Key Points
Concurrent RAS pathway and TP53 mutations predict a dismal outcome in human AML.
p53−/− synergizes with enhanced oncogenic Nras signaling to transform MEPs to AML-initiating cells.
Abstract
Somatic mutations in TP53 and NRAS are associated with transformation of human chronic myeloid diseases to acute myeloid leukemia (AML). Here, we report that concurrent RAS pathway and TP53 mutations are identified in a subset of AML patients and confer an inferior overall survival. To further investigate the genetic interaction between p53 loss and endogenous NrasG12D/+ in AML, we generated conditional NrasG12D/+p53−/− mice. Consistent with the clinical data, recipient mice transplanted with NrasG12D/+p53−/− bone marrow cells rapidly develop a highly penetrant AML. We find that p53−/− cooperates with NrasG12D/+ to promote increased quiescence in megakaryocyte-erythroid progenitors (MEPs). NrasG12D/+p53−/− MEPs are transformed to self-renewing AML-initiating cells and are capable of inducing AML in serially transplanted recipients. RNA sequencing analysis revealed that transformed MEPs gain a partial hematopoietic stem cell signature and largely retain an MEP signature. Their distinct transcriptomes suggests a potential regulation by p53 loss. In addition, we show that during AML development, transformed MEPs acquire overexpression of oncogenic Nras, leading to hyperactivation of ERK1/2 signaling. Our results demonstrate that p53−/− synergizes with enhanced oncogenic Nras signaling to transform MEPs and drive AML development. This model may serve as a platform to test candidate therapeutics in this aggressive subset of AML.
Introduction
Myelodysplastic syndromes (MDSs) and myelodysplastic syndrome/myeloproliferative neoplasms (MDS/MPNs) are chronic myeloid neoplasms recently defined as distinct entities by the World Health Organization. MDS represents a preleukemic state of ineffective hematopoiesis hallmarked by bone marrow dysplasia with peripheral blood cytopenias,1 whereas patients with MDS/MPNs have additional proliferative features that share clinical similarities with MPNs.2,3 The natural history of both entities is heterogeneous. Some patients have a relatively benign natural history that does not require therapy, whereas others succumb within 12 months.4,5 One major cause of mortality in both cases is transformation to acute myeloid leukemia (AML). The risk of transformation is highly variable in different subtypes of MDSs ranging from 5% to 65%, whereas AML transformation occurs in approximately 30% of patients with MDS/MPNs.4,5 Although there are clinical and genetic models that are able to refine the leukemia transformation rate in cohorts of patients,4,6 the mechanisms driving AML transformation remain largely unknown.
TP53 and NRAS mutations are 2 somatic events that have been clinically associated with AML transformation.6,7 TP53 regulates a wide range of biological processes to suppress tumorigenesis, including cell cycle progression, apoptosis, autophagy, and cell differentiation.8 TP53 mutations are prevalent in patients with AML or MDS who have a complex karyotype, but they are rare in chronic diseases with largely normal karyotype, such as chronic myelomonocytic leukemia (CMML, an MDS/MPN disease) and MPNs.9-11 Clinically, TP53 mutations are significantly associated with erythroid abnormalities12,13 and severe thrombocytopenia in MDSs.6 Furthermore, TP53 has been identified as being among the strongest negative predictors of survival and relapse after allogeneic transplant in MDSs. Of note, deficiency in and mutations of p53 in mice leads to T- and/or B-cell malignancies but not myeloid diseases,14,15 suggesting that p53 mutations require additional genetic events to promote myeloid malignancies. The recent identification of TP53 mutation as a driver in promoting transformation of JAK2V617F-positive MPNs to AML supports this hypothesis.11
Mutations in NRAS and other RAS pathway genes are frequently identified in CMML and its juvenile counterpart JMML. In contrast to JMML, in which activating RAS pathway mutations are obligatory founding events, RAS mutations in CMML have been identified as initiating or secondary events.2,16-20 Interestingly, oncogenic RAS mutations are particularly enriched in the proliferative CMML subtype, which is phenotypically more consistent with JMML than with MDSs.21 In our previous report,22 which was in agreement with human studies, we showed that recipients transplanted with NrasG12D/+ bone marrow cells developed a highly penetrant CMML-like disease after a prolonged latency. However, none of the recipients with CMML spontaneously transformed to AML. Although acquired uniparental disomy (UPD) or homozygosis of the oncogenic NRAS allele was observed in CMML patients as well as in our CMML mice,22,23 it seemed to promote CMML progression but was insufficient to induce malignant transformation to AML on its own.11 Collectively, this suggests that, like TP53, additional genetic events are required to promote transformation of oncogenic NRAS-driven chronic myeloid neoplasms.
Previous studies have identified that NRAS and TP53 mutations are concurrent in a subset of MDS patients (3 of 43),12 and deletion of NF1 and/or SPRY4 (2 negative regulators of the RAS pathway) co-occur with deletion and/or mutation of TP53 in a subset of AML patients from public data in The Cancer Genome Atlas.24 Here, we report that sequencing analysis of 1238 cases of myeloid neoplasms identified 8 patients with mutations in both TP53 and RAS pathway genes, the majority of which were secondary AML with a dismal prognosis. We thus investigated the genetic interaction of p53 loss and endogenous oncogenic Nras signaling in AML. Our study identified a strong synergy between them in regulating megakaryocyte-erythroid progenitor (MEP) quiescence and leukemogenic transformation of MEPs to promote AML.
Materials and methods
Mice
All mouse lines were maintained in a pure C57BL/6 genetic background (backcrossed for more than 10 generations). Mice bearing a conditional oncogenic Nras (NrasLox-stop-Lox (LSL) G12D/+)25 were crossed to Mx1-Cre mice26 to generate mice carrying both alleles (NrasLSL G12D/+Mx1-Cre). The p53 conditional knockout mice (p53fl/fl) were obtained from The Jackson Laboratory (Stock No. 008462)27 and crossed with Mx1-Cre to generate p53fl/flMx1-Cre mice. NrasLSL G12D/+p53fl/fl mice were crossed to p53fl/flMx1-Cre to generate compound mice NrasLSL G12D/+p53fl/flMx1-Cre. Genotyping of NrasG12D/+ and Mx1-Cre was done as previously described.22 Genotyping of p53fl/fl was performed per instructions from The Jackson Laboratory. CD45.1+ congeneic recipient mice were purchased from the National Cancer Institute.
To induce Cre expression, 5- to 7-week-old mice were injected intraperitoneally with 100 μg of polyinosinic-polycytidylic acid (pI-pC; GE Healthcare) every other day for 3 times. The day of the first pI-pC injection was defined as day 1. All experiments were performed on day 12. The polymerase chain reaction analysis of recombination efficiency at the p53 locus was performed as previously described.27 Additional methods are described in the supplemental Data, available on the Blood Web site.
