Key Points
The intensified standard-of-care regimens for younger patients with MCL do not overcome the deleterious effects of TP53 mutations.
MCLs with TP53 mutations should be considered for alternative frontline treatment.
Abstract
Despite recent advances in lymphoma treatment, mantle cell lymphoma (MCL) remains incurable, and we are still unable to identify patients who will not benefit from the current standard of care. Here, we explore the prognostic value of recurrent genetic aberrations in diagnostic bone marrow (BM) specimens from 183 younger patients with MCL from the Nordic MCL2 and MCL3 trials, which represent current standard-of-care regimens. In the univariate model, mutations of TP53 (11%) and NOTCH1 (4%), and deletions of TP53 (16%) and CDKN2A (20%), were significantly associated with inferior outcomes (together with MIPI, MIPI-c, blastoid morphology, and Ki67 > 30%); however, in multivariate analyses, only TP53 mutations (HR, 6.2; P < .0001) retained prognostic impact for overall survival (OS), whereas TP53 mutations (HR, 6.9; P < .0001) and MIPI-c high-risk (HR, 2.6; P = .003) had independent prognostic impact on time to relapse. TP53-mutated cases had a dismal outcome, with a median OS of 1.8 years, and 50% relapsed at 1.0 years, compared to a median OS of 12.7 years for TP53-unmutated cases (P < .0001). TP53 mutations were significantly associated with Ki67 > 30%, blastoid morphology, MIPI high-risk, and inferior responses to both induction- and high-dose chemotherapy. In conclusion, we show that TP53 mutations identify a phenotypically distinct and highly aggressive form of MCL with poor or no response to regimens including cytarabine, rituximab, and autologous stem-cell transplant (ASCT). We suggest patients with MCL should be stratified according to TP53 status, and that patients with TP53 mutations should be considered for experimental frontline trials exploring novel agents.
Introduction
Mantle cell lymphoma (MCL) is a rare type of non-Hodgkin lymphoma (NHL), which accounts for 5% to 8% of all NHLs. Historically, MCL was associated with a dismal outcome, with a median overall survival (OS) of only 3 to 5 years1,2 ; however, during the past decades, the outcome, especially for younger patients, has improved substantially by an intensified frontline regimen including cytarabine, rituximab, and consolidation with high-dose therapy and autologous stem-cell transplant (ASCT).1,3,4 Hence, in the recent long-term follow-up of such a regimen, the Nordic MCL2 trial, we observed a median OS and progression-free survival (PFS) of 12.7 and 8.5 years, respectively.5 Despite this marked improvement, MCL remains an incurable, albeit heterogeneous, disease with a wide span of early and late relapses. However, none of the existing risk stratification systems has yet been incorporated into clinical decision making.4,5
The most frequently used clinical prognosticator, the MCL International Prognostic Index (MIPI), has been thoroughly validated to define 3 groups of patients with diverse prognoses, and has recently been refined to include Ki67 expression (then called MIPI-c) with the 10% to 13% of patients who are high-risk, showing an exceedingly poor outcome.6-8 Also, blastoid cases of MCL have recurrently been associated with more aggressive disease and poorer outcome.7,9
Molecular studies of MCL demonstrate recurrent aberrations in genes involved in the regulation of the cell cycle, DNA repair, and epigenetics.10-13 In recent years, next-generation sequencing has led to comprehensive mutational characterization of MCL; however, most studies have been performed in either small or diverse patient cohorts, limiting the translational impact of the findings.10,14,15 Nonetheless, TP53 mutations have recurrently demonstrated negative prognostic impact for the outcome of patients with MCL, and more recently, NOTCH1 and NOTCH2 mutations have been associated with inferior outcomes.14-18 Furthermore, del(17p13) and del(9p21) (harboring TP53 and CDKN2A, respectively) have recurrently been associated with poor outcome.12,17,19-21 Recently, Delfau-Larue et al22 validated this in a large homogenous cohort of patients from the European-MCL Younger trial, but with no information about TP53 mutations.
Here we aim to describe the prognostic effect of the most common genomic alterations of MCLs in the large, homogenously treated cohorts from 2 Nordic trials, MCL2 and MCL3. Both regimens represent current standard-of-care regimens for younger patients, and thus hold important information for the daily handling of patients with MCL.
