common myeloid progenitor; Ctl, control; EP, erythroid progenitor (Ter119+CD71+); FACS, fluorescence-activated cell sorter; GMP, granulocyte-macrophage progenitor; Gran, granulocyte (Mac1+Gr1+); LSK, Lineage–Sca1+cKit+; MEP, megakaryocyte-erythroid progenitor; MP, myeloid progenitor; qRT-PCR, quantitative reverse-transcription polymerase chain reaction; SP, single positive; WT, wild-type.
Jarid1b is highly expressed in primitive hematopoietic populations; however, its function is unclear. To determine how loss of Jarid1b would impact steady-state hematopoiesis, we characterized the hematopoietic system of the constitutive Jarid1b knockout (KO) mouse, in which exon 6 was deleted, resulting in a frameshift and subsequent termination mutation.10 Loss of Jarid1b (supplemental Figure 2) did not affect auxiliary hematopoietic organs (Figure 1C-E) or T cells within the thymus (Figure 1F). qRT-PCR confirmed that Jarid1b was expressed in wild-type littermate controls in a manner similar to C57/Bl6 mice (supplemental Figure 3A and Figure 1A-B). Peripheral blood (PB) analysis also did not reveal any difference in blood composition (Figure 1G-I). Analysis of BM progenitor compartments did not reveal any differences in the frequencies of MPs or the Lineage–Sca1+cKit+ (LSK) compartment, which contains MPPs and HSCs (Figure 1J). Further characterization of primitive BM progenitor compartments did not reveal any differences in the frequencies of HSCs, MPPs, CMPs, GMPs, MEPs, or CLPs as a result of Jarid1b loss (Figure 1K-M and supplemental Figure 3B-C). Characterization of the aged hematopoietic system in Jarid1b KO mice was not possible due to increased adult morbidity and mortality in >20-week-old constitutive Jarid1b KO mice (supplemental Figure 4A). BM analysis of 13- to 18-week-old Jarid1b KO mice showed no change to the frequency of HSCs within the LSK compartment or to other BM progenitor compartments relative to 9- to 12-week-old mice or to wild-type mice (supplemental Figure 5A-B). Thus, despite high expression within primitive hematopoietic compartments, Jarid1b does not appear to be required for maintenance of steady-state hematopoiesis. It is possible, however, that redundancy or compensatory mechanisms masked any phenotype after constitutive Jarid1b deletion.
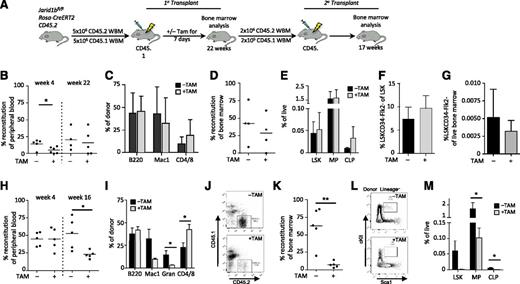
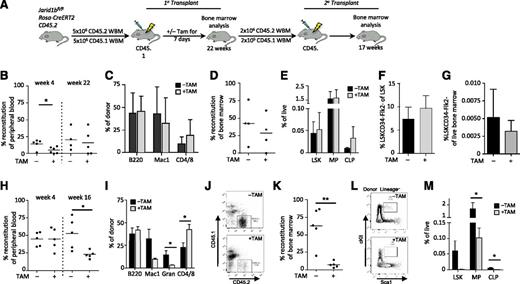
Self-renewal phenotypes as a result of genetic deletion of Jarid1b may have been masked in the constitutive system. To more stringently explore stem cell phenotypes, we competitively transplanted BM from Jarid1bfl/flRosaCreERT211 mice to allow for inducible deletion of Jarid1b through in vivo tamoxifen treatment (Figure 2A). Deletion of Jarid1b was confirmed through quantitative genomic PCR (supplemental Figure 6A-C). Jarid1b KO BM (+TAM; Figure 2B) showed significantly decreased engraftment by 4 weeks compared with the untreated control (−TAM; Figure 2B); however, by 22 weeks, no difference in the PB engraftment or lineage distribution of Jarid1b KO BM (+TAM) or control (−TAM) BM was observed (Figure 2B-C). Consistent with this finding, BM analysis at 24 weeks showed no change in the level of donor reconstitution (Figure 2D) or in the BM progenitor and HSC compartments as a result of Jarid1b deletion (Figure 2E-G). To assess the effect of Jarid1b deletion on HSC self-renewal potential, secondary transplants were performed (Figure 2A). PB analysis at 4 weeks showed no difference in short-term engraftment of Jarid1b KO BM compared to the untreated BM; however, by 16 weeks, Jarid1b KO BM showed significantly decreased donor PB chimerism (Figure 2H). PB lineage analysis at 16 weeks revealed a significant increase in CD4/8+ T cells and a significant decrease in granulocytes (Figure 2I). BM analysis of the secondary graft revealed a significant 8.4-fold decrease in donor chimerism and a significant corresponding depletion of BM progenitor compartments, including LSK, MPs, and CLPs, as a result of Jarid1b deletion (Figure 2J-M). Further analysis of the MP compartment revealed that CMPs, but not GMPs or MEPs, were significantly reduced as a result of Jarid1b deletion (supplemental Figure 7). Although loss of Jarid1b increased T cells and decreased myeloid cells within the PB, loss of Jarid1b did not appear to induce lineage bias within the BM (Figure 2I, M). It should be noted that BM from wild-type mice was not used to control for the effects of tamoxifen within these experiments; thus, it is possible that some of these effects may have resulted from tamoxifen exposure. This caveat aside, our results suggest that Jarid1b may also regulate hematopoietic differentiation in addition to regulating HSC self-renewal; however, whether Jarid1b regulates myeloid or lymphoid differentiation will require further investigation.