Results
RAS pathway and TP53 mutations co-occur in clinical AML samples
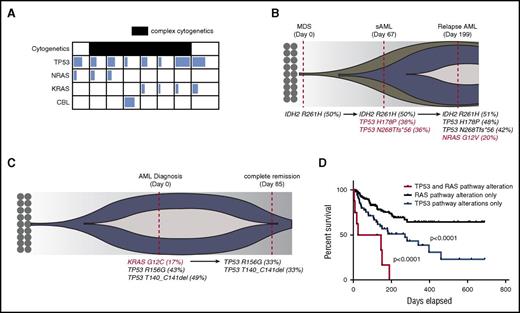
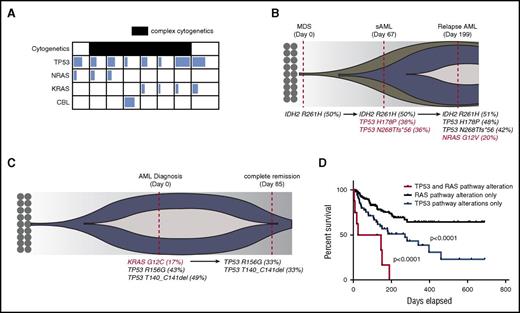
To determine how concurrent RAS pathway and TP53 mutations influence the prognosis of human myeloid leukemia, we screened patients with myeloid neoplasms (including MPNs, MDSs, MPN/MDS, and AML) who were sequenced for 54 genes recurrently mutated in myeloid leukemia. Of 1238 patients reviewed, 74 (5.9%) were found to have single TP53 mutations, 212 (17%) were found to have single RAS pathway mutations (KRAS, NRAS, or CBL), and 8 patients with AML (0.6%) had both TP53 and RAS pathway mutations (Figure 1A; supplemental Table 1). The majority of these patients harbored a complex karyotype and were classified as either secondary AML or AML with MDS-related changes as defined by the World Health Organization classification system (supplemental Table 1), suggesting that these mutations may cooperate to promote disease transformation in humans. In fact, two patients in whom serial sequencing was available demonstrated the acquisition and loss of RAS mutations during disease progression and remission, respectively (Figure 1B-C). Although we could not definitively determine whether these mutations co-occur in the same cells, the fact that TP53 mutations (most likely heterozygote), have a variant allele frequency close to 50% suggests that the estimated leukemia cell proportion with TP53 mutation is close to 100%. Similarly, estimated leukemia cell proportion with a RAS mutation is above 30%. Therefore, RAS and TP53 mutations would be present in at least a fraction of leukemia cells. The mean white blood cell count was 12.7 × 103/dL (range, 1.68-43 × 103/dL), mean peripheral blast percentage was 22% (range, 3%-87%), mean hemoglobin was 8.6 g/dL (range, 7.5-10.1 g/dL), and mean platelet count was 97 × 103/dL (range, 26-316 × 103/dL) at the time of genetic testing. The median overall survival of this genetic cohort was 85 days (range, 5-151 days), which was significantly shorter than that for patients with TP53 mutations alone (271 days) and patients with RAS pathway mutations whose median survival was not reached in our data set (P < .001) (Figure 1D).
TP53 and RAS pathway mutations co-occur in human AML and predict an inferior outcome. (A) Variant allele frequency (VAF) of RAS pathway mutations and TP53 mutations in co-mutated patients. The degree of shading in each box corresponds to the VAF of that mutation. Patients with complex cytogenetics are shaded in black. Details of complex cytogenetics and VAF information are included in supplemental Table 1. (B) Fish plot of a representative case with sequential sampling that acquired TP53 mutations at the time of AML transformation and then an NRAS mutation during AML relapse. Red-colored mutations highlight acquired genetic events. (C) Fish plot of a representative patient with sequential sampling who lost a KRAS mutation at the time of clinical remission with induction chemotherapy. Red-colored mutation highlights the lost genetic event. (D) Overall survival of TP53 and RAS pathway mutated cohort (n = 8) vs the TP53 mutated cohort (n = 55 patients with survival data) vs the RAS pathway mutated cohort (n = 154 patients with survival data). P values were determined by the log-rank test. sAML, secondary AML.
TP53 and RAS pathway mutations co-occur in human AML and predict an inferior outcome. (A) Variant allele frequency (VAF) of RAS pathway mutations and TP53 mutations in co-mutated patients. The degree of shading in each box corresponds to the VAF of that mutation. Patients with complex cytogenetics are shaded in black. Details of complex cytogenetics and VAF information are included in supplemental Table 1. (B) Fish plot of a representative case with sequential sampling that acquired TP53 mutations at the time of AML transformation and then an NRAS mutation during AML relapse. Red-colored mutations highlight acquired genetic events. (C) Fish plot of a representative patient with sequential sampling who lost a KRAS mutation at the time of clinical remission with induction chemotherapy. Red-colored mutation highlights the lost genetic event. (D) Overall survival of TP53 and RAS pathway mutated cohort (n = 8) vs the TP53 mutated cohort (n = 55 patients with survival data) vs the RAS pathway mutated cohort (n = 154 patients with survival data). P values were determined by the log-rank test. sAML, secondary AML.
To further validate the concurrence of TP53 and RAS pathway mutations in human AML, we interrogated The Cancer Genome Atlas AML data set by using the cBioPortal web tool.28,29 Of 191 samples evaluated, 6 patients (3.1%) harbored a TP53 mutation or deletion and RAS pathway mutation or deletion of negative regulator(s): 1 with TP53 mutation/deletion and NF1 deletion; 1 with TP53 mutation, NRAS mutation, NF1 fusion, and SPRY4 deletion; 1 with TP53 mutation and NRAS mutation; 1 with TP53 mutation and NF1 deletion; and 2 with TP53 mutation and SPRY4 deletion (supplemental Figure 1A; supplemental Table 2). Because amplification does not always translate to increase messenger RNA or gene activity, we considered the patient with TP53 mutation/amplification and MEK amplification as TP53 alone. Their median overall survival was 3 months, which was short compared with 7 months with TP53 (n = 11) and 19 months with RAS pathway mutation or deletion alone (n = 29) (supplemental Figure 1B). Collectively, these data suggest that TP53 and RAS pathway mutations co-occur at very low frequencies in human AML and may predict a dismal overall survival.
Loss of p53 synergizes with oncogenic Nras to induce a highly penetrant AML
To investigate the genetic interactions between p53 deficiency and oncogenic Nras signaling in leukemogenesis, we generated Mx1-Cre, p53fl/flMx1-Cre, NrasLSL G12D/+Mx1-Cre, and NrasLSL G12D/+p53fl/flMx1-Cre mice (supplemental Figure 2A). At 5 to 7 weeks old, these mice were given pI-pC as described in “Materials and methods” to induce oncogenic Nras expression from its endogenous locus and simultaneously delete p53 expression. The day of the first pI-pC injection was defined as day 1, and all mice were euthanized on day 12 for various analyses. We refer to the mice treated with pI-pC compound as p53−/−, NrasG12D/+, and NrasG12D/+p53−/− mice and Mx1-Cre mice treated with pI-pC as control mice throughout this study. We evaluated p53 deletion efficiency in whole bone marrow cells of NrasG12D/+p53−/− mice on day 12. A majority of whole bone marrow cells lost the floxed p53 allele (supplemental Figure 2B).
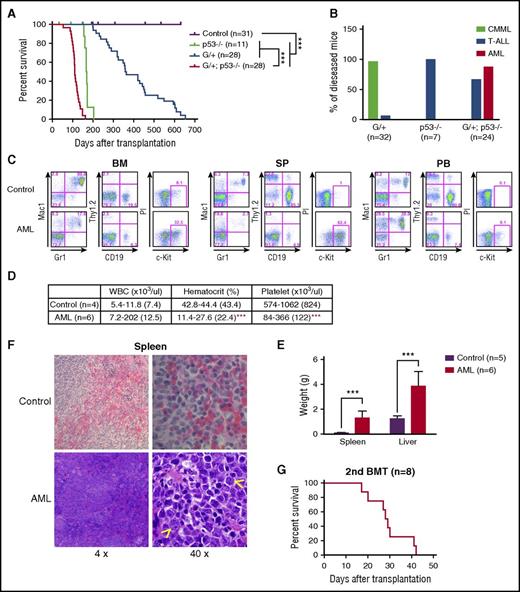
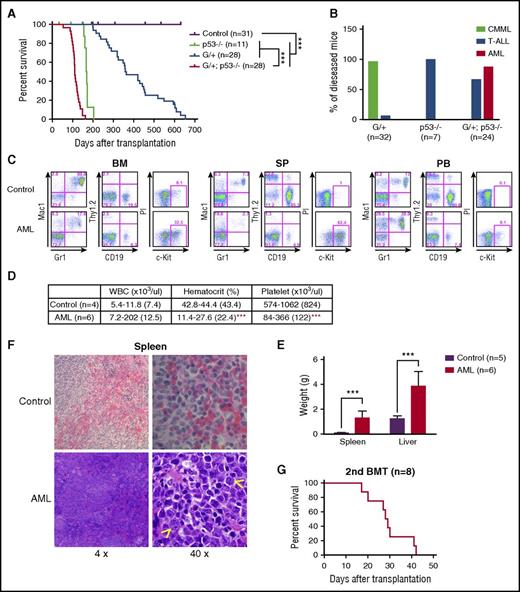
To investigate the cell-autonomous interaction between p53 loss and NrasG12D/+, we transplanted 2.5 × 105 bone marrow cells (CD45.2+) isolated from control, p53−/−, NrasG12D/+, and NrasG12D/+p53−/− mice along with same number of congenic competitor cells (CD45.1+) into individual lethally irradiated recipient mice (CD45.1+). Consistent with our previous reports, more than 90% of the recipient mice transplanted with NrasG12D/+ cells developed CMML-like phenotypes with a median survival of 360 days,22 whereas all the recipients with p53−/− cells developed acute T-cell lymphoblastic leukemia/lymphoma (T-ALL) with a median survival of 168 days (Figure 2A-B). In contrast, about 88% of the recipients that received NrasG12D/+p53−/− cells developed AML, and about 67% of the recipients developed T-ALL (some mice developed both diseases) with a median survival of 111 days, significantly shorter than that in other groups of recipients (Figure 2A-B).