Materials and methods
Patients
Three hundred twenty patients were included in the Nordic MCL2 and MCL3 trials from 2000 to 2005 and 2005 to 2009, respectively.3,23 Patients were younger than 66 years, had stage II-IV MCL, and were considered fit for ASCT. Diagnostic specimens underwent central pathological review according to World Health Organization criteria. All samples were required to express Cyclin-D1 or to carry the t(11;14) translocation. Both of the MCL2 and MCL3 regimens consisted of an induction phase of alternating R-maxi-CHOP (cyclophosphamide, doxorubicin, vincristine, and prednisone) and R-high-dose cytarabine, followed by high-dose chemotherapy with BEAM (bis-chloroethylnitrosourea, etoposide, cytarabine, and melphalan) or BEAC (bis-chloroethylnitrosourea, etoposide, cytarabine, and cyclophosphamide) and ASCT.3,23 Patients with an available minimal residual disease (MRD) marker by nested polymerase chain reaction (PCR) for translocation t(11;14) or clonal immunoglobulin heavy chain rearrangement were monitored during follow-up and treated preemptively with rituximab on MRD-positivity without concurrent clinical relapse.24 Patients from the MCL3 protocol in complete remission unconfirmed (CRu)/partial remission pre-ASCT received Zevalin to optimize the induction response; however, this did not alter the outcome, and thus the combined cohorts are here considered as one.3,23 The studies were approved by local ethical committees, and all patients signed informed consent.
One hundred eighty-three patients from the combined MCL2 and MCL3 cohort, with available DNA from diagnostic bone marrow (BM) samples, were included in the genomic studies. Of these, deletions were analyzed in 177 patients, and mutations in 176. Both were analyzed in 170 patients.
Next-generation sequencing
DNA was extracted from fresh frozen pretreatment BM specimens using QIAprep Miniprep (Qiagen, Valencia, CA). Samples were analyzed using a custom-designed multiplex Ion Ampliseq panel (Ampliseq designer, Thermo Fischer Scientific, Waltham, MA) including selected coding regions, splice sites, and untranslated regions of the following 8 genes: ATM, KMT2D, CCND1, TP53, WHSC1, BIRC3, NOTCH1, and NOTCH2 (supplemental Table 1, available on the Blood Web site). The gene panel was constructed on the basis of previous whole-exome and whole-genome studies.10,14 Initial DNA quantification was carried out using Qubit broad range assay. Ion Ampliseq technology was used for library construction, and quantification of the final library was performed by qPCR, using a TaqMan Ion library quantification kit. Template preparation was automatically carried out on the Ion Chef Instrument, using Hi-Q technology and reagents. Seven samples with unique barcodes were simultaneously loaded on a 318 Ion Chip. Subsequent sequencing was carried out on the Ion PGM System using Hi-Q technology. All steps were carried out according to manufacturer’s instructions, and reagents and equipment were manufactured by Thermo Fischer Scientific.
Variant calling
Cutoff for calling a somatic variant was 5%. Median coverage for all samples was 2900×, and the limit for calling a variant was set to 400×. Variants were carefully investigated using the IGV software (Broad Institute). Common single nucleotide polymorphisms (SNPs; as reported >1% by dbSNP database) were excluded from analyses. We report only nonsynonymous mutations and mutations in splice site region or untranslated regions that were not reported in the SNP databases. Eight variants have been reported both as rare SNPs and as missense mutations, and are reported here as mutations, but with references to both COSMIC-ID and dbSNP-ID shown in supplemental Table 2. Seven mutations were verified by Sanger sequencing because of coverage below 400×.
Three TP53 mutations with variant allele frequencies of 3% to 5% were detected. Because of the apparent effect of TP53 mutations, these 3 mutations were validated in a specific TP53 panel on the Ion Torrent platform, and hence were included in all subsequent analyses (supplemental Table 2).
Deletion analysis
TP53 and CDKN2A deletions were identified by droplet digital PCR, using the QX200 system (Bio-Rad Laboratories, Hercules, CA). Fifty nanograms of DNA were analyzed using PrimePCR droplet digital PCR copy number variation (CNV) assays with RPP30 as reference locus, according to the manufacturer’s instructions (Bio-Rad). CNs were calculated using the QuantaSoft software. The dynamic range of the instrument and threshold settings were determined by analysis of blood DNA from 6 healthy donors. CN loss was defined as CN < 1.95. Each sample was analyzed at least twice and scored manually. The person who performed the data analysis and evaluation was blinded for patient characteristics.