Jarid1b deletion compromises HSC potential. (A) Experimental design for transplantation. Five million BM cells isolated from Jarid1bfl/flRosaCreERT2 mice were competitively transplanted (donor-to-recipient ratio of 1:1) into recipient mice, followed by exposure to tamoxifen (+TAM) or vehicle (−TAM) for 7 days to induce Jarid1b deletion from the donor BM. Primary transplant analysis of PB donor reconstitution at 4 and 22 weeks; *P = .0346, unpaired Student t test (B); PB donor lineage distribution at week 22 (C); BM donor reconstitution at week 24 (D); and frequency of LSK, MP, and CLP (Lin–Flk2+IL7Rα+cKitmidScalmid) populations within BM (E). Frequency of HSCs (LSKCD34−Flk2−) within the LSK compartment (F) and within the live BM (G). (H-M) Secondary transplant analysis. Using FACS, 2 million donor cells were isolated from those primary transplants showing chimerism >10% and transplanted competitively (donor-to-recipient ratio of 10:1) into secondary recipients. (H) PB donor reconstitution at 4 and 16 weeks. *P = .0205; unpaired Student t test. (I) PB donor lineage distribution at week 16. CD4/CD8: *P = .0277; Gran (granulocytes): *P = .047. (J) Representative flow cytometry plots showing donor (CD45.2) vs recipient (CD45.1) BM from mice treated with (+) or without (−) TAM. (K) BM donor reconstitution at week 17. **P = .0019; unpaired Student t test. (L) Representative flow plots of LSK gates in mice transplanted with BM treated with (+) or without (−) TAM. (M) Frequency of LSK, MP, and CLP populations within BM. MP: *P = .0104; CLP: *P = .0182; unpaired Student t test. Error bars ± standard deviation.
Jarid1b deletion compromises HSC potential. (A) Experimental design for transplantation. Five million BM cells isolated from Jarid1bfl/flRosaCreERT2 mice were competitively transplanted (donor-to-recipient ratio of 1:1) into recipient mice, followed by exposure to tamoxifen (+TAM) or vehicle (−TAM) for 7 days to induce Jarid1b deletion from the donor BM. Primary transplant analysis of PB donor reconstitution at 4 and 22 weeks; *P = .0346, unpaired Student t test (B); PB donor lineage distribution at week 22 (C); BM donor reconstitution at week 24 (D); and frequency of LSK, MP, and CLP (Lin–Flk2+IL7Rα+cKitmidScalmid) populations within BM (E). Frequency of HSCs (LSKCD34−Flk2−) within the LSK compartment (F) and within the live BM (G). (H-M) Secondary transplant analysis. Using FACS, 2 million donor cells were isolated from those primary transplants showing chimerism >10% and transplanted competitively (donor-to-recipient ratio of 10:1) into secondary recipients. (H) PB donor reconstitution at 4 and 16 weeks. *P = .0205; unpaired Student t test. (I) PB donor lineage distribution at week 16. CD4/CD8: *P = .0277; Gran (granulocytes): *P = .047. (J) Representative flow cytometry plots showing donor (CD45.2) vs recipient (CD45.1) BM from mice treated with (+) or without (−) TAM. (K) BM donor reconstitution at week 17. **P = .0019; unpaired Student t test. (L) Representative flow plots of LSK gates in mice transplanted with BM treated with (+) or without (−) TAM. (M) Frequency of LSK, MP, and CLP populations within BM. MP: *P = .0104; CLP: *P = .0182; unpaired Student t test. Error bars ± standard deviation.
Our results contrast with a recent report that used short hairpin RNA knockdown to study the role of Jarid1b in primitive hematopoietic cells.16 Cellot et al3 suggested that Jarid1b acts as a negative regulator of HSC self-renewal; however, our results using formal genetics strongly suggest that Jarid1b is required for HSC self-renewal. Our finding is consistent with studies showing the importance of epigenetic regulation to HSC potential.4 It will be interesting to investigate the functional role of JARID1B in human HSCs and within the leukemic stem cell and non–stem cell compartments of AML; overexpression of JARID1B in AML subtypes suggests that deregulation of JARID1B may contribute to hematopoietic malignancy. Development of specific small molecule inhibitors of JARID1B would allow for further functional determination of the role of JARID1B in human hematopoiesis and leukemia and may lead to novel therapeutic modalities.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors sincerely thank A. Zguro and A. Sankar for animal husbandry and technical assistance, as well as all members of the T.E., K.H., and D.J.R. laboratories.
This work was supported by junior research fellowships from Kay Kendall Leukaemia Fund and Merton College Oxford (M.H.S.), an EMBO long-term postdoctoral fellowship (M.A.), a grant from the Danish Medical Research Council (K.H.), and program grants from Leukaemia and Lymphoma Research and Cancer Research UK (T.E.).
Authorship
Contribution: M.H.S. designed and performed transplant experiments and wrote the manuscript; M.A. generated and characterized Jarid1b−/− and Jarid1b+/− mice and provided insightful discussion; P.S., V.A.C., and K.H. provided Jarid1bfl/flRosaCreERT2 BM and insightful discussion; Y.G. provided experimental support; D.J.R. provided discussion; and T.E. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for Mareike Albert is Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany.
Correspondence: Tariq Enver, Stem Cell Group, UCL Cancer Institute, University College London, 72 Huntley St, Paul O’Gorman Building, London WC1E 6BT, United Kingdom; e-mail: t.enver@ucl.ac.uk.