p53 deletion and NrasG12D/+ induce a highly penetrant AML. (A-F) Lethally irradiated mice were transplanted with 2.5 × 105 total bone marrow cells from control, p53−/−, NrasG12D/+, or NrasG12D/+p53−/− mice along with same number of competitor cells. (A) Kaplan-Meier comparative survival analysis of reconstituted mice. Cumulative survival was plotted against days after transplantation. P value was determined by the log-rank test. (B) Disease incidence in recipient mice transplanted with p53−/−, NrasG12D/+, or NrasG12D/+p53−/− cells. (C) Representative flow cytometry analysis of bone marrow (BM), spleen (SP), and peripheral blood (PB) cells from moribund AML NrasG12D/+p53−/− and age-matched control mice. Note: red blood cells were lysed by using ammonium chloride treatment before flow analysis. (D) Complete blood count was performed on peripheral blood samples collected from moribund AML NrasG12D/+p53−/− and age-matched control mice. The results are presented as range (median). (E) Spleen and liver weight of moribund AML NrasG12D/+p53−/− and control mice. (F) Representative spleen histologic hematoxylin and eosin–stained sections from moribund AML NrasG12D/+p53−/− and control mice. Arrowheads indicate immature blast cells. (G) 1 × 106 bone marrow or 5 × 106 spleen cells from moribund AML NrasG12D/+p53−/− mice were transplanted into sublethally irradiated mice. Kaplan-Meier comparative survival curve was plotted against days after transplantation. The results are presented as mean ± standard deviation (SD). ***P < .001. BMT, bone marrow transplantation.
p53 deletion and NrasG12D/+ induce a highly penetrant AML. (A-F) Lethally irradiated mice were transplanted with 2.5 × 105 total bone marrow cells from control, p53−/−, NrasG12D/+, or NrasG12D/+p53−/− mice along with same number of competitor cells. (A) Kaplan-Meier comparative survival analysis of reconstituted mice. Cumulative survival was plotted against days after transplantation. P value was determined by the log-rank test. (B) Disease incidence in recipient mice transplanted with p53−/−, NrasG12D/+, or NrasG12D/+p53−/− cells. (C) Representative flow cytometry analysis of bone marrow (BM), spleen (SP), and peripheral blood (PB) cells from moribund AML NrasG12D/+p53−/− and age-matched control mice. Note: red blood cells were lysed by using ammonium chloride treatment before flow analysis. (D) Complete blood count was performed on peripheral blood samples collected from moribund AML NrasG12D/+p53−/− and age-matched control mice. The results are presented as range (median). (E) Spleen and liver weight of moribund AML NrasG12D/+p53−/− and control mice. (F) Representative spleen histologic hematoxylin and eosin–stained sections from moribund AML NrasG12D/+p53−/− and control mice. Arrowheads indicate immature blast cells. (G) 1 × 106 bone marrow or 5 × 106 spleen cells from moribund AML NrasG12D/+p53−/− mice were transplanted into sublethally irradiated mice. Kaplan-Meier comparative survival curve was plotted against days after transplantation. The results are presented as mean ± standard deviation (SD). ***P < .001. BMT, bone marrow transplantation.
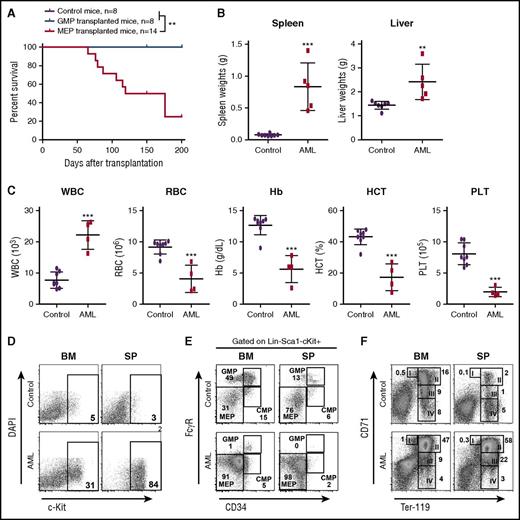
The recipient mice with AML displayed prominent monocytosis, anemia, and a reduced platelet count in the peripheral blood, expansion of myeloid compartment in the spleen, and marked hepatosplenomegaly (Figure 2C-E). Both spleen and liver were significantly infiltrated with immature blast cells (Figure 2F; Jingfang Zhang, unpublished data). The diagnosis of AML was primarily based on the presence of sheets of immature blast cells in the spleen (Figure 2F), which was often associated with a high percentage of c-Kit+ blast cells (Figure 2C). The AML disease was transplantable to secondary recipients; 7 of 8 recipients rapidly developed a lethal AML-like disease (Figure 2G; supplemental Figure 3). To our surprise, flow cytometric analysis of AML cells demonstrated that in about 90% of the AML mice, the expression of CD45 was significantly downregulated. Genotyping of AML cells showed that p53 expression was completely deleted, indicating the donor-derived origin of these leukemia cells (supplemental Figure 2B). To confirm that these CD45–/low AML cells were transformed from hematopoietic cells, we purified CD45+ bone marrow cells from NrasG12D/+p53−/− mice for transplantation. Indeed, the CD45+ hematopoietic cells generated CD45–/low AML cells in recipients (Table 1). We also generated a small cohort of primary NrasG12D/+p53−/− mice (supplemental Figure 4). They all died within 70 days after the last pI-pC injection with an AML-like disease (1 mouse also simultaneously developed T-ALL), similar to that in the recipient mice described above. Our results demonstrate that p53−/− cooperates with oncogenic Nras to promote AML development.
Loss of p53 induces myeloid progenitor (MP) expansion and increases MP re-plating capability in NrasG12D/+ mice
To determine the mechanisms underlying NrasG12D/+p53−/−-induced AML, we analyzed the hematopoietic compartment of control, p53−/−, NrasG12D/+, and NrasG12D/+p53−/− mice on day 12. After acute induction of Cre expression, p53−/− mice were grossly indistinguishable from control mice (supplemental Figure 5A-C). Consistent with previous reports,30,31 NrasG12D/+ mice showed significantly enlarged spleens and a significant increase of granulocytes and monocytes in spleens compared with control and p53−/− mice (supplemental Figure 5A,C). NrasG12D/+p53−/− mice were generally comparable to NrasG12D/+ mice except for moderate granulocytic/monocytic hyperplasia in peripheral blood (supplemental Figure 5C).
Because NrasG12D/+ hematopoietic stem cells (HSCs) serve as CMML-initiating cells,31 we investigated how p53 deficiency affects NrasG12D/+ HSC function. Consistent with previous reports,31,32 our data showed that HSC and Lin–Sca1+c-Kit+ (LSK) compartments in p53−/− and NrasG12D/+ mice were moderately but significantly expanded (supplemental Figure 6A). However, we observed significant expansion of the multipotent progenitor (MPP; defined as Lin–CD41–CD48–c-Kit+Sca1+CD150–) compartment only in NrasG12D/+ mice (supplemental Figure 6B-C). Loss of p53 did not significantly alter the size of HSC and MPP compartments in NrasG12D/+ mice but further expanded their LSK compartment (supplemental Figure 6A-C). Cell cycle profiling using Ki-67 and 4′,6-diamidino-2-phenylindole staining did not reveal significant changes in p53−/− HSCs, MPPs, and LSKs compared with control cells (supplemental Figure 6D-F). Although NrasG12D/+p53−/− HSCs were hyperproliferative in a manner similar to that of NrasG12D/+ HSCs, it seemed that NrasG12D/+p53−/− MPPs and LSKs were moderately but significantly more proliferative than NrasG12D/+ cells (supplemental Figure 6D-F). Our data suggest that p53 is largely dispensable for NrasG12D/+-induced HSC proliferation, but its deficiency might lead to significant changes in downstream NrasG12D/+ progenitors, which are enriched in LSKs.