Statistics
Statistical analyses were performed in SPSS 22.0 for Windows and GraphPad Prism 7.02 for Windows (GraphPad). To compare demographics, laboratory tests, and mutational status between the 2 groups, we used Pearson’s χ-squared test or Fischer’s exact test for dichotomous variables. Overall survival (OS), progression-free survival (PFS), and cumulative incidence of relapse (CIR) were used as outcomes for all prognostic analyses. Starting point for all 3 was date of treatment start, and endpoint for OS was date of death of any cause, endpoint for PFS was date of documented relapse or progression or death of any cause, and endpoint for CIR was date of documented relapse or progression of MCL. The log-rank test was used to compare the outcome of groups in univariate analyses, and Cox regression analysis was used for multivariate analyses. Differences were considered statistically significant when P < .05.
Results
Patient characteristics
Three hundred twenty patients were enrolled in the MCL2 and MCL3 trials.3,23 One patient from the MCL2 trial has since been withdrawn because of a change of diagnosis.5 Of the remaining 319 patients, the median age was 57 years (range 29-65), 76% were male, 51% were MIPI high- or intermediate-risk, 18% had blastoid morphology, and 41% had Ki67 ≥ 30%.
With a median follow-up time of 9.2 years for all 319 patients, median OS and PFS were 12.5 and 8.2 years, respectively, and 50% of the patients had relapsed at 10.2 years. In line with previous reports, blastoid morphology, Ki67 ≥ 30%, and higher MIPI and MIPI-c risk groups were significantly associated with poorer outcomes (supplemental Figure 1; supplemental Table 3). Importantly for the subsequent genetic analyses, we did not observe any differences in outcome among patients with or without BM involvement by morphological review (supplemental Table 3).
Genetic findings
Of the combined cohort of 319 patients, DNA was available for 191, and 183 samples were of sufficient quality for the subsequent genetic analyses. The study cohort was selected only by availability of DNA and did not differ from the entire MCL2 and MCL3 cohort in terms of baseline characteristics (Table 1). However, a significantly higher proportion of patients with available DNA achieved a complete remission after induction therapy (P = .004), and there was a trend toward superior OS, but no differences in PFS and CIR.
Deletions.
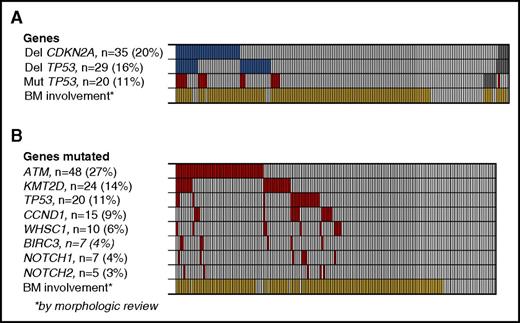
On the basis of the findings in a recent study, we investigated the presence of TP53 and CDKN2A deletions, which had both shown independent, prognostic impact.22 Deletions of CDKN2A were detected in 35 (20%) of 177 patients, and deletions of TP53 in 29 (16%) of 176, and both deletions were detected in 12 (7%) of 176 patients. In total, deletion of either gene was detected in 52 patients (30%) (Figure 1A). The applied method did not discriminate between homo- and heterozygous deletions.
Deletions and point mutations in MCL2 and MCL3 patients. (A) Deletions of CDKN2A and TP53 (blue) and mutations of TP53 for comparison (red), according to BM involvement of MCL by morphologic review (yellow). Gray boxes indicate samples not tested. (B) Mutation frequency of 8 selected genes. *BM involvement by morphologic assessment.
Deletions and point mutations in MCL2 and MCL3 patients. (A) Deletions of CDKN2A and TP53 (blue) and mutations of TP53 for comparison (red), according to BM involvement of MCL by morphologic review (yellow). Gray boxes indicate samples not tested. (B) Mutation frequency of 8 selected genes. *BM involvement by morphologic assessment.
Mutations.
By targeted sequencing of 8 genes recurrently mutated in MCL, we detected a total of 154 mutations in the 176 patient samples examined, with ATM (27%), KMT2D (14%), TP53 (11%), and CCND1 (9%) being the most frequently mutated genes. Ninety (51%) patients carried at least 1 mutated gene, and 33 (19%) carried more than 1 mutated gene (Figure 1B).
