Key Points
Autosomal-dominant SAMD9L gain-of-function mutations predispose to myeloid malignancies involving chromosome 7 aberrations.
Hematopoietic reversions frequently occur postnatally and are associated with milder disease manifestations.
Abstract
Several monogenic causes of familial myelodysplastic syndrome (MDS) have recently been identified. We studied 2 families with cytopenia, predisposition to MDS with chromosome 7 aberrations, immunodeficiency, and progressive cerebellar dysfunction. Genetic studies uncovered heterozygous missense mutations in SAMD9L, a tumor suppressor gene located on chromosome arm 7q. Consistent with a gain-of-function effect, ectopic expression of the 2 identified SAMD9L mutants decreased cell proliferation relative to wild-type protein. Of the 10 individuals identified who were heterozygous for either SAMD9L mutation, 3 developed MDS upon loss of the mutated SAMD9L allele following intracellular infections associated with myeloid, B-, and natural killer (NK)–cell deficiency. Five other individuals, 3 with spontaneously resolved cytopenic episodes in infancy, harbored hematopoietic revertant mosaicism by uniparental disomy of 7q, with loss of the mutated allele or additional in cisSAMD9L truncating mutations. Examination of 1 individual indicated that somatic reversions were postnatally selected. Somatic mutations were tracked to CD34+ hematopoietic progenitor cell populations, being further enriched in B and NK cells. Stimulation of these cell types with interferon (IFN)-α or IFN-γ induced SAMD9L expression. Clinically, revertant mosaicism was associated with milder disease, yet neurological manifestations persisted in 3 individuals. Two carriers also harbored a rare, in trans germ line SAMD9L missense loss-of-function variant, potentially counteracting the SAMD9L mutation. Our results demonstrate that gain-of-function mutations in the tumor suppressor SAMD9L cause cytopenia, immunodeficiency, variable neurological presentation, and predisposition to MDS with −7/del(7q), whereas hematopoietic revertant mosaicism commonly ameliorated clinical manifestations. The findings suggest a role for SAMD9L in regulating IFN-driven, demand-adapted hematopoiesis.
Introduction
Myelodysplastic syndrome (MDS) is a heterogeneous group of clonal hematopoietic stem and progenitor cell (HSPC) disorders that are characterized by impaired hematopoiesis, which may progress to acute myeloid leukemia (AML).1,2 The risk of developing MDS increases with age, and MDS represents one of the most common cancers of the elderly. Acquired hematopoietic stem cell (HSC) cytogenetic abnormalities are one of the main risk factors.3,4 In adults, del(5q) represents the most common cytogenetic aberration, followed by the complete or partial loss of chromosome 7, or −7/del(7q).5,6 Pediatric MDS is rare, accounting for only 9% of hematological malignancies, and is most frequently associated with monosomy 7 (−7).7,8
Pediatric MDS is often associated with inherited bone marrow failure syndromes caused by mutations in genes required for DNA repair, chromosomal stability, and telomere elongation, which result in an increased risk of acquiring somatic mutations. For instance, 10% to 30% of patients with Fanconi anemia or telomeropathy develop MDS/AML.9-12 Furthermore, several rare, monogenic causes of familial MDS/AML have recently been uncovered, including autosomal-dominant CEBPA, DDX41, RUNX1, ANKRD26, ETV6, SRP72, and GATA2 mutations.13-19 These genetic conditions have common characteristics such as cytopenia and immunodeficiency, yet differ in their clinical manifestations, age at diagnosis of MDS, and associated somatic tumor aberrations. Interestingly, germ line heterozygous GATA2 mutations are associated with a wide spectrum of clinical manifestations, including immunodeficiency, lymphedema, and bone marrow failure, as well as MDS/AML.20,21 Following HSPC attrition, patients develop monocyte, dendritic, and B- and natural killer (NK)-cell deficiencies. Alveolar proteinosis and elevated Fms-like tyrosine kinase 3 ligand (FLT-3L) levels are also indicative of GATA-2 haploinsufficiency.21 In GATA-2 haploinsufficiency–associated MDS, −7 and trisomy 8 represent recurrent cytogenetic aberrations.20 In children, germ line GATA2 mutations explain 6% of primary MDS cases, but up to 37% of −7 cases.22 Notably, −7 is associated with a high risk of progression to AML.23
The incidence of hereditary MDS may be underestimated.24 Notably, genetic causes of syndromes involving chromosome 7 aberrations remain poorly characterized. Ataxia-pancytopenia syndrome (ATXPC; Mendelian Inheritance in Man [MIM] no. 159550) is an autosomal-dominant disorder that is associated with prominent neurological features, including ataxia and nystagmus, as well as hematologic cytopenias and predisposition to myeloid leukemia involving −7/del(7q).25 Myelodysplasia and leukemia syndrome with −7 (MIM no. 252270) is defined by at least 2 siblings presenting with MDS/AML with −7.26 These syndromes suggest that additional genetic predispositions to MDS exist.
Here we report clinical, genetic, and functional investigations in 2 families with early-onset MDS −7/del(7q), identifying heterozygous germ line gain-of-function mutations in SAMD9L and a variable degree of hematological and neurological symptoms. We also uncovered carriers who displayed distinct hematopoietic revertant mosaicism associated with milder clinical presentation.
Methods
Subjects and samples
This study was approved by the ethic committees of the participating institutions. Informed consents from the individuals included in the study were obtained according to the Declaration of Helsinki. DNA was extracted by standard procedure from peripheral blood, skin fibroblasts, buccal swabs, or sorted blood cell populations. For F1:III-2, DNA was extracted from neonatal screening–derived dried blood spot (Guthrie card) samples. Medical files were reviewed to collect clinical and immunological data.
Sequencing, dPCR, and SNP arrays
Whole-exome sequencing (WES) was performed on peripheral blood DNA from individuals I-4, II-2, II-4, III-1, and III-2 from family 1 (exome capture with Agilent SureSelect [version 5; Agilent Genomics] and sequencing on Illumina HiSeq2000 [Illumina]). The sequence was mapped to the human genome build GRCh37. Detailed information on WES, Sanger sequencing, and digital polymerase chain reaction (dPCR) is provided in the supplemental Methods, available on the Blood Web site. To determine somatic loss of heterozygosity (LOH), DNA was analyzed using single nucleotide polymorphism (SNP) arrays (Infinium OmniExpressExome; Illumina), as detailed in the supplemental Methods.
Flow cytometry and immunological investigations
Absolute lymphocyte counts and assays of cytotoxic lymphocyte phenotype and function were performed according to standard procedures, as described in the supplemental Methods.
To investigate allelic frequency of mutant SAMD9L in cell subsets, cells were sorted by flow cytometry, as detailed in the supplemental Methods. HSCs, multipotent progenitor (MPP), granulocyte/macrophage progenitor, and common lymphoid progenitor (CLP) cells were sorted from bone marrow, whereas differentiated immune-cell subsets were sorted from peripheral blood mononuclear cells.
Functional assessment of SAMD9L variants
HEK 293FT cells were transiently transfected (Lipofectamine 2000; Thermo Fisher Scientific) with constructs encoding SAMD9L variants. For cell proliferation assays, cells were labeled for 20 minutes with 1 µM of CellTrace Violet (Thermo Fisher Scientific) in prewarmed phosphate-buffered saline, washed, and transfected after 3 hours. The cells were harvested 72 hours after transfection, fixed, and analyzed by flow cytometry (LSR Fortessa; BD Biosciences), as detailed in the supplemental Methods.