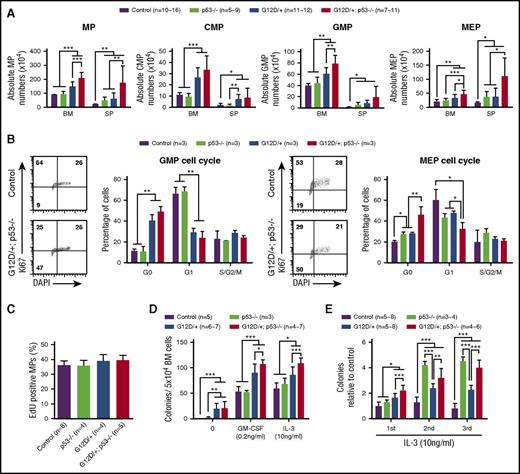
To determine how p53 deficiency affected NrasG12D/+ MP function, we analyzed the MP (defined as Lin–IL-7Rα–Sca1–c-Kit+) compartment in control, p53−/−, NrasG12D/+, and NrasG12D/+p53−/− mice on day 12. Compared with control mice, loss of p53 alone induced MP expansion in spleen but not in bone marrow (Figure 3A). Consistent with our previous results,30 the MP compartment in NrasG12D/+ mice was significantly expanded in both bone marrow and spleen compared with control and/or p53−/− mice. In addition, deletion of p53 resulted in a markedly increased number of MPs in NrasG12D/+ mice (Figure 3A). Importantly, the expansion of the MP compartment in NrasG12D/+p53−/− mice was not the result of increased cell proliferation (Figure 3B-C). Rather, NrasG12D/+p53−/− granulocyte-macrophage progenitors (GMPs) and MEPs showed significantly increased quiescence in G0 stage (Figure 3B). Of note, although NrasG12D/+ significantly promoted quiescence in GMPs, p53−/− did not seem to affect GMP function (Figure 3B). In contrast, NrasG12D/+ alone and p53−/− alone moderately but significantly induced MEP quiescence, whereas their simultaneous presence promoted a more profound phenotype in NrasG12D/+p53−/− MEPs (Figure 3B). These results suggest that NrasG12D/+ and p53−/− cooperate to regulate the function of MEPs.
p53 deficiency leads to further expansion, increased quiescence, and re-plating capability of NrasG12D/+myeloid progenitors. Control, p53−/−, NrasG12D/+, and NrasG12D/+p53−/− mice were treated with pI-pC and euthanized on day 12 for analysis as described in “Materials and methods.” (A) Quantification of MPs, CMPs, GMPs, and MEPs in BM and SP. (B) Cell cycle analysis of bone marrow GMPs and MEPs using Ki-67/4′,6-diamidino-2-phenylindole (DAPI) staining. (C) A 16-hour pulse of 5′-ethynyl-2′-deoxyuridine (EdU) to quantify proliferating bone marrow MPs. (D) 5 × 104 bone marrow cells isolated from control, p53−/−, NrasG12D/+, or NrasG12D/+p53−/− mice were plated in semisolid medium without cytokines or with 0.2 ng/mL of murine GM-CSF or 10 ng/mL murine interleukin-3 (IL-3). Colonies were counted 7 to 10 days after culture. (E) Methylcellulose culture of 5 × 104 bone marrow cells with 10 ng/mL murine IL-3 over 3 rounds of re-plating. Data are presented as mean ± SD. *P < .05; **P < .01; ***P < .001. n = 10∼16, a range of 10-16 mice.
p53 deficiency leads to further expansion, increased quiescence, and re-plating capability of NrasG12D/+myeloid progenitors. Control, p53−/−, NrasG12D/+, and NrasG12D/+p53−/− mice were treated with pI-pC and euthanized on day 12 for analysis as described in “Materials and methods.” (A) Quantification of MPs, CMPs, GMPs, and MEPs in BM and SP. (B) Cell cycle analysis of bone marrow GMPs and MEPs using Ki-67/4′,6-diamidino-2-phenylindole (DAPI) staining. (C) A 16-hour pulse of 5′-ethynyl-2′-deoxyuridine (EdU) to quantify proliferating bone marrow MPs. (D) 5 × 104 bone marrow cells isolated from control, p53−/−, NrasG12D/+, or NrasG12D/+p53−/− mice were plated in semisolid medium without cytokines or with 0.2 ng/mL of murine GM-CSF or 10 ng/mL murine interleukin-3 (IL-3). Colonies were counted 7 to 10 days after culture. (E) Methylcellulose culture of 5 × 104 bone marrow cells with 10 ng/mL murine IL-3 over 3 rounds of re-plating. Data are presented as mean ± SD. *P < .05; **P < .01; ***P < .001. n = 10∼16, a range of 10-16 mice.
Consistent with our quantification analysis using flow cytometry, in vitro colony assay in semisolid medium showed that NrasG12D/+p53−/− MPs formed a significantly higher number of colonies in the presence of murine granulocyte-macrophage colony-stimulating factor (mGM-CSF) or murine interleukin-3 than control, p53−/−, and NrasG12D/+ MPs, although spontaneous colony formation of NrasG12D/+p53−/− MPs in the absence of cytokines was indistinguishable from NrasG12D/+ MPs (Figure 3D). To evaluate the re-plating capability of MPs, we re-plated bone marrow cells in culture for additional rounds in vitro. NrasG12D/+ MPs showed moderately increased self-renewal vs control MPs, whereas p53−/− and NrasG12D/+p53−/− MPs displayed much higher capability to re-plate in vitro (Figure 3E). Together, our data indicate that loss of p53 significantly enhances re-plating of NrasG12D/+ MPs in vitro and leads to their further expansion in vivo.
MEPs deficient for p53 and expressing oncogenic Nras are transformed to AML-initiating cells
Because recipients of NrasG12D/+p53−/− cells developed a highly penetrant AML and NrasG12D/+p53−/− MPs acquired increased re-plating capability in vitro, we investigated whether these MPs were transformed to leukemia-initiating cells, supporting AML development in vivo. As expected, transplantation of 30 highly purified NrasG12D/+p53−/− HSCs and MPPs in irradiated recipients led to AML and T-ALL, whereas transplantation of 10 000 NrasG12D/+p53−/− MPs resulted in AML only (Table 1). Although p53−/− MPs showed re-plating capability comparable to that of NrasG12D/+p53−/− MPs in vitro, they failed to initiate AML in vivo (supplemental Table 3), suggesting that the increased self-renewal in p53−/− MPs was not leukemogenic. To test for the in vivo self-renewal potential of AML-initiating cells, we further purified CD45.1–Lin–c-Kit+ cells from primary recipients receiving NrasG12D/+p53−/− MPs and transplanted them into secondary and tertiary recipients. In both cases, these cells could efficiently induce AML formation (Jingfang Zhang, unpublished data). These results indicate that NrasG12D/+p53−/− MPs are transformed to AML-initiating cells.