Bone marrow involvement
By the original morphologic assessment, 144 (79%) patients had BM involvement at diagnosis. In the mutational analyses, morphological BM involvement was significantly associated with the presence of mutations in any of the genes tested (P < .0001); however, mutations were detected in 8 patients with no morphological BM involvement (Figure 1B; supplemental Table 2). In addition, we identified TP53 and/or CDKN2A deletions in 45 (33%) of 138 (33%) patients with morphologic BM involvement compared with 7 (18%) of 38 patients with no morphologic BM involvement (P = .09; Figure 1A).
Prognostic relevance of genetic aberrations
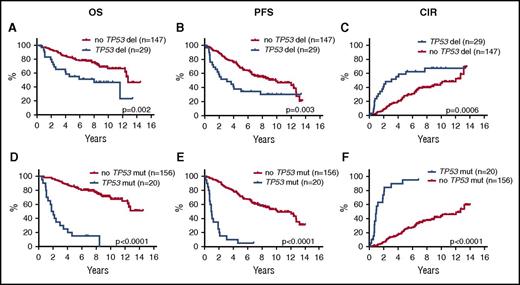
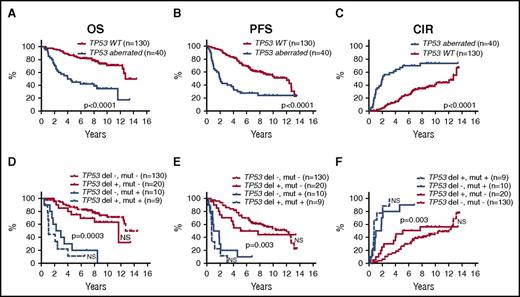
In univariate log-rank analyses, TP53 deletions, CDKN2A deletions, TP53 mutations, and NOTCH1 mutations were each significantly associated with poorer outcome, and WHSC1 mutations were associated with shorter OS, but were not significant for PFS or CIR (Figure 2; supplemental Figure 2; supplemental Table 4). Likewise, the combined TP53 aberrations (23%) (mutations and deletions) and the combined NOTCH1 and NOTCH2 mutations (7%) were associated with poorer outcome (Figure 3A-C; supplemental Table 4).
Prognostic impact of TP53 deletions and mutations on OS, PFS, and CIR. Kaplan-Meier estimates of OS, PFS, and CIR stratified by the presence or absence of TP53 deletions (A-C) and TP53 mutations (D-F).
Prognostic impact of TP53 deletions and mutations on OS, PFS, and CIR. Kaplan-Meier estimates of OS, PFS, and CIR stratified by the presence or absence of TP53 deletions (A-C) and TP53 mutations (D-F).
Prognostic impact of TP53 aberrations. Kaplan-Meier estimates of OS, PFS, and CIR according to the presence of the either TP53 aberrations (deletions and/or mutations) compared with WT TP53 (A-C), and stratified into 4 groups by the presence or absence of TP53 deletions and mutations, respectively (D-F). P values indicate log-rank tests of adjacent curves. NS, not significant.
Prognostic impact of TP53 aberrations. Kaplan-Meier estimates of OS, PFS, and CIR according to the presence of the either TP53 aberrations (deletions and/or mutations) compared with WT TP53 (A-C), and stratified into 4 groups by the presence or absence of TP53 deletions and mutations, respectively (D-F). P values indicate log-rank tests of adjacent curves. NS, not significant.
The presence of TP53 mutations was significantly associated with NOTCH1 mutations (5 of 7 NOTCH1 mutations; P = .0002), deletions of CDKN2A (11 of 35 CDKN2A deletions; P = .0002), and TP53 deletions (9 of 29 TP53 deletions; P = .001). Of 169 patients, 39 (23%) carried TP53 mutations and/or deletions, and of these, 9 (5%) carried both aberrations (Figures 1A and 3).
In the multivariate Cox regression analyses (n = 147), only TP53 mutations (HR, 6.2; P < .0001) showed independent prognostic effect for OS, whereas for PFS and CIR, both TP53 mutations (PFS: HR, 6.8 [P < .0001]; CIR: HR, 6.9 [P < .0001]) and MIPI-c high-risk (PFS: HR, 2.2 [P = .01]; CIR: HR, 2.6 [P < .003]) had significant prognostic impact (Table 2). The MIPI-c index was included as a bimodal variable (high-risk or not) to obtain the highest effect (