Results
Clinical history
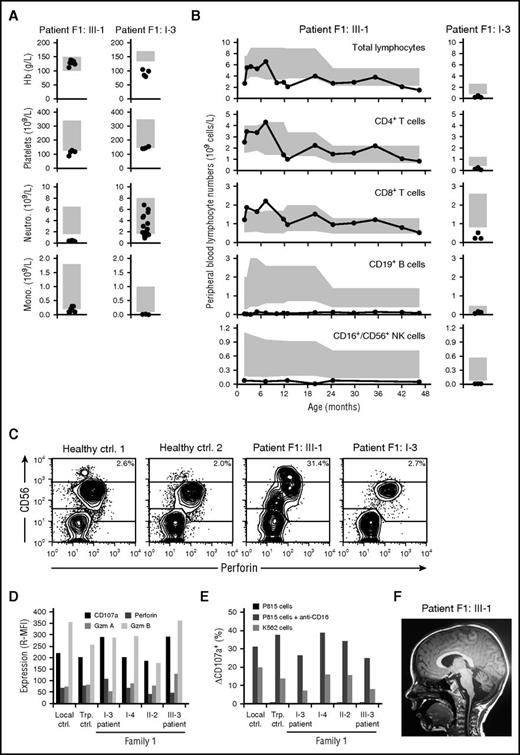
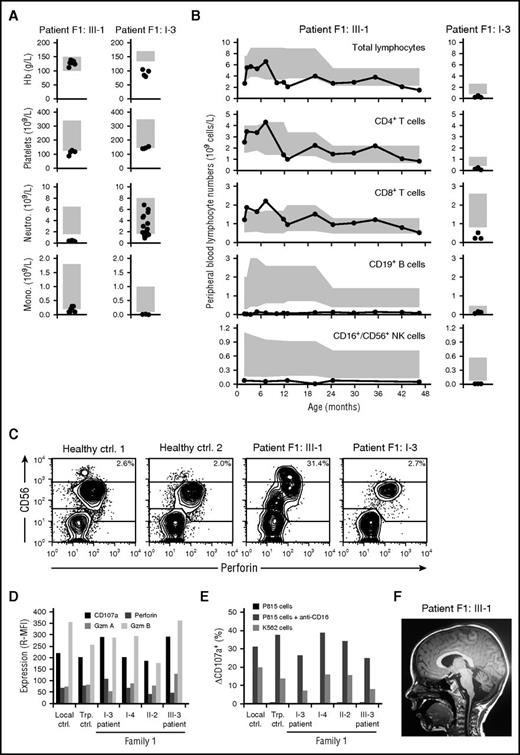
Family 1, of Swedish origin, is a 3-generation family with 2 patients with MDS (Table 1). The index case (F1:III-1) had a history of congenital cytomegalovirus infection and trilineage cytopenia (Figure 1A) and was diagnosed with MDS −7 at the age of 4 years. Cellular immunodeficiency with decreased peripheral blood B and NK cell numbers and borderline monocyte numbers was consistently observed prior to the diagnosis of MDS (Figure 1A-B). Immunoglobulin (Ig) levels were also low (IgG, 3.36 g/L; IgA, 0.37 g/L; IgM, 0.49 g/L). Failure to control cytomegalovirus suggested an NK-cell deficiency, yet CD56bright to CD56dim NK-cell differentiation as well as killing capacity appeared normal in the patient (Figure 1C-E), arguing against known primary immunodeficiencies associated with NK-cell dysfunction.27,28 Upon diagnosis of MDS, the patient underwent a matched unrelated HSC transplantation (HSCT) at the age of 4.5 years with myeloablative conditioning, according to the protocol of the European Working Group of MDS and JMML in Childhood. Following HSCT, neurological symptoms were noted (supplemental Table 1). Magnetic resonance imaging of the brain showed pronounced bilateral white substance changes and progressive cerebellar degeneration (Figure 1F).
Cellular deficiencies and neurological manifestations in family 1. (A) Peripheral blood hemoglobin (Hb), platelet, neutrophil (Neutro.), and monocyte (Mono.) counts as well as (B) lymphocyte subset counts in F1:III-1 and F1:I-3, who subsequently were diagnosed with childhood and adult MDS, respectively. Each patient was repeatedly assessed, with individual measurements denoted by circles. Gray boxes depict age-related reference values. (C) Plots show perforin versus CD56 expression among gated CD3− lymphocytes, as assessed by flow cytometry. (D) Bar graph depicts the relative median fluorescence intensity (R-MFI) of CD107a, perforin, granzyme A (Gzm A), and granzyme B (Gzm B), as indicated, in gated CD3−CD56dim NK cells. (E) Bar graph depicts the frequency of exocytosing CD3−CD56dim NK cells, as evaluated by induction of surface CD107a expression (ΔCD107a), by target cells, as indicated. (F) Sagittal magnetic resonance image of F1:III-1 revealing bilateral white substance changes and cerebellar degeneration. Ctrl, control; Trp, transport control.
Cellular deficiencies and neurological manifestations in family 1. (A) Peripheral blood hemoglobin (Hb), platelet, neutrophil (Neutro.), and monocyte (Mono.) counts as well as (B) lymphocyte subset counts in F1:III-1 and F1:I-3, who subsequently were diagnosed with childhood and adult MDS, respectively. Each patient was repeatedly assessed, with individual measurements denoted by circles. Gray boxes depict age-related reference values. (C) Plots show perforin versus CD56 expression among gated CD3− lymphocytes, as assessed by flow cytometry. (D) Bar graph depicts the relative median fluorescence intensity (R-MFI) of CD107a, perforin, granzyme A (Gzm A), and granzyme B (Gzm B), as indicated, in gated CD3−CD56dim NK cells. (E) Bar graph depicts the frequency of exocytosing CD3−CD56dim NK cells, as evaluated by induction of surface CD107a expression (ΔCD107a), by target cells, as indicated. (F) Sagittal magnetic resonance image of F1:III-1 revealing bilateral white substance changes and cerebellar degeneration. Ctrl, control; Trp, transport control.
The grandfather of the index patient, F1:I-3, presented at the age of 54 years with a severe Legionella infection, cytopenia, and mild dysplastic megakaryocytopoiesis in the bone marrow. Two years later, the bone marrow was classified as MDS. Peripheral blood NK cell and monocyte numbers were consistently low (Figure 1B). The patient also suffered from alveolar proteinosis and died as a result of respiratory failure at the age of 58 years. A very mild balance impairment was noted. His brother, F1:I-2, developed balance impairment at 15 years and cytopenia at 61 years of age. The younger sister (F1:III-2) of the index patient and 1 aunt (F1:II-4) developed thrombocytopenia and trilineage cytopenia, respectively, in infancy, but both spontaneously recovered. The mother of the index patient (F1:II-2) has a very mild balance impairment, whereas the other aunt (F1:II-1) is healthy, although with somewhat low monocyte numbers.
Family 2, of Finnish origin, is a 2-generation family whose clinical phenotype is consistent with ATXPC (Table 1; supplemental Table 1).25 The index patient (F2:II-4) initially presented at 6 weeks of age with transient thrombocytopenia after bronchiolitis, experienced severe varicella infection, and developed thrombocytopenia and neutropenia at 13 months of age. The patient was diagnosed with MDS −7 at the age of 18 months and underwent an HSCT at the age of 2 years. The patient gradually developed neurological symptoms, and at the age of 8 years, displayed nystagmus, balance problems, stiffness, and brisk reflexes of the lower limbs, as well as attention deficit hyperactivity disorder. The patient's growth and development are normal. An older brother, F2:II-1, was diagnosed with severe aplastic anemia at 14 months of age and recovered spontaneously after 2 months. Both brothers displayed recurrent otitis. F2:II-1 also suffers neurologic problems. The mother, F2:I-2, has nystagmus, memory problems, and muscle weakness. A brain MRI at the age of 32 years showed cerebellar atrophy and signal intensities around the posterior part of the lateral ventricles.
In summary, members of both families presented with cytopenias in combination with immunodeficiency, neurological symptoms, and susceptibility to MDS −7/del(7q), suggesting an underlying genetic syndrome.
Identification of heterozygous SAMD9L mutations
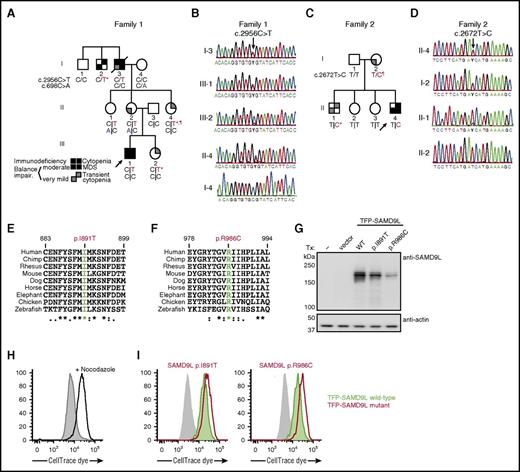
Genetic investigations were first performed on family 1. In light of herpes virus infection, cellular deficiencies, and alveolar proteinosis, GATA-2 haploinsufficiency (MIM no. 614172) was considered. However, contrasting GATA-2 haploinsufficiency, in which CD3–CD56bright NK cells are lost,29 adaptive NK cells predominate,30,31 and FLT-3L levels are very high,21 F1:III-1 displayed, if anything, increased CD3–CD56bright NK cells, whereas canonical CD3–CD56dim NK cells dominated, FLT-3L levels were only moderately elevated (supplemental Figure 1), and no mutations were identified by targeted sequencing of GATA2 exons and conserved intronic elements in F1:I-3. Moreover, both alleles of GATA2 were equally transcribed (data not shown). Therefore, another inherited disorder was suspected.