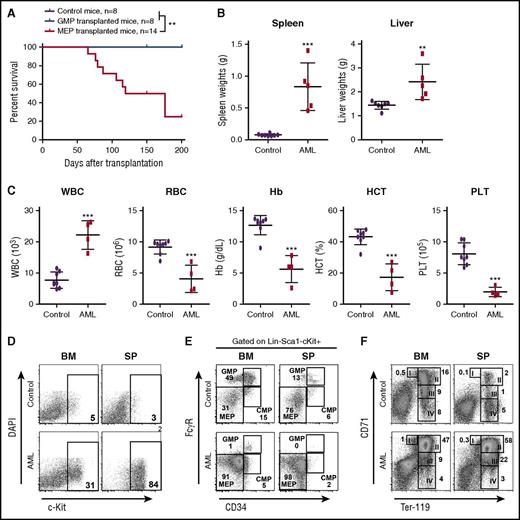
To identify which specific population of MPs was transformed to AML-initiating cells, we performed RNA sequencing (RNA-Seq) analysis of control MPs and AML MPs (defined as CD45.1–Lin–c-Kit+). Volcano analysis revealed that 2453 genes were significantly upregulated and 3604 genes were significantly downregulated in AML MPs (fold change >2 and false discovery rate [FDR] <0.05) (supplemental Figure 7A; supplemental Table 4). Gene set enrichment analysis showed that genes regulating erythroid development were significantly upregulated in AML MPs (supplemental Figure 7B), whereas biological processes involved in immune response and leukocyte function were significantly enriched in the downregulated genes (Jingfang Zhang, unpublished data). The latter observation was affirmed by gene ontology analysis of 496 upregulated and 1697 downregulated genes (fold change >16 and FDR <0.001) (supplemental Figure 7C). Therefore, we isolated GMPs and MEPs from NrasG12D/+p53−/− mice and transplanted them to irradiated recipients (Figure 4). In proportion to 10 000 MPs in our earlier experiment, we transplanted 4000 GMPs or 2000 MEPs in each recipient (Table 1). Consistent with our molecular and cellular characterizations, only NrasG12D/+p53−/− MEPs but not GMPs induced AML formation in vivo (Figure 4A; Table 1). The AML phenotypes (Figure 4B-D) were similar to those described in Figure 2. Consistent with the donor cell type, we found that AML MPs immunophenotypically resembled MEPs (Figure 4E). Despite the expansion of MEPs, erythroid differentiation was blocked at the CD71+TER119+ stage in both bone marrow and spleen (Figure 4F). Similar to p53−/− MPs, none of the p53−/− MEPs and GMPs could initiate AML in recipient mice (supplemental Table 3). Our data demonstrate that NrasG12D/+p53−/− MEPs are transformed to leukemia-initiating cells and support AML development in vivo.
MEPs deficient for p53 and expressing oncogenic Nras are transformed to AML-initiating cells. Control and NrasG12D/+p53−/− mice were treated with pI-pC and euthanized on day 12 as described in “Materials and methods.” GMPs and MEPs were isolated from NrasG12D/+p53−/− bone marrow and transplanted to lethally irradiated mice as described in Table 1. Control recipients were transplanted with 2.5 × 105 total bone marrow cells (CD45.2+) from control mice and the same number of congenic bone marrow cells (CD45.1+). (A) Kaplan-Meier comparative survival analysis of recipient mice. Cumulative survival was plotted against days after transplantation. P value was determined by the log-rank test. (B) Quantification of spleen and liver weights in moribund AML mice transplanted with NrasG12D/+p53−/− MEPs and age-matched control recipients. (C) Complete blood count analysis of peripheral blood samples collected from moribund AML mice transplanted with NrasG12D/+p53−/− MEPs and age-matched control recipients. (D-F) Analysis of donor-derived (CD45.1–) (D) blast cells (c-Kit+), (E) myeloid progenitors, and (F) erythroid differentiation in BM and SP of moribund AML mice transplanted with NrasG12D/+p53−/− MEPs and age-matched control recipients. Data are presented as mean ± SD. **P < .01; ***P < .001. Hb, hemoglobin; HCT, hematocrit; PLT, platelet; RBC, red blood cell; WBC, white blood cell.
MEPs deficient for p53 and expressing oncogenic Nras are transformed to AML-initiating cells. Control and NrasG12D/+p53−/− mice were treated with pI-pC and euthanized on day 12 as described in “Materials and methods.” GMPs and MEPs were isolated from NrasG12D/+p53−/− bone marrow and transplanted to lethally irradiated mice as described in Table 1. Control recipients were transplanted with 2.5 × 105 total bone marrow cells (CD45.2+) from control mice and the same number of congenic bone marrow cells (CD45.1+). (A) Kaplan-Meier comparative survival analysis of recipient mice. Cumulative survival was plotted against days after transplantation. P value was determined by the log-rank test. (B) Quantification of spleen and liver weights in moribund AML mice transplanted with NrasG12D/+p53−/− MEPs and age-matched control recipients. (C) Complete blood count analysis of peripheral blood samples collected from moribund AML mice transplanted with NrasG12D/+p53−/− MEPs and age-matched control recipients. (D-F) Analysis of donor-derived (CD45.1–) (D) blast cells (c-Kit+), (E) myeloid progenitors, and (F) erythroid differentiation in BM and SP of moribund AML mice transplanted with NrasG12D/+p53−/− MEPs and age-matched control recipients. Data are presented as mean ± SD. **P < .01; ***P < .001. Hb, hemoglobin; HCT, hematocrit; PLT, platelet; RBC, red blood cell; WBC, white blood cell.
AML MPs gain partial HSC signature and largely retain MEP signature
We reasoned that if NrasG12D/+p53−/− MEPs were transformed to self-renewing AML MPs, their transcriptome should be reprogrammed to reflect this phenotype. To test this hypothesis, we defined an HSC gene signature as a set of differentially expressed (DE) genes identified between long term-HSCs and MPPs by using previously published microarray data.33 The HSC signature genes were then hierarchically clustered on the basis of their expression in AML MPs (Figure 5A). Our results show that AML MPs display an HSC-like signature.
Myeloid progenitors isolated from AML mice display partial HSC signature and largely maintain MEP identity. RNA-Seq–based gene expression comparison between control myeloid progenitors (Ctrl-MP) and myeloid progenitors isolated from AML mice (AML-MP). (A) Heat map depicting relative expression of HSC signature (genes DE in long-term HSCs [LT-HSCs] vs MPPs, previously described33 ) in AML MPs. Genes in blocks a1 and a2 show similar expression profiles between AML MPs and LT-HSCs. (B) Heat map showing relative expression of MEP signature (genes differentially expressed in MEPs vs LSKs, CMPs, and GMPs, previously described34 ) in Ctrl- and AML-MPs. Genes in blocks b1 and b2 demonstrate similar expression profiles between AML-MPs and MEPs. (C) Expression levels of Gata1 and Gata2 in Ctrl- and AML-MPs were quantified using reads per kilobase of transcript per million mapped reads (RPKM). (D) Venn diagram illustrating the overlap among DE genes in AML-MPs and DE genes in Gata2−77−/− CMPs (Gata2).
Myeloid progenitors isolated from AML mice display partial HSC signature and largely maintain MEP identity. RNA-Seq–based gene expression comparison between control myeloid progenitors (Ctrl-MP) and myeloid progenitors isolated from AML mice (AML-MP). (A) Heat map depicting relative expression of HSC signature (genes DE in long-term HSCs [LT-HSCs] vs MPPs, previously described33 ) in AML MPs. Genes in blocks a1 and a2 show similar expression profiles between AML MPs and LT-HSCs. (B) Heat map showing relative expression of MEP signature (genes differentially expressed in MEPs vs LSKs, CMPs, and GMPs, previously described34 ) in Ctrl- and AML-MPs. Genes in blocks b1 and b2 demonstrate similar expression profiles between AML-MPs and MEPs. (C) Expression levels of Gata1 and Gata2 in Ctrl- and AML-MPs were quantified using reads per kilobase of transcript per million mapped reads (RPKM). (D) Venn diagram illustrating the overlap among DE genes in AML-MPs and DE genes in Gata2−77−/− CMPs (Gata2).
Next, we defined an MEP signature as DE genes in MEP compared with LSK, common MP (CMP), and GMP (P < .001) by using another set of published microarray data.34 The MEP signature genes were then hierarchically clustered on the basis of their expression in control and AML MPs (Figure 5B). We found that the MEP signature is largely maintained in AML MPs.
Consistent with the MEP signature in AML MPs, we found that Gata1 and Gata2 (2 master transcription factors involved in megakaryocyte and erythroid development) were significantly upregulated in AML MPs (Figure 5C). Recently, we discovered a Gata2 enhancer (–77) that confers Gata2 expression selectively in myeloid progenitor cells.35 Targeted deletion of −77 in mice significantly reduced the production of MEPs. RNA-Seq analysis of −77+/+ and −77−/− CMPs identified 133 differentially expressed genes. Comparison between −77-regulated genes in CMPs and genes differentially expressed between control MPs and AML MPs revealed 86 overlapping genes (Figure 5D). These 86 overlapping genes include genes that encode crucial regulators for megakaryocyte and/or erythroid development, such as Gata2,36 Gata1,37 Epor,38 Fog1,39,40 Sox6,41 and Klf1,42 which are all downregulated in −77−/− CMPs but upregulated in AML MPs.
