Abstract
We have previously reported that hypermethylationof the GADD45A promoter (GADD45AmeHI) occurs frequently in AML at a specific CpG residue (CpG1) and associates with poor overall survival for patients on standard chemotherapy (Perugini et al, Leukemia 2013). Sequenom multiplex analysis of 195 AML patients revealed a co-occurrence of GADD45AmeHI with recurrent mutations at conserved residues in IDH1 and IDH2 (p<0.0001, Fisher's exact test). These mutations in IDH1 and IDH2 result in enzyme isoforms that produce high levels of the onco-metabolite 2-hydroxyglutarate with a wide-range of effects including inhibition of α-KG-dependent dioxygenases and association with a profound DNA hypermethylation phenotype in AML (Figueroa et al, Cancer Cell 2010). Furthermore these mutations are found in pre-leukemic AML clones (Shlush et al, Nature 2014) and lead to pre-leukaemic phenotypes in mouse models (Sasaki et al, Nature 2012, Kats et al, Cell Stem Cell 2014, Ogawara et al, Cancer Research 2015). Here we investigated the relationship between hypermethylation at GADD45A CpG1, IDH1/2 mutation status, global methylation patterns and patient survival.
We performed survival analysis to determine disease-free survival (DFS) and relapse-free survival (RFS) for AML patients with GADD45AmeHIor IDH1/2-mutations. This showed that GADD45AmeHI is a significant independent predictor of poor DFS and RFS, particularly in normal karyotype AML (Cox regression analysis, NK-AML DFS, P=0.009 HR=2.55, RFS, P=0.003 HR=2.75). Despite the co-association of GADD45AmeHI with mutations in IDH1 and IDH2, the mutation status of IDH1/2 did not predict DFS or RFS in these patients. To examine further the relationship between GADD45AmeHI and IDH1/2-mutation, and to investigate how this might influence tumour cell biology in AML, we determined global methylation patterns for a panel of AML diagnosis (Dx) samples (base-pair-resolution analysis using enhanced reduced representation bisulfite sequencing; ERRBS) in which both GADD45AmeHI and IDH mutation status has been determined. Unsupervised analyses of global methylation patterns grouped the AML Dx samples into three clusters including cluster 1 (n=12) which was associated with GADD45AmeHI samples with IDH- mutations, cluster 2 (n=13) which was enriched for GADD45AmeHIlacking IDH- mutations, and cluster 3 (n=9) which was associated with GADD45AmeLO(low CpG1 methylation) IDH-WT AML. We propose that this CpG in the GADD45A promoter may be subject to alternative events affecting DNA methylation in AML pathogenesis, including events distinct from IDH1/2 mutation. Finally, in GADD45AmeHI AML we detected hypermethylated regions compared to CD34+ normal bone marrow controls within 2016 gene promoters, 848 of which were unique to the GADD45AmeHI samples and not present in IDH1/2-mutant samples. We hypothesize that these differentially methylated genes may contribute mechanistically to the poor survival observed for this subtype.
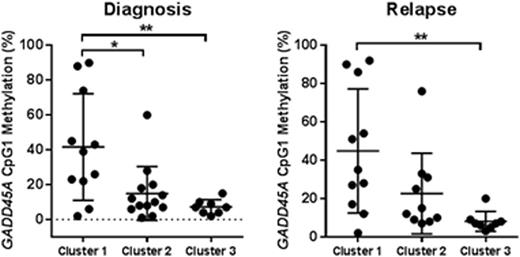
To determine how GADD45AmeHI status might associate with disease progression, DNA methylation assessment was performed on the patient panel-matched relapse samples (Rx). While GADD45AmeHI occurs frequently in both cluster 1 and 2 there is a significant difference in level of GADD45A CpG1 methylation at Dx and Rx for samples in cluster 1 vs cluster 2 and 3 (Figure 1), consistent with mutant IDH1/2 activity influencing methylation levels at this CpG site. Given that GADD45A has an established basal role in the maintenance of genomic stability (Liebermann & Hoffman, Springer 2013), and is a determinant of HSC self-renewal and response to genotoxic insult (Wingert et al, Stem Cells 2016, Chen et al, Blood 2014) we are also investigating whether GADD45A methylation and silencing plays a direct role in determining aggressiveness and response to chemotherapy for GADD45AmeHIAML.
In conclusion this data suggests that methylation at this specific CpGof the GADD45A promoter, in combination with IDH1/2 mutation status, associate with varying global methylation phenotypes. Importantly, we demonstrate that GADD45AmeHI better predicts poorer prognosis than IDH1/2 mutation status, despite the significant co-association of these characteristics in AML.
SES and FEGB contributed equally to this work.
GADD45A CpG1 methylation in patient cluster 1-3 at diagnosis and relapse. * P<0.05, ** P<0.01.
GADD45A CpG1 methylation in patient cluster 1-3 at diagnosis and relapse. * P<0.05, ** P<0.01.
Guzman:Cellectis: Research Funding. Roboz:Agios, Amgen, Amphivena, Astex, AstraZeneca, Boehringer Ingelheim, Celator, Celgene, Genoptix, Janssen, Juno, MEI Pharma, MedImmune, Novartis, Onconova, Pfizer, Roche/Genentech, Sunesis, Teva: Consultancy; Cellectis: Research Funding. Levine:Qiagen: Membership on an entity's Board of Directors or advisory committees; Novartis: Consultancy. Melnick:Janssen: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.