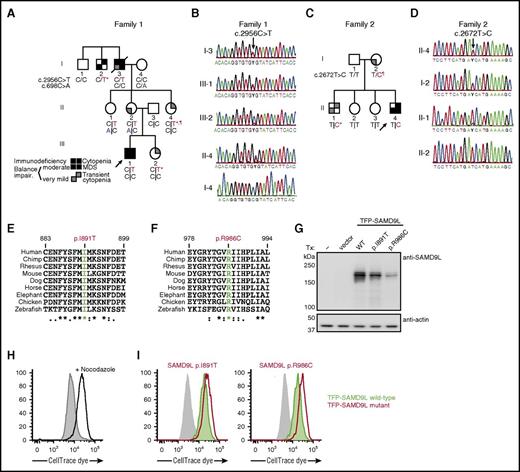
Aiming at a molecular diagnosis in this family, WES of DNA samples from F1:I-3, F1:I-4, F1:II-2, F1:III-1, and F1:III-2 were analyzed. Rare coding variants present in the affected individuals fitting an X-linked recessive or autosomal-dominant inheritance model were considered. Located on 7q21.2, a heterozygous missense variant c.2956C>T (p.Arg986Cys) in SAMD9L (transcript ENST00000318238) was detected (Figure 2A). The variant, predicted to be damaging, was not listed in the Exome Aggregation Consortium (ExAC) database.32 The 2 affected individuals, F1:III-1 and F1:I-3, as well as F1:II-4, who experienced transient cytopenia in infancy, were heterozygous carriers of this variant, as confirmed by Sanger sequencing (Figure 2B). In F1:III-2, who also experienced transient cytopenia in infancy, the SAMD9L c.2956C>T variant was identified in only 53 of 434 (12%) exome sequencing reads from peripheral blood–derived DNA, and was underrepresented in Sanger traces (Figure 2B). Two mildly affected or unaffected individuals, F1:II-1 and F1:II-2, were also carriers of the SAMD9L c.2956C>T variant. Notably, these 2 individuals inherited a rare SAMD9L c.698C>A (p.Thr233Asn) missense variant from F1:I-4, reported in 0.00003% of individuals in the ExAC database (Figure 2A; supplemental Figure 2).32
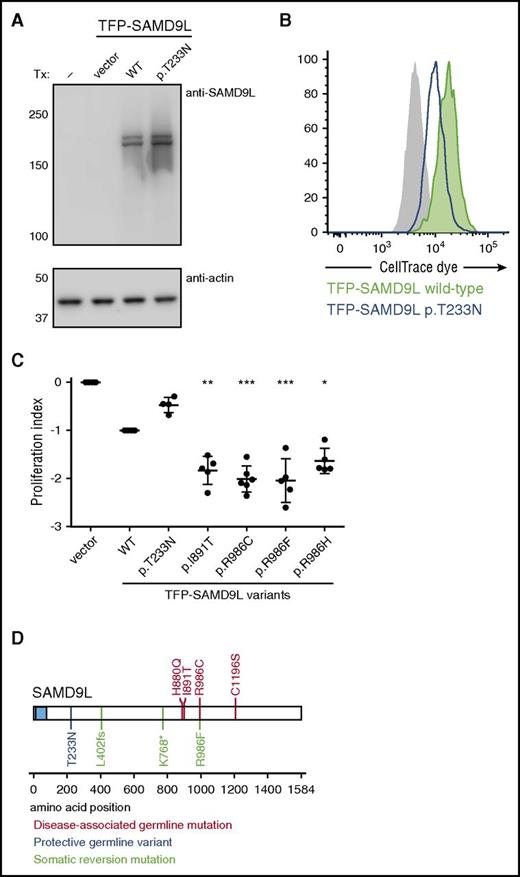
Heterozygous SAMD9L gain-of-function mutations associated with cytopenia, susceptibility to MDS with chromosome 7 aberrations, immunodeficiency, and ataxia. (A) Pedigree of family 1. Segregation of the SAMD9L c.2956C>T mutation and the rare SAMD9L c.698C>A variant is shown. Filled quadrants depict the clinical manifestations, as illustrated in the legend. Individuals with somatic in vivo reversion are indicated by asterisk (*) showing UPD(7q), or paragraph symbol (¶) showing second-site mutation. Genotypes are indicated with a vertical line (|) or forward slash (/), depending on whether phase information is known or unknown, respectively. (B) Sanger traces from family 1 for the SAMD9L c.2956C>T mutation (arrow). Individuals I-3, III-1, III-2, II-4, I-4 are shown. (C) Pedigree of family 2. Segregation of the SAMD9L c.2672T>C mutation is shown. Individuals with somatic in vivo reversion are indicated by asterisk (*) showing UPD(7q), or paragraph symbol (¶), showing second-site mutation. (D) Sanger traces from family 2 for the c.2672T>C mutation (arrow). Individuals II-4, I-2, II-1, and II-2 are shown. (E-F) Multispecies evolutionary conservation of the amino acid residues (E) Ile891 and (F) Arg986 mutated in family 2 and family 1, respectively. The sequence alignment was performed with Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). The asterisk (*) indicates positions which have a single, fully conserved residue; the colon (:) and period (.) indicate conservation between groups of strongly similar properties, scoring >0.5 in the Gonnet PAM 250 matrix and ≤0.5 in the Gonnet PAM 250 matrix, respectively. (G-I) 293FT cells were transiently transfected (Tx) with TFP-SAMD9L wild-type (WT) or patient-derived mutants, as indicated. (G) Western blots of recombinant SAMD9L variant expression, as indicated. (H-I) Cell proliferation in 293FT cells assessed by dye dilution assays. CellTrace (Thermo Fisher Scientific) dye-labeled 293FT cells were transfected with TFP or the TFP-SAMDL variants indicated, cultured for 72 hours, and analyzed by flow cytometry. (H) Effect of proliferation/microtube inhibitor nocodazole (10 µg/mL) on 293FT cell proliferation. (I) Dye dilution assays in TFP-SAMD9L–transfected 293FT cells. Dye levels were monitored in TFP− cells (filled gray histograms) and compared with cells expressing uniformly intermediate levels of TFP-SAMD9L variants, as indicated. (G-I) Single, representative experiments of 5 experiments are shown. Impair, impairment.
Heterozygous SAMD9L gain-of-function mutations associated with cytopenia, susceptibility to MDS with chromosome 7 aberrations, immunodeficiency, and ataxia. (A) Pedigree of family 1. Segregation of the SAMD9L c.2956C>T mutation and the rare SAMD9L c.698C>A variant is shown. Filled quadrants depict the clinical manifestations, as illustrated in the legend. Individuals with somatic in vivo reversion are indicated by asterisk (*) showing UPD(7q), or paragraph symbol (¶) showing second-site mutation. Genotypes are indicated with a vertical line (|) or forward slash (/), depending on whether phase information is known or unknown, respectively. (B) Sanger traces from family 1 for the SAMD9L c.2956C>T mutation (arrow). Individuals I-3, III-1, III-2, II-4, I-4 are shown. (C) Pedigree of family 2. Segregation of the SAMD9L c.2672T>C mutation is shown. Individuals with somatic in vivo reversion are indicated by asterisk (*) showing UPD(7q), or paragraph symbol (¶), showing second-site mutation. (D) Sanger traces from family 2 for the c.2672T>C mutation (arrow). Individuals II-4, I-2, II-1, and II-2 are shown. (E-F) Multispecies evolutionary conservation of the amino acid residues (E) Ile891 and (F) Arg986 mutated in family 2 and family 1, respectively. The sequence alignment was performed with Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). The asterisk (*) indicates positions which have a single, fully conserved residue; the colon (:) and period (.) indicate conservation between groups of strongly similar properties, scoring >0.5 in the Gonnet PAM 250 matrix and ≤0.5 in the Gonnet PAM 250 matrix, respectively. (G-I) 293FT cells were transiently transfected (Tx) with TFP-SAMD9L wild-type (WT) or patient-derived mutants, as indicated. (G) Western blots of recombinant SAMD9L variant expression, as indicated. (H-I) Cell proliferation in 293FT cells assessed by dye dilution assays. CellTrace (Thermo Fisher Scientific) dye-labeled 293FT cells were transfected with TFP or the TFP-SAMDL variants indicated, cultured for 72 hours, and analyzed by flow cytometry. (H) Effect of proliferation/microtube inhibitor nocodazole (10 µg/mL) on 293FT cell proliferation. (I) Dye dilution assays in TFP-SAMD9L–transfected 293FT cells. Dye levels were monitored in TFP− cells (filled gray histograms) and compared with cells expressing uniformly intermediate levels of TFP-SAMD9L variants, as indicated. (G-I) Single, representative experiments of 5 experiments are shown. Impair, impairment.
In light of clinical similarities to family 1, the coding regions of SAMD9L were Sanger sequenced in family 2 (Figure 2C). In the index case (F2:II-4), a novel heterozygous SAMD9L c.2672T>C (p.Ile891Thr) missense variant, predicted to be damaging, was identified. Segregation studies identified this variant in the affected mother (F2:I-2) and brother (F2:II-1). In F2:II-1, underrepresentation of the variant in Sanger traces was noted (Figure 2D).