Differentially expressed genes in AML MPs are enriched for potential p53 target genes
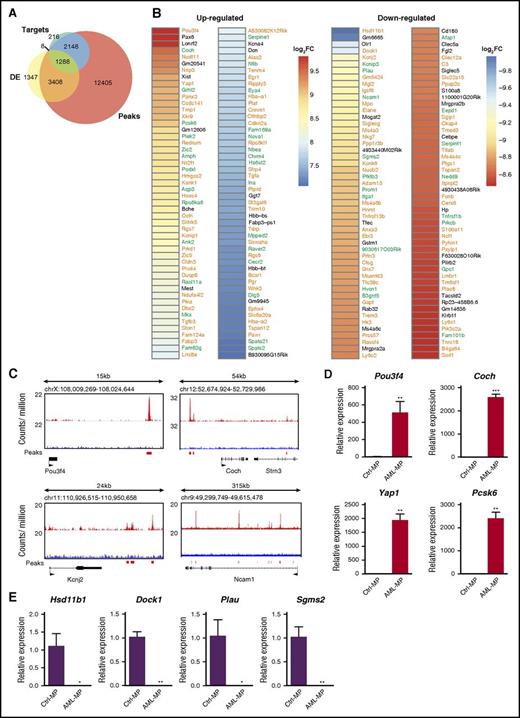
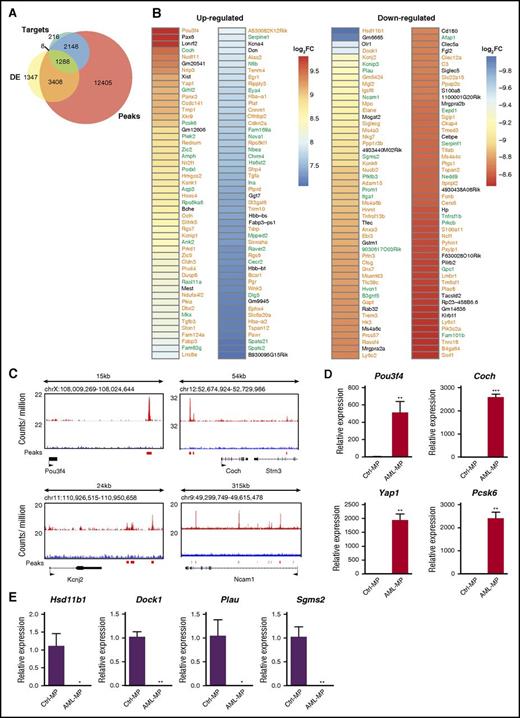
Because p53 is a master transcription factor that regulates proliferation and differentiation of stem and progenitor cells,43,44 we investigated whether the transcriptome characteristic of AML MPs could be regulated by p53 loss. By using previously published chromatin immunoprecipitation sequencing data45 that were collected from mouse embryonic stem cells treated with doxorubicin (a widely used DNA damage agent to upregulate p53), we identified 19 257 genes with significant p53 binding peaks within 25 Kb of their loci (upstream and downstream) as potential p53 target genes (“peaks” in Figure 6A). In the same published study, 3697 genes were identified as p53 direct target genes that are involved in DNA damage response (“targets” in Figure 6A). The genes included in “peaks” and “targets” were further compared with DE genes in AML MPs derived from our AML mice (fold change >2 and FDR <0.05) (DE genes in Figure 5A). We found that about 78% of the DE genes are potential p53 target genes (Figure 6A). Among them, 1288 DE genes belonged to both “targets” and “peaks,” whereas 3408 DE genes were overlapped with “peaks” only. Consistent with this observation, in the top 100 up- and downregulated DE genes based on fold change, 85 and 75 of them were potential p53 target genes, respectively (Figure 6B). Significant p53 binding peaks were confirmed in a few genes randomly selected from the list described in Figure 6B (Figure 6C). In addition, we validated the differential expression of 8 genes described in Figure 6B using quantitative reverse transcription polymerase chain reaction (Figure 6D-E). These data suggest that p53 loss may play a significant role in driving AML MP formation.
Genesdifferentially expressed in AML MPs are enriched for potential p53 target genes. (A) Venn diagrams illustrating the overlap among DE genes in AML MPs, p53 target genes involved in DNA damage response (“targets”), and the genes with p53 binding peaks close to their loci (“peaks”). “Targets” and “peaks” have been described previously.45 (B) Heat map displaying statistically significant (FDR <0.001), top 100 up- and downregulated genes in AML MPs (based on fold change). Genes in green are p53 target genes involved in DNA damage response, whereas genes in orange are genes with p53 binding peaks close to their loci but not regulated by p53 in DNA damage response. (C) Genomic views of p53 binding peaks at 4 representative gene loci. (D-E) Validation of differentially expressed genes in AML MPs vs control MPs using quantitative reverse transcriptase polymerase chain reaction (qRT-PCR). Total RNAs were extracted from sorted control and AML MPs using RNEasy Mini Kit (Qiagen). Complementary DNAs (cDNAs) were synthesized using iScript cDNA Synthesis Kit (Bio-Rad). qRT-PCR was performed by using PrimeTime qPCR Assay (ITD; Integrated DNA Technologies) and Maxima Probe qPCR Master Mix (Thermo Scientific) on a CFX96 Real-Time System (Bio-Rad) according to the manufacturer’s instructions. Quantification results of (D) 4 up- and (E) 4 downregulated genes in AML MPs vs control MPs are shown here. Data are presented as mean ± SD. *P < .05; **P < .01; ***P < .001. Log2FC, fold change expressed as 2-based log units.
Genesdifferentially expressed in AML MPs are enriched for potential p53 target genes. (A) Venn diagrams illustrating the overlap among DE genes in AML MPs, p53 target genes involved in DNA damage response (“targets”), and the genes with p53 binding peaks close to their loci (“peaks”). “Targets” and “peaks” have been described previously.45 (B) Heat map displaying statistically significant (FDR <0.001), top 100 up- and downregulated genes in AML MPs (based on fold change). Genes in green are p53 target genes involved in DNA damage response, whereas genes in orange are genes with p53 binding peaks close to their loci but not regulated by p53 in DNA damage response. (C) Genomic views of p53 binding peaks at 4 representative gene loci. (D-E) Validation of differentially expressed genes in AML MPs vs control MPs using quantitative reverse transcriptase polymerase chain reaction (qRT-PCR). Total RNAs were extracted from sorted control and AML MPs using RNEasy Mini Kit (Qiagen). Complementary DNAs (cDNAs) were synthesized using iScript cDNA Synthesis Kit (Bio-Rad). qRT-PCR was performed by using PrimeTime qPCR Assay (ITD; Integrated DNA Technologies) and Maxima Probe qPCR Master Mix (Thermo Scientific) on a CFX96 Real-Time System (Bio-Rad) according to the manufacturer’s instructions. Quantification results of (D) 4 up- and (E) 4 downregulated genes in AML MPs vs control MPs are shown here. Data are presented as mean ± SD. *P < .05; **P < .01; ***P < .001. Log2FC, fold change expressed as 2-based log units.
AML MPs acquire overexpression of oncogenic Nras
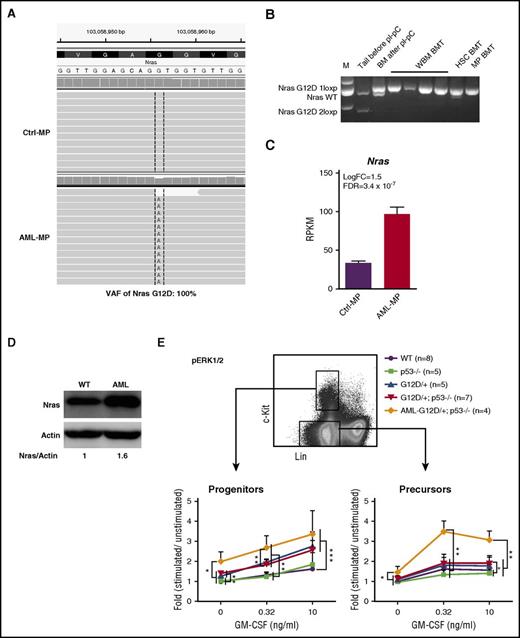
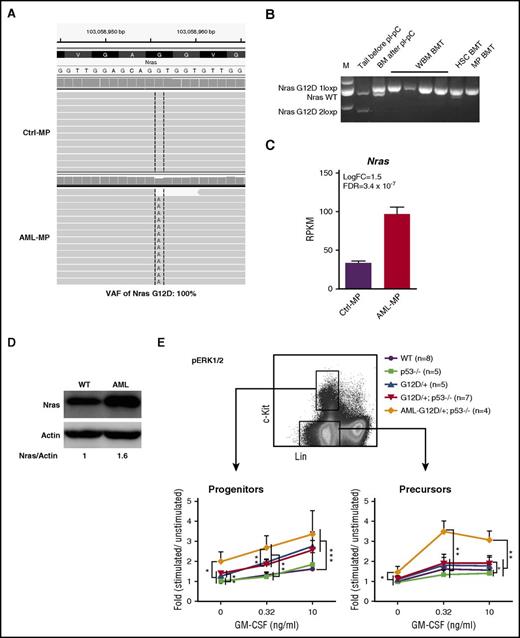
Although p53 deletion contributed to the transformation of MEPs to AML MPs, p53−/− MPs failed to induce AML in vivo, suggesting that additional genetic mutations, such as oncogenic Nras mutation, synergize with p53−/− to promote AML development. To uncover potential cooperating genetic mutations, we re-analyzed RNA-Seq data from AML MPs using compiled RNA-Seq results from control MPs as a baseline. Our analysis revealed that AML MPs isolated from 4 AML mice expressed only NrasG12D mutation without wild-type (wt) Nras (variant allele frequency, 100%; Figure 7A). No additional structural or genetic alterations were identified. We further confirmed this observation in multiple AML samples isolated from independent recipients (Figure 7B). Moreover, oncogenic Nras was significantly overexpressed in AML MPs at both the transcriptional (Figure 7C) and protein (Figure 7D) levels. These results suggest that overexpression of oncogenic Nras cooperates with p53 deficiency to promote AML.
Overexpression of oncogenic Nras leads to hyperactivation of ERK1/2 signaling in AML MPs. (A) Representative sequencing results from RNA-Seq analysis show UPD of NrasG12D mutation in AML MPs. (B) Genotyping analysis of the Nras locus in multiple AML samples isolated from independent recipients (spleen weight, >1 g). (C) RPKM of Nras in control and AML MPs. (D) Western blot analysis of Nras expression levels in AML and wt spleens. (E) Whole bone marrow cells isolated from control, p53−/−, NrasG12D/+, and NrasG12D/+p53−/− mice on day 12 and moribund AML NrasG12D/+p53−/− recipients were serum and cytokine starved for 2 hours and stimulated with various concentrations of murine GM-CSF (0, 0.32, and 10 ng/mL) at 37°C for 10 minutes. Levels of phosphorylated ERK1/2 (p-ERK1/2) were measured using phosphor-specific flow cytometry. Non-neutrophil bone marrow cells were gated for data analysis. Myeloid progenitors are enriched in Lin–/lowc-Kit+ cells, whereas myeloid precursors are enriched in Lin–/lowc-Kit– cells. To quantify the activation of ERK1/2, median intensities of p-ERK1/2 at different GM-CSF concentrations are compared with their respective control cells at 0 ng/mL, which is arbitrarily set at 1. Data are presented as mean ± SD. *P < .05; **P < .01; ***P < .001.
Overexpression of oncogenic Nras leads to hyperactivation of ERK1/2 signaling in AML MPs. (A) Representative sequencing results from RNA-Seq analysis show UPD of NrasG12D mutation in AML MPs. (B) Genotyping analysis of the Nras locus in multiple AML samples isolated from independent recipients (spleen weight, >1 g). (C) RPKM of Nras in control and AML MPs. (D) Western blot analysis of Nras expression levels in AML and wt spleens. (E) Whole bone marrow cells isolated from control, p53−/−, NrasG12D/+, and NrasG12D/+p53−/− mice on day 12 and moribund AML NrasG12D/+p53−/− recipients were serum and cytokine starved for 2 hours and stimulated with various concentrations of murine GM-CSF (0, 0.32, and 10 ng/mL) at 37°C for 10 minutes. Levels of phosphorylated ERK1/2 (p-ERK1/2) were measured using phosphor-specific flow cytometry. Non-neutrophil bone marrow cells were gated for data analysis. Myeloid progenitors are enriched in Lin–/lowc-Kit+ cells, whereas myeloid precursors are enriched in Lin–/lowc-Kit– cells. To quantify the activation of ERK1/2, median intensities of p-ERK1/2 at different GM-CSF concentrations are compared with their respective control cells at 0 ng/mL, which is arbitrarily set at 1. Data are presented as mean ± SD. *P < .05; **P < .01; ***P < .001.
To further validate our genetic findings, we measured oncogenic Nras-mediated GM-CSF signaling from different groups of animals. Consistent with previous reports,22,46 GM-CSF signaling was largely normal in NrasG12D/+ cells on day 12 (Figure 7E). Similar to our colony assay result, GM-CSF signaling in p53−/− cells was comparable to that in control cells, and GM-CSF signaling in NrasG12D/+p53−/− cells was indistinguishable from that in NrasG12D/+ cells (Figure 7E). In contrast, compared with NrasG12D/+ and NrasG12D/+p53−/− cells, ERK1/2 signaling at the basal level and evoked by GM-CSF was significantly hyperactivated in AML MPs that overexpressed oncogenic Nras (Figure 7E). Together, our data suggest that overexpression of oncogenic Nras, through UPD of the oncogenic Nras allele and possibly additional epigenetic regulation, leads to hyperactivation of ERK1/2 signaling and synergizes with p53−/− to induce AML.
Discussion
In this study, we showed that RAS pathway and TP53 mutations co-occur in clinical AML samples and confer a dismal overall outcome. Consistent with this clinical observation, primary NrasG12D/+p53−/− mice and recipients transplanted with NrasG12D/+p53−/− cells rapidly develop a highly penetrant AML. NrasG12D/+ cooperates with p53−/− to promote increased quiescence in MEPs but not in GMPs. Consequently, NrasG12D/+p53−/− MEPs are transformed to AML-initiating cells. These cells acquire an HSC-like signature while largely maintaining an MEP signature. During AML development, transformed MEPs gain overexpression of oncogenic Nras, which leads to hyperactivation of ERK1/2 signaling in AML cells. Therefore, loss of p53 synergizes with oncogenic Nras to promote AML development (supplemental Figure 8).
Although our donor cells began with NrasG12D/+, the wt Nras allele was lost in most of the AML samples (Figure 7A-B). Moreover, oncogenic Nras is overexpressed in AML MPs (Figure 7C-D). We believe that both genetic and epigenetic mechanisms contribute to the overexpression of oncogenic Nras. Although we cannot definitively conclude that loss of the Nras wt allele is the result of UPD of the oncogenic allele, we certainly favor this possibility on the basis of several observations. First, UPD of the oncogenic Nras allele has been documented in both human and mouse CMML.22,23 Second, homozygocity of the oncogenic Ras allele promotes human and mouse JMML/CMML progression,30,47 whereas loss of wt Nras does not have an impact on oncogenic Nras-induced leukemogenesis.48 Of note, although UPD of the oncogenic Nras allele is identified in human and mouse CMML, it promotes only CMML development but fails to induce AML on its own.22 Similarly, p53−/− alone does not confer leukemogenic self-renewal of MPs (supplemental Table 1). Consistent with a previous report showing that p53 loss enables aberrant self-renewal of KrasG12D/+ MPs,49 our study identified a significant synergy between p53−/− and enhanced oncogenic Nras signaling in AML development.