Both SAMD9L c.2672T>C and c.2956C>T variants affect evolutionarily conserved amino acid residues (Figure 2E-F), and the gene is located in a region of chromosome 7 that is commonly deleted in myeloid malignancies.33 In mice, Samd9l deficiency causes development of MDS with age, implying that SAMD9L is a tumor suppressor.34 Thus, the identified SAMD9L variants potentially represented autosomal-dominant, disease-causing mutations.
SAMD9L p.Ile891Thr and p.Arg986Cys mutants decrease cell proliferation
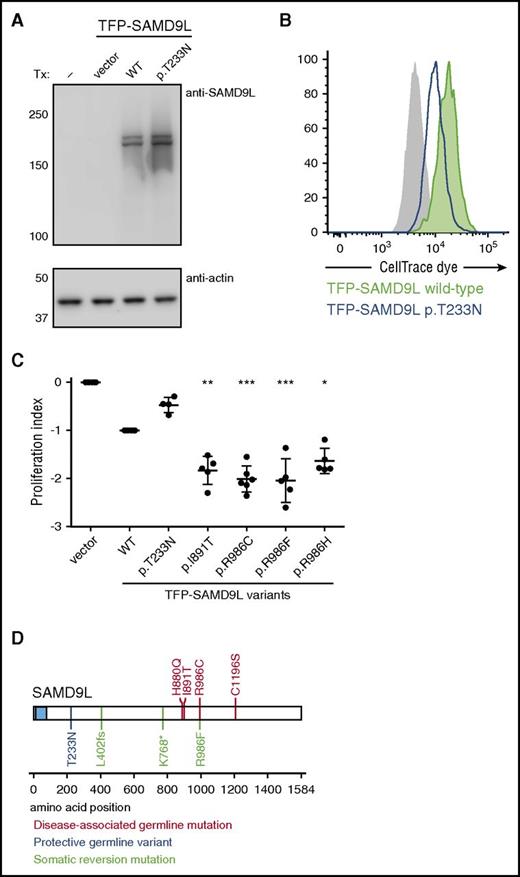
Other than a report demonstrating that murine Samd9l localizes to endosomes and inhibits growth factor signaling,34 little is known regarding the structure and function of SAMD9L. To evaluate the effect of patient-derived SAMD9L mutations on protein expression, wild-type or mutant N-terminal fluorescent-protein–tagged SAMD9L was transiently expressed in 293FT cells. Whereas endogenous SAMD9L was not detected, both wild-type and mutant teal fluorescent protein (TFP)–SAMD9L was expressed (Figure 2G). To determine the impact of patient-derived SAMD9L mutations on cell proliferation, cells were dye labeled and subsequently transfected with wild-type or mutant TFP-SAMD9L. Following transfection, cells were monitored by flow cytometry for TFP-SAMD9L expression as well as dye dilution, indicating cell division. Nocodazole, an inhibitor of microtubule polymerization that arrests the cell cycle, prevented dye dilution (Figure 2H). Ectopic expression of TFP-SAMD9L wild type impaired cellular proliferation, whereas expression of TFP-SAMD9L p.Ile891Thr or p.Arg986Cys mutants strikingly halted proliferation (Figure 2I). As such, the patient-derived SAMD9L p.Arg986Cys and p.Ile891Thr variants augment the growth-suppressing activity of SAMD9L and thus represent gain-of-function mutations.
Myelodysplastic cells lack the mutant SAMD9L allele
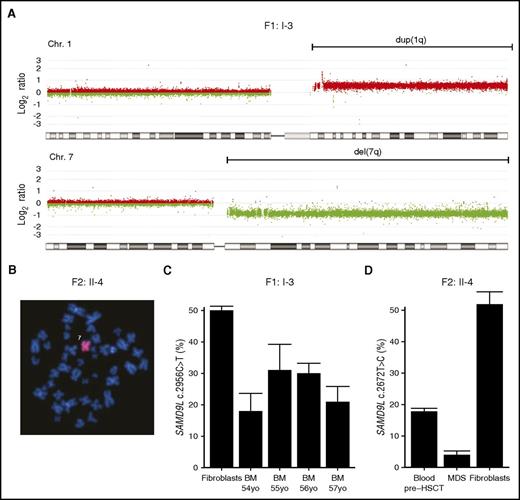
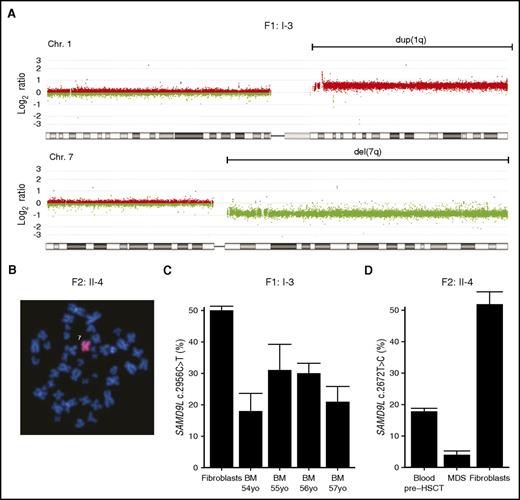
Given the strong proliferative inhibition by patient-derived SAMD9L mutations, the patients’ observed susceptibility to MDS might appear paradoxical. In all 3 patients who developed MDS, cytogenetic aberrations involved chromosome 7, with −7 in the 2 pediatric cases and der(1;7)(q10;p10) in F1:I-3 (Figure 3A-B), leading to a loss of the 7q arm. Mutation-specific dPCR assays demonstrated decreased frequencies of the SAMD9L mutations in DNA from the myelodysplastic bone marrow of F1:I-3 and F2:II-4, indicating that the loss of chromosome 7 material eliminated the SAMD9L mutations (Figure 3C-D). Seemingly, the SAMD9L mutations were not conducive to transformation and tumor cell propagation. Rather, cytogenetic events eradicating the allele carrying mutant SAMD9L and leading to −7/del(7q)/der(1;7)(q10;p10) may explain MDS predisposition.
Loss of the SAMD9L gain-of-function–mutated allele in myelodysplastic cells. (A) Microarray-based comparative genomic hybridization of bone marrow–derived DNA of F1:I-3 showing duplication of 1q (in red) and deletion of 7q (in green), consistent with the der(1;7)(q10;p10) finding by karyotype. (B) Fluorescence in situ hybridization analysis with chromosome 7 painting reveals monosomy 7 in bone marrow cells from F2:II-4. (C) Frequency of SAMD9L c.2956C>T mutation relative to wild type assessed by mutation-specific droplet-dPCR in fibroblasts and serial bone marrow samples of F1:I-3. (D) Frequency of SAMD9L c.2672T>C mutation relative to wild type assessed by mutation-specific dPCR in DNA derived from MDS, peripheral blood, and fibroblasts of F2:II-4. For each sample, the data displayed is the combination of at least 2 dPCR chips. Error bars show 95% confidence levels.
Loss of the SAMD9L gain-of-function–mutated allele in myelodysplastic cells. (A) Microarray-based comparative genomic hybridization of bone marrow–derived DNA of F1:I-3 showing duplication of 1q (in red) and deletion of 7q (in green), consistent with the der(1;7)(q10;p10) finding by karyotype. (B) Fluorescence in situ hybridization analysis with chromosome 7 painting reveals monosomy 7 in bone marrow cells from F2:II-4. (C) Frequency of SAMD9L c.2956C>T mutation relative to wild type assessed by mutation-specific droplet-dPCR in fibroblasts and serial bone marrow samples of F1:I-3. (D) Frequency of SAMD9L c.2672T>C mutation relative to wild type assessed by mutation-specific dPCR in DNA derived from MDS, peripheral blood, and fibroblasts of F2:II-4. For each sample, the data displayed is the combination of at least 2 dPCR chips. Error bars show 95% confidence levels.
Revertant mosaicism in healthy carriers of SAMD9L mutations
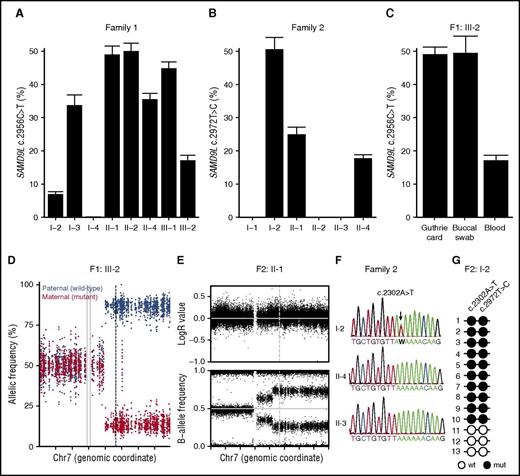
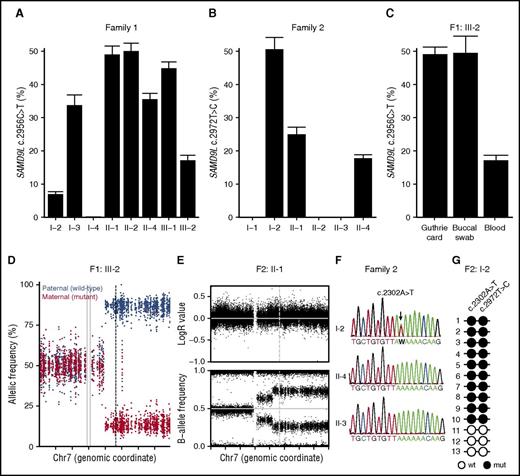
In addition to the development of MDS in 3 patients, clinical heterogeneity was observed among carriers of either SAMD9L mutation (Table 1). Prompted by findings in a few carriers that indicated underrepresentation of the SAMD9L c.2672T>C and c.2956C>T mutations compared with wild-type mutations, we accurately quantified frequencies of the mutated allele by mutation-specific dPCR assays on peripheral blood–derived DNA. In agreement with the WES data, the allele frequency of the SAMD9L c.2956C>T mutation in F1:III-2 was reduced to 17% (Figure 4A). A reduced mutant allele frequency was also detected in F1:I-2, F1:I-3, F1:II-4, and to a lesser extent in F1:III-1 (Figure 4A). Of note, although Sanger traces appeared to be wild type in F1:I-2, a variant allele frequency of ∼6% was detected by dPCR. In family 2, a reduced mutant allele frequency in F2:II-1 as well as the index patient was detected, confirming the Sanger traces (Figure 4B). The reduced allele frequency was therefore present in peripheral blood from individuals who either developed MDS or had a history of transient cytopenia in infancy or predominant neurologic problems.
Somatic revertant mosaicism of the SAMD9L mutations. (A-B) Quantification of the frequency of the (A) SAMD9L c.2956C>T in family 1 and (B) SAMD9L c.2672T>C in family 2 relative to the wild-type allele in peripheral blood–derived DNA, as assessed by mutation-specific dPCR assay. (C) Additional quantification of the frequency of the SAMD9L c.2956C>T in DNA from a Guthrie card dried blood spot and a buccal swab from F1:III-2. For each sample, values represent the mean of at least 2 chips. Error bars denote 95% confidence levels. (D) Allele frequency of phased heterozygous variants on chromosome 7 of F1:III-2, as calculated from the allelic read-depth from WES data. The variants are color-coded on the basis of the parental origin, as indicated. The gray vertical lines indicate the centromere of chromosome 7, and the dashed line indicates the position of the SAMD9L c.2956C>T mutation. (E) Log R ratio of intensity signal and B-allele frequency for SNP on chromosome 7 of F2:II-1, as determined by SNP array. The dashed vertical line indicates the position of the SAMD9L c.2972T>C mutation. (F) Sanger traces from members of family 2, as indicated, for the SAMD9L c.2302A>T nonsense mutation identified in F2:I-2 (arrow). (G) Schematic representation of the genotype of PCR-derived SAMD9L clones spanning the SAMD9L c.2302A>T and c.2972T>C mutations. Open circles represent wild-type (wt) nucleotide sequence, whereas filled circles represent mutated (mut) nucleotide sequence.
Somatic revertant mosaicism of the SAMD9L mutations. (A-B) Quantification of the frequency of the (A) SAMD9L c.2956C>T in family 1 and (B) SAMD9L c.2672T>C in family 2 relative to the wild-type allele in peripheral blood–derived DNA, as assessed by mutation-specific dPCR assay. (C) Additional quantification of the frequency of the SAMD9L c.2956C>T in DNA from a Guthrie card dried blood spot and a buccal swab from F1:III-2. For each sample, values represent the mean of at least 2 chips. Error bars denote 95% confidence levels. (D) Allele frequency of phased heterozygous variants on chromosome 7 of F1:III-2, as calculated from the allelic read-depth from WES data. The variants are color-coded on the basis of the parental origin, as indicated. The gray vertical lines indicate the centromere of chromosome 7, and the dashed line indicates the position of the SAMD9L c.2956C>T mutation. (E) Log R ratio of intensity signal and B-allele frequency for SNP on chromosome 7 of F2:II-1, as determined by SNP array. The dashed vertical line indicates the position of the SAMD9L c.2972T>C mutation. (F) Sanger traces from members of family 2, as indicated, for the SAMD9L c.2302A>T nonsense mutation identified in F2:I-2 (arrow). (G) Schematic representation of the genotype of PCR-derived SAMD9L clones spanning the SAMD9L c.2302A>T and c.2972T>C mutations. Open circles represent wild-type (wt) nucleotide sequence, whereas filled circles represent mutated (mut) nucleotide sequence.
The detection of an ∼50% SAMD9L c.2956C>T frequency in DNA from a buccal swab of F1:I-2 (not shown) as well as F1:III-2 (Figure 4C) suggested that the mosaicism specifically occurred in hematopoietic cells. Moreover, the detection of a bona fide germ line SAMD9L mutation in DNA extracted from the neonatal dried blood spot of F1:III-2 substantiated the hypothesis of postnatal somatic reversion (Figure 4C).
To understand mechanisms by which the reversion had occurred, we used WES data to evaluate the allelic frequencies of all phased heterozygous sites on chromosome 7 in F1:III-2. Analyses uncovered 7q LOH, including the SAMD9L locus (Figure 4D). Validated by SNP arrays (supplemental Figure 3A), findings established a mosaic copy-neutral (CN)–LOH caused by segmental uniparental disomy (UPD) of the paternal chromosome in ∼80% of blood cells. Similarly, 7q CN-LOH was identified in F1:I-2, F1:II-4, and F2:II-1, but not in F1:II-1 or F1:II-2 (Figure 4E; supplemental Figure 3B-C). In F1:I-2 and F2:II-1, the presence of 2 different levels of CN-LOH, with the overall highest level of LOH encompassing SAMD9L, suggested occurrence of at least 2 independent events of acquired UPD involving the same parental chromatid. Interestingly, UPD(7q) was also detected in an estimated 70% of peripheral blood cells of patient F2:II-4 (Figure 4B; supplemental Figure 3D), when −7 had already been detected in the bone marrow. Thus, somatic revertant mosaicism by mitotic recombination was recurrent in both families.
In F1:II-4, the mutation-specific dPCR revealed the presence of a double population with a distinct affinity for the mutation-binding probe (supplemental Figure 4A), suggesting a second-site mutation. Congruently, WES indicated a subclonal c.2957G>T variant, adjacent and in cis to the SAMD9L c.2956C>T mutation in 10% of sequencing reads, resulting in a subclone of cells with SAMD9L p.Arg986Phe as a result of the multinucleotide variant at codon 986 (CGT>TTT) (supplemental Figure 4B). Moreover, a SAMD9L c.1204_1208delCTCAT frameshift variant was also detected in 26% of reads and Sanger validated (supplemental Figure 4C). Sequencing of SAMD9L in all carriers for which WES data were not available revealed that neurologically affected F2:I-2, who lacked any evidence for mitotic recombination, carried a SAMD9L c.2302A>T (p.Lys768Ter) nonsense mutation in ∼80% of blood cells (Figure 4F). Cloning of a PCR amplicon that included both mutations revealed a cis configuration of the SAMD9L c.2302A>T and c.2956C>T mutations (Figure 4G), yet the c.2302A>T mutation was absent in the children carrying the SAMD9L c.2956C>T mutation (Figure 4G), indicating a somatic origin. Together, these findings demonstrate that somatic revertant mosaicism could occur through mechanisms other than mitotic recombination leading to UPD(7q).
In summary, revertant mosaicism was therefore identified in 5 of the 7 carriers of the SAMD9L mutations, who hitherto have not developed MDS. Apparent reversions occurred by both mitotic recombination and second-site nonsense mutation, with the coexistence of distinct reverted clones. Notably, no revertant mosaicism was detected in the 2 carriers, F1:II-1 and F1:II-2, who also harbored the SAMD9L variant c.689C>A in trans (data not shown).
Disease-modifying SAMD9L variants and revertant mutants represent loss-of-function variants
Intriguingly, F1:II-1, who was healthy, and F1:II-2, who had only very mild imbalance problems, carried the disease-causing SAMD9L c.2956C>T gain-of-function mutation in trans with the rare SAMD9L c.689C>A variant. We thus hypothesized that SAMD9L c.689C>A (p.Thr233Asn) could represent a disease-modifying, loss-of-function variant. Congruently, ectopic expression of TFP-SAMD9L p.Thr233Asn in 293FT cells attenuated cell proliferation to a lesser extent than that of TFP-SAMD9L wild type (Figure 5A-C).
Functional evaluation of disease-modifying and somatic reversion SAMD9L variants. (A-C) 293FT cells were transiently transfected (Tx) with TFP-SAMD9L wild-type (WT) or potentially disease-modifying p.Thr233Asn variant, as indicated. (A) Western blots of recombinant SAMD9L variant expression, as indicated. (B) Cell proliferation in 293FT cells assessed by dye dilution assays of TFP-SAMD9L–transfected 293FT cells. Dye levels were monitored in TFP− cells (filled gray histograms) and compared with cells expressing uniformly intermediate levels of TFP-SAMD9L variants, as indicated. A single representative experiment is shown. (C) Cumulative data from independent experiments on growth inhibition associated with specific TFP-SAMD9L variants, as indicated. According to the index, 0 denotes growth of vector-transfected cells, whereas −1 approximates TFP-SAMD9L wild-type transfected cells. Significance was determined by 1-way analysis of variance (*P < .05; **P < .005; ***P < .0005. (D) Overview of SAMD9L structure, including SAM domain (blue). Positions of identified disease-associated germ line SAMD9L gain-of-function mutations (red), germ line loss-of-function variants (blue), and somatic reversion mutations (green) are indicated. Disease-associated germ line SAMD9L mutations reported by Chen and colleagues38 are included (red).
Functional evaluation of disease-modifying and somatic reversion SAMD9L variants. (A-C) 293FT cells were transiently transfected (Tx) with TFP-SAMD9L wild-type (WT) or potentially disease-modifying p.Thr233Asn variant, as indicated. (A) Western blots of recombinant SAMD9L variant expression, as indicated. (B) Cell proliferation in 293FT cells assessed by dye dilution assays of TFP-SAMD9L–transfected 293FT cells. Dye levels were monitored in TFP− cells (filled gray histograms) and compared with cells expressing uniformly intermediate levels of TFP-SAMD9L variants, as indicated. A single representative experiment is shown. (C) Cumulative data from independent experiments on growth inhibition associated with specific TFP-SAMD9L variants, as indicated. According to the index, 0 denotes growth of vector-transfected cells, whereas −1 approximates TFP-SAMD9L wild-type transfected cells. Significance was determined by 1-way analysis of variance (*P < .05; **P < .005; ***P < .0005. (D) Overview of SAMD9L structure, including SAM domain (blue). Positions of identified disease-associated germ line SAMD9L gain-of-function mutations (red), germ line loss-of-function variants (blue), and somatic reversion mutations (green) are indicated. Disease-associated germ line SAMD9L mutations reported by Chen and colleagues38 are included (red).
Altogether, 4 SAMD9L mutation carriers experienced UPD(7q), resulting in cells with biallelic wild-type SAMD9L. Furthermore, F1:II-4 and F2:I-2 displayed somatic cis truncating mutations, most likely representing loss-of-function rescues. In addition, at a lower frequency than the truncating mutations, F1:II-4 harbored a somatic SAMD9L c.2957G>T reversion mutation in cis. In cellular assays, the SAMD9L p.Arg986Phe reversion mutant displayed a similar level of inhibition as the SAMD9L p.Arg986Cys disease-causing mutant (Figure 5C). Notably, a rare SAMD9L c.2957G>A (p.Arg986His) variant is present at a frequency of 0.00003% in the ExAC database,32 displaying significantly more inhibition of cell proliferation than wild-type TFP-SAMD9L in cellular assays (Figure 5C). It is therefore possible that this variant may also be associated with disease, possibly with lower penetrance. Disease-causing loss-of-function variants described here and by Chen and colleagues38 map to the C-terminal half of SAMD9L (Figure 5D).
Occurrence of somatic SAMD9L reversion mutations in hematopoietic progenitor cells and further selection in hematopoietic cell lineages
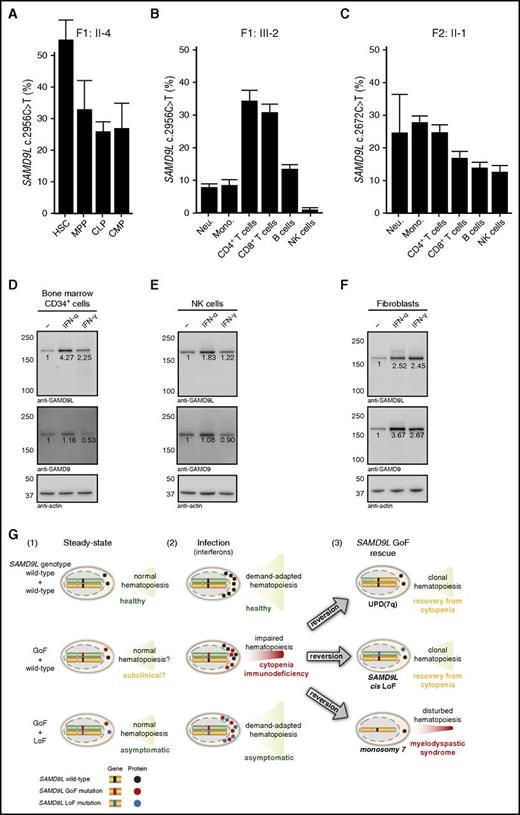
In order to delineate the cell types and lineages, in which the reversions occur and potentially are selected, specific cell subpopulations were sorted from bone marrow and peripheral blood. For F1:II-4, DNA was isolated from HSC, MPP, CLP, and CMP cells. Notably, whereas HSCs harbored the SAMD9L c.2956C>T mutation at the expected frequency of 50%, a lower frequency of the mutation was detected in MPP, CLP, and CMP cells from F1:II-4 (Figure 6A), demonstrating that reversions may initially arise upon differentiation of HSCs.
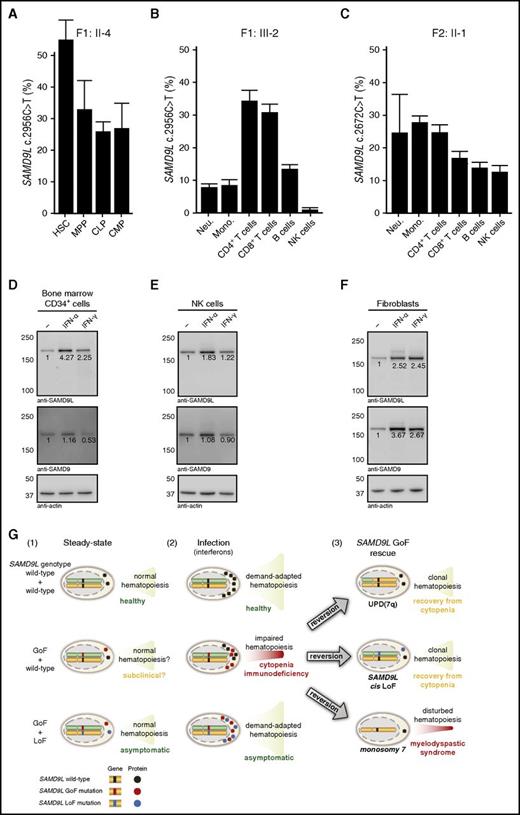
Frequency of SAMD9L mutation in hematopoietic stem, progenitor, and differentiated cell populations and interferon-stimulated expression of SAMDL. (A) Quantification of the frequency of the SAMD9L c.2956C>T in distinct bone marrow–derived HSPC populations from F1:II-2, as indicated. (B-C) Quantification of the frequency of the SAMD9L (B) c.2956C>T and (C) c.2672T>C mutations in specific peripheral blood–derived immune cell populations from F1:III-2 and F2:II-1, respectively, as indicated. For each sample, values represent the mean of at least 2 chips. Error bars denote 95% confidence levels. (D-F) Evaluation of SAMD9L and SAMD9 expression in (D) bone marrow-derived CD34+ HSPCs, (E) peripheral blood–derived NK cells, and (F) fibroblasts stimulated with interferon (IFN)-α or IFN-γ, as indicated. (G) Hypothetical model of the pathophysiology of germ line SAMD9L gain-of-function (GoF) mutations in relation to HSPC proliferation and differentiation. Healthy individuals, with 2 wild-type SAMD9L copies (top panel), have (1) normal, steady-state hematopoiesis, and (2) increased cellular output upon infection-induced, demand-adapted hematopoiesis. In contrast, carriers of heterozygous SAMD9L GoF mutations (middle panel) may (1) display grossly normal (and perhaps subclinical) hematopoiesis for some time, but (2) experience cytopenias and immunodeficiency upon infection early in life. In this setting, interferons can promote SAMD9L expression, with SAMD9L GoF mutants acting as potent suppressors of cell proliferation, dramatically impairing hematopoiesis and immunity. The ensuing hematopoietic crisis can facilitate (3) selection and expansion of revertant mutants, by UPD(7q), SAMD9L loss-of-function (LoF) mutations in cis, or monosomy 7. Whereas UPD(7q) and in cisSAMD9L LoF mutations can support clonal hematopoiesis and recovery from cytopenia, monosomy 7 is associated with development of MDS. Finally, carriers of combined SAMD9L GoF mutation and rare LoF variants in trans (bottom panel) are asymptomatic, suggesting they have normal (1) steady-state and (2) demand-adapted hematopoiesis. As such, pathogenic effects of SAMD9L GoF mutations may be balanced by SAMD9L LoF mutations. Mono, monocyte; Neu, neutrophil.
Frequency of SAMD9L mutation in hematopoietic stem, progenitor, and differentiated cell populations and interferon-stimulated expression of SAMDL. (A) Quantification of the frequency of the SAMD9L c.2956C>T in distinct bone marrow–derived HSPC populations from F1:II-2, as indicated. (B-C) Quantification of the frequency of the SAMD9L (B) c.2956C>T and (C) c.2672T>C mutations in specific peripheral blood–derived immune cell populations from F1:III-2 and F2:II-1, respectively, as indicated. For each sample, values represent the mean of at least 2 chips. Error bars denote 95% confidence levels. (D-F) Evaluation of SAMD9L and SAMD9 expression in (D) bone marrow-derived CD34+ HSPCs, (E) peripheral blood–derived NK cells, and (F) fibroblasts stimulated with interferon (IFN)-α or IFN-γ, as indicated. (G) Hypothetical model of the pathophysiology of germ line SAMD9L gain-of-function (GoF) mutations in relation to HSPC proliferation and differentiation. Healthy individuals, with 2 wild-type SAMD9L copies (top panel), have (1) normal, steady-state hematopoiesis, and (2) increased cellular output upon infection-induced, demand-adapted hematopoiesis. In contrast, carriers of heterozygous SAMD9L GoF mutations (middle panel) may (1) display grossly normal (and perhaps subclinical) hematopoiesis for some time, but (2) experience cytopenias and immunodeficiency upon infection early in life. In this setting, interferons can promote SAMD9L expression, with SAMD9L GoF mutants acting as potent suppressors of cell proliferation, dramatically impairing hematopoiesis and immunity. The ensuing hematopoietic crisis can facilitate (3) selection and expansion of revertant mutants, by UPD(7q), SAMD9L loss-of-function (LoF) mutations in cis, or monosomy 7. Whereas UPD(7q) and in cisSAMD9L LoF mutations can support clonal hematopoiesis and recovery from cytopenia, monosomy 7 is associated with development of MDS. Finally, carriers of combined SAMD9L GoF mutation and rare LoF variants in trans (bottom panel) are asymptomatic, suggesting they have normal (1) steady-state and (2) demand-adapted hematopoiesis. As such, pathogenic effects of SAMD9L GoF mutations may be balanced by SAMD9L LoF mutations. Mono, monocyte; Neu, neutrophil.
To determine if HSC-derived mutations were further selected, DNA was isolated from peripheral blood neutrophils, monocytes, B cells, CD4+ and CD8+ T cells, as well as NK cells in F1:III-2, F1:II-4, F2:II-1, and F2:I-2. In F1:III-2, the lowest frequency of the SAMD9L c.2956C>T mutation was observed in NK cells, B cells, and neutrophils (Figure 6B). Similarly, in F1:II-4 and F2:II-1, the lowest frequency of the SAMD9L mutation was observed in NK cells and B cells (Figure 6C; supplemental Figure 5A), whereas this effect was not observed in F2:I-2, in whom the SAMD9L c.2972T>C c.2956C>T mutation was linked to a truncating reversion mutation (supplemental Figure 5B). Together, these data imply a role for SAMD9L in regulating HSC renewal and differentiation, as well as in differentiation of mature leukocyte subsets, most notably B cells and NK cells. These cellular effects are in line with the observations of cellular deficiencies in F1:III-1, in whom reversion mutations had not arisen.
Inducible expression of SAMD9L
The strong antiproliferative effect of disease-causing germ line SAMD9L mutations could appear at odds with the normal frequencies of the mutation discovered at birth or at the time of hematopoietic crises that first occur postnatally, with the selection of revertant mutants. Interestingly, SAMD9L is reportedly an IFN-stimulated gene in human CD4+ T cells.35 IFNs can play an important role in activating HSC proliferation.36 We therefore hypothesized that expression of SAMD9L might be induced by IFNs in HSCs as well as other IFN-responsive cells. Indeed, stimulation of CD34+ HSPCs with IFN-α or IFN-γ induced SAMD9L expression (Figure 6D). Similarly, these antiviral factors also induced SAMD9L expression in NK cells and fibroblasts (Figure 6E-F). Our results demonstrate that SAMD9L expression is induced by IFN-stimulation in a variety of cell types, including HSPCs, potentially explaining how immune responses may induce cytopenias and provide a basis for selection of SAMD9L reversion mutants.
Discussion
Several rare germ line syndromes that predispose individuals to develop MDS/AML have recently been delineated; these syndromes represent a separate category in the revised World Health Organization classification of myeloid neoplasms.1 Still, most familial cases remain unexplained.37 We describe 2 families with cytopenia, immunodeficiency, predisposition to MDS, and progressive cerebellar dysfunction, resembling ATXPC syndrome and myelodysplasia and leukemia syndrome with −7.25,26 In 1 family, a cellular immunodeficiency somewhat resembling GATA2 haploinsufficiency was characterized. Heterozygous SAMD9L missense gain-of-function mutations were identified as a cause of disease in both families. Several carriers displayed mild, variable expressivity, explained by hematopoietic somatic revertant mosaicism or a compensatory effect of a germ line in transSAMD9L loss-of-function variant. Our findings confirm and extend work by Chen and colleagues,38 who recently identified heterozygous SAMD9L missense mutations in the ATXPC index family.
SAMD9L is located on 7q21, head to tail with SAMD9, constituting a homologous, evolutionarily conserved gene pair.33,39 Both proteins negatively regulate cell proliferation.34 Reports of downregulated SAMD9 and SAMD9L transcription in neoplastic and malignant cells are consistent with tumor suppressor activity.33,40 The SAMD9L Ile891 and Arg986 amino acid residues are conserved and synonymous in SAMD9. We found that disease-causing SAMD9L mutations identified by us as well as those identified by Chen and colleagues38 increased suppression of cell proliferation (supplemental Figure 6), ie, they represent gain-of-function mutations. Similarly, de novo germ line SAMD9 gain-of-function mutations were recently associated with a syndrome of myelodysplasia, infection, restriction of growth, adrenal hypoplasia, genital phenotypes, and enteropathy (known as the MIRAGE syndrome; MIM no. 617053).41 Autosomal-recessive SAMD9 loss-of-function mutations are associated with normophosphatemic familial tumoral carcinosis, with no reported effect in heterozygous carriers (MIM no. 610455).42,43
Inherited cancer syndromes have greatly contributed to basic concepts of tumor biology, with germ line mutations in the tumor suppressors RB1 and APC postulating the “2-hit” hypothesis by Knudson44 and providing the concept of “multistep” carcinogenesis by Vogelstein and Kinzler.45 Our study and that by Chen and colleagues38 indicate that heterozygous SAMD9L missense mutations are causative of familial MDS. Notably, mutated SAMD9L alleles were lost in MDS cells. Similarly, 2 of 11 children carrying germ line SAMD9 gain-of-function mutations developed MDS −7, lacking the SAMD9 mutations in MDS cells.41 Instead of representing malignancy-predisposing mutations in the classical sense of tumor suppressor genes, gain-of-function mutations in SAMD9 or SAMD9L together provide the first human examples of “adaptation by aneuploidy.”46 HSPCs that eliminate SAMD9 or SAMD9L gain-of-function mutations through aneuploidy gain a competitive advantage, simultaneously predisposing to MDS (Figure 6G). Other than ATXPC and MSML7, mutations in SAMD9L may also cause juvenile myelomonocytic leukemia (JMML). In 25% of patients with JMML, −7 is detected.47 Moreover, 7q21.3 microdeletions encompassing SAMD9 and SAMD9L have been described in JMML.48 Notably, several cases of transient −7 have been reported,49-53 suggesting that clones carrying such somatic aberrations may gain transient competitive advantage and subsequently recede.
Given the high prevalence of −7/del(7q) cytogenetic aberrations in both primary and sporadic forms of MDS and their high risk of progression to AML,23,54 responsible genes have been sought by high-resolution genotyping and sequencing. Commonly deleted regions have been identified at 7q21-22, 7q34, and 7q35-36.48,55,56 Whereas 7q21-22 encompasses SAMD9 and SAMD9L, somatic mutations in CUX1 (7q22.1) and EZH2 (7q35-36) have been reported.55,56 In mice, Samd9l haploinsufficiency is associated with age-related MDS development,34 whereas somatic SAMD9L or SAMD9 mutations were not found in 43 examined human patients with MDS/AML.34 In humans, SAMD9 may compensate for SAMD9L loss-of-function mutations. Intriguingly, we found that an additional in transSAMD9L loss-of-function variant was associated with lack of hematopoietic reversion and protection from disease. As opposed to GATA2, heterozygous SAMD9 and SAMD9L loss-of-function mutations are present in the general population, suggesting that haploinsufficiency may be tolerated.32 In settings of SAMD9L mutation–triggered aneuploidy, haploinsufficiency of additional genes on 7q likely contributes to MDS development.57,58 Still, to what extent heterozygous germ line or somatic SAMD9L loss-of-function mutations may predispose to cancer warrants further investigation.
Somatic revertant mosaicism has been reported in bone marrow failure syndromes, primary immunodeficiency syndromes, and inherited skin diseases.59 Often ameliorating disease, it can occur through distinct genetic mechanisms. Revealing strong selective advantages of hematopoietic wild-type over mutant SAMD9L cells, we detected blood revertant mosaicism in 5 of 10 individuals carrying the SAMD9L mutations. Following hematopoietic crisis in infancy, reversions were derived by mitotic recombination as well as second-site mutations, including upstream loss-of-function mutations in cis. In 2 individuals, several reversion events were detected in peripheral blood. Such reversions were traced to HSPCs. A similar event of clonal hematopoietic reversion with clinical improvement was recently described in a patient with an autosomal-recessive syndrome of cytopenia, immunodeficiency, and developmental abnormalities caused by MYSM1 mutation, a gene required for HSPC quiescence.60 Age-related clonal hematopoiesis is associated with an increased risk of malignancy.61-64 Although somatic HSPC revertant mosaicism can rescue the effect of damaging germ line SAMD9L mutations, these clonal selection processes may thus predispose to cancer.
The apparent tolerance of SAMD9L haploinsufficiency, yet evolutionary conservation of SAMD9 and SAMD9L, raises questions regarding the function of these proteins. Both SAMD9 and SAMD9L have been identified as IFN-stimulated genes,35,65 and can restrict orthopox and West Nile virus infection.66,67 We demonstrate that IFNs trigger their expression in several hematopoietic cell types. Infections increase demand on hematopoiesis.68 Both IFN-α and IFN-γ activate quiescent HSCs.36,69 It can thus be speculated that SAMD9 and SAMD9L represent critical antiviral gatekeepers regulating demand-adapted, IFN-driven hematopoiesis (Figure 6G). In these settings, reversion mutants are frequently selected to further support hematopoiesis. Furthermore, we observed increasing degrees of SAMD9L reversion in B and NK cells, suggesting that SAMD9L may regulate differentiation of diverse immune cell lineages. It remains to be determined to what extent the reservoir of mutated HSCs represents a risk for later MDS/AML development, potentially instigated by inflammation.
Our findings have several important clinical implications, because the majority of SAMD9L mutation carriers may not present with overt clinical manifestation. Variable expressivity and revertant mosaicism of SAMD9L gain-of-function mutations may lead to ascertainment bias, thus frustrating efforts to identify disease-causing mutations. SAMD9L mutation screening should be considered in all pediatric patients with MDS, AML, or JMML with chromosome 7 aberrations, even in the absence of neurological symptoms or a family history of myeloid malignancies. Analyses should include sequencing of DNA from peripheral blood as well as nonhematopoietic tissue. The coexistence of rare inherited missense mutations in SAMD9L and UPD(7q) or additional somatic mutations is highly suggestive of pathogenic germ line mutations. Additionally, potential HLA-identical sibling donors have to be carefully evaluated, given that UPD(7q) can disguise carriers of SAMD9L mutations. The established functional assay may provide a key to identifying disease-causing SAMD9L missense variants. SAMD9L gain-of-function mutation carriers frequently develop severe neurological manifestations, the cellular basis of which remains unclear. Elucidating the pathogenesis of SAMD9L gain-of-function mutations may provide therapeutic options.
In conclusion, our results indicate an important role of SAMD9L in regulation of hematopoietic cell proliferation and differentiation, and highlight SAMD9L gain-of-function mutations as a likely cause of ATXPC and MSLM7 syndromes. Efforts aimed at identifying SAMD9L mutations should be considered in pediatric patients with myeloid −7/del7(q) neoplasms, as well as unexplained cytopenia or ataxia syndromes. Acquired hematopoietic reversion mutations are commonly observed in carriers of SAMD9L gain-of-function mutations, and further studies are required to establish the long-term risk of diverse disease manifestations in these carriers.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all family members for their participation. The authors also thank Marios Dimitriou for critical comments on the manuscript, Henrik Lillebjörn for acquisition of patient material from the biobank, Teona Roschupkina for assisting with bone marrow sorts, Vasilios Zachariadis for fruitful discussions, Fulya Taylan for guidance on dPCR experiments, the MedH Core Flow Cytometry facility (Karolinska Institutet [KI], supported by KI/Stockholm Läns Landsting [SLL]) for providing cell sorting services, and the Clinical Genomics unit at SciLifeLab for whole-exome sequencing as well as the SNP and SEQ Technology Platform in Uppsala for SNP genotyping.
This work was supported by a grant from the European Research Council (ERC) under the European Union's Seventh Framework Programme (FP/2007-2013; ERC Grant Agreement 311335), the Swedish Research Council, Norwegian Research Council, Swedish Foundation for Strategic Research, Wallenberg Foundation, Swedish Cancer Foundation, Swedish Childhood Cancer Foundation, as well as the Stockholm County Council and Karolinska Institutet Center for Innovative Medicine (Y.T.B.), and the Hemato-Linné, Cancerfonden and Barncancerfonden (J.C.). B.T. and J.D. are supported by doctoral student scholarships from the Board of Postgraduate Studies at Karolinska Institute and by the Swedish Society for Medical Research and Skåne University Hospital, respectively. M.V. held postdoctoral scholarships from the Wenner-Gren Foundation and EMBO (ALTF 206-2015, cofunded by the European Commision [LTFCOFUND2013, GA-2013-609409]) and received support from the Histiocytosis Association and the Swedish Society for Medicine. Computations were performed on resources provided by Swedish National Infrastructure for Computing (SNIC) through the Uppsala Multidisciplinary Center for Advanced Computational Science under Project SNIC b2012204.
Authorship
Contribution: B.T. designed experiments, performed WES analyses on family 1 and targeted sequencing on family 2, identified disease-causing and disease-modifying SAMD9L variants, identified somatic reversions, performed and analyzed dPCR assays, analyzed SNP arrays, and wrote the manuscript; J.D. analyzed and compiled clinical data on family 1, designed experiments, performed WES analyses on family 1 and genotyped siblings of F1:I-3, analyzed FLT-3L serum levels, and performed and analyzed dPCR assays; M.V. designed experiments, generated SAMD9L expression constructs, developed and performed SAMD9L functional assay, performed western blot analyses, and analyzed data; E.R. cared for family 2, and analyzed and compiled clinical data; T.D.H. sorted peripheral blood mononuclear cell populations; S.C.C.C. performed phenotypic and functional analyses of cytotoxic lymphocytes; J.K.-E. neurologically examined family 2 and made the clinical diagnosis; S.G. neurologically examined family 1; A.R.N. sorted bone marrow HSPC populations; T.R. excluded GATA2 mutations in patient F1:I-3; H.K. performed cytogenetic analyses on F2:II-4; D.B. supervised research; T.F. provided scientific expertise, genetic counseling, and DNA from family F1; J.-I.H. supervised research; M.M. performed hematological investigations of family 2; R.N. performed hematological investigations of family 2; L.N. cared for patient F1:I-3, and analyzed and compiled clinical data; C.J.P. advised on sorting of bone marrow cell populations; A.P. neurologically examined family 1; H.Q. provided CD34+ HSPCs for western blot experiments; J.U. supervised neurological investigations of family 2; J.M. supervised clinical and genetic investigations of family 2, and analyzed and compiled clinical data; U.T. cared for the index patients F1:III-1 and F1:III-2, initiated genetic investigations of family 1, and analyzed and compiled clinical data; J.C. designed experiments, analyzed data, and contributed to drafting of the manuscript; Y.T.B designed experiments, analyzed data, and wrote the manuscript; and all authors commented on and approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bianca Tesi, Center for Molecular Medicine L8:02, Karolinska University Hospital Solna, 17176, Stockholm, Sweden; e-mail: bianca.tesi@ki.se; and Yenan T. Bryceson, Center for Hematology and Regenerative Medicine, Department of Medicine, Karolinska Institutet, Karolinska University Hospital Huddinge, 14186, Stockholm, Sweden; e-mail: yenan.bryceson@ki.se.
References
Author notes
B.T., J.D., and M.V. are joint first authors.
J.C. and Y.T.B. are joint senior authors.