It was hypothesized that p53 loss synergizes with oncogenic Ras during tumorigenesis because overexpression of oncogenic Ras leads to p53-mediated cell senescence, and p53 loss overcomes this cellular defect to promote tumor formation.50 However, expression of oncogenic Ras from its endogenous locus does not induce cell senescence in vivo or in vitro.51,52 Therefore, the synergy between p53 loss and oncogenic Ras must be through other mechanisms. In our NrasG12D/+p53−/−-induced AML model, we believe that oncogenic Nras signaling synergizes with p53 loss to promote leukemogenic transformation of MEPs. We found that although NrasG12D/+ significantly promotes quiescence in GMPs, p53−/− does not seem to affect GMP function (Figure 3B). This observation is consistent with the high mutation frequency of NRAS and rare TP53 mutations identified in CMML patients.10 In contrast, NrasG12D/+ alone and p53−/− alone moderately but significantly induce MEP quiescence, whereas their simultaneous presence promotes a more profound phenotype in NrasG12D/+p53−/− MEPs (Figure 3B). In support of our conclusion, a recent study of leukemic transformation of MPNs to AML demonstrated that overexpression of JAK2V617F mutant in p53−/− bone marrow cells transforms MEPs to AML-initiating cells.11
Although we could not definitely show that MEPs are transformed in human AML patients with concurrent RAS pathway and TP53 mutations, the observation that transformed MEPs led to anemia and thrombocytopenia in our model (Figures 2D and 4F) is highly consistent with the clinical findings in humans that TP53 mutations are significantly associated with erythroid abnormalities in MDSs and CMML12,13 and that NRAS and TP53 mutations are closely associated with severe thrombocytopenia in MDSs.6 Our clinical data demonstrate that patients with TP53 and RAS pathway mutations have an aggressive clinical course that is likely more adverse than that with either mutation alone (Figure 1). The murine model reported herein is highly consistent with this clinical observation and may serve as a tool to preclinically credential candidate therapeutics for this genetic subgroup of patients in future studies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the University of Wisconsin Carbone Comprehensive Cancer Center for use of its Shared Services (Flow Cytometry Core and Histology Laboratory) to complete this research.
This work was supported by startup funds from University of Wisconsin-Madison and a National Science Foundation CAREER award (MCB-1552455) (Xuehua Zhong), National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases grant no. DK68634 (E.H.B.), the American Society of Hematology Scholar Award and MDS Evans Foundation Fellow award (E.P.), and grant nos. R01CA152108 and R01HL113066, and a Scholar Award from the Leukemia & Lymphoma Society (Jing Zhang) This work was also supported in part by National Institutes of Health National Cancer Institute (NCI) Comprehensive Cancer Center Support grant no. P30 CA014520-UW. The patient sample (CMML-OSU) for this study was provided by the Ohio State University Comprehensive Cancer Center Leukemia Tissue Bank, supported by the NCI grant no. P30 CA016058.
Authorship
Contribution: Jingfang Zhang, E.P., and Jing Zhang conceived and designed the study; Jingfang Zhang and G.K. acquired the mouse data; Jingfang Zhang, G.K., L.L., Xinmin Zhang, E.A.R., A.R., X.G., Y.L., E.H.B., Xuehua Zhong, and Jing Zhang analyzed and interpreted the mouse data; J.S., M.H., and E.P. analyzed the human clinical data; Jingfang Zhang, L.L., E.A.R., X.G., E.B., Xuehua Zhong, E.P., and Jing Zhang wrote, reviewed, and/or revised manuscript; Y.L., J.W., Y.-I.C., K.D.J., Y.Z., D.Y., B.B., D.M.L., and Y.L. provided technical or material support; and E.P. and Jing Zhang supervised the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Eric Padron, MRCWEST, 12902 Magnolia Dr, Tampa, FL 33629; e-mail: eric.padron@moffitt.org; and Jing Zhang, University of Wisconsin-Madison, McArdle Laboratory for Cancer Research, Room 7453, Wisconsin Institutes for Medical Research II, 1111 Highland Ave, Madison, WI 53705; e-mail: zhang@oncology.wisc.edu.
References
Author notes
Jingfang Zhang and G.K. contributed equally to this study.

![Figure 5. Myeloid progenitors isolated from AML mice display partial HSC signature and largely maintain MEP identity. RNA-Seq–based gene expression comparison between control myeloid progenitors (Ctrl-MP) and myeloid progenitors isolated from AML mice (AML-MP). (A) Heat map depicting relative expression of HSC signature (genes DE in long-term HSCs [LT-HSCs] vs MPPs, previously described33) in AML MPs. Genes in blocks a1 and a2 show similar expression profiles between AML MPs and LT-HSCs. (B) Heat map showing relative expression of MEP signature (genes differentially expressed in MEPs vs LSKs, CMPs, and GMPs, previously described34) in Ctrl- and AML-MPs. Genes in blocks b1 and b2 demonstrate similar expression profiles between AML-MPs and MEPs. (C) Expression levels of Gata1 and Gata2 in Ctrl- and AML-MPs were quantified using reads per kilobase of transcript per million mapped reads (RPKM). (D) Venn diagram illustrating the overlap among DE genes in AML-MPs and DE genes in Gata2−77−/− CMPs (Gata2).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/129/3/10.1182_blood-2016-06-719237/4/m_blood719237f5.jpeg?Expires=1769290118&Signature=HG2mG3z-8qNTGCR-krOZa0Rix7ErFQfEpzqjvxH8vCKJ010wbGny7k4UqN4mm3wwHdr7Iy3uEszWxzSqeRmJHnE8A1OqKHMWEhFupJYwj0sGb4gzwCNRfuj0a7XE-ag4ESX7H53ZI6w1JkMy0VM1ceuRuyjnyoF2wORel0XF4NsUhNw4zR1GFap7-kUR-hZ7ajFh6HkqIOxpYyu1fGyLvtBh1cnq4MELMuDFHKEVWGcJe29NeTOP~PufigvmE-cbAZ4LryHWrNs86nldbc49V6ijjkUHLy14771cS7RVJ3A2YMMg3G6baL4D0ORcg-mJ9JF2HLJCs1rA-sxAvNbgDA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 5. Myeloid progenitors isolated from AML mice display partial HSC signature and largely maintain MEP identity. RNA-Seq–based gene expression comparison between control myeloid progenitors (Ctrl-MP) and myeloid progenitors isolated from AML mice (AML-MP). (A) Heat map depicting relative expression of HSC signature (genes DE in long-term HSCs [LT-HSCs] vs MPPs, previously described33) in AML MPs. Genes in blocks a1 and a2 show similar expression profiles between AML MPs and LT-HSCs. (B) Heat map showing relative expression of MEP signature (genes differentially expressed in MEPs vs LSKs, CMPs, and GMPs, previously described34) in Ctrl- and AML-MPs. Genes in blocks b1 and b2 demonstrate similar expression profiles between AML-MPs and MEPs. (C) Expression levels of Gata1 and Gata2 in Ctrl- and AML-MPs were quantified using reads per kilobase of transcript per million mapped reads (RPKM). (D) Venn diagram illustrating the overlap among DE genes in AML-MPs and DE genes in Gata2−77−/− CMPs (Gata2).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/129/3/10.1182_blood-2016-06-719237/4/m_blood719237f5.jpeg?Expires=1769376680&Signature=1xFYTiOdTWl4V8WmpjY~nX3evuMCAqoTawM7OrAkZLsz9qk0cml4RE2EfI-djBYiOiC4cybG3VP3XOexdQcYZo8TxsOqebPKSR8bxzpaT3SIQF6Mx1s6mTZPiWV0S3S9F9suabqnhnfJaSyAslvfHCbEijkLU5aeFiyuzt~TfCS8dVBTRKc2cLljnfWXMKlpjxnFrRsJzFTgla4uzKKgbP2WVVTZtAbSMlwNkAHFTCZgQH5WI0Wxz7qdOwM8u9-vdJMOoxLqsKUwErwkY24Jh~FTzoD~upmby-Fn5QTV17ujlNywK1cvA5DJep~Q3eMOqBAev-PBpKo~PEABSCf-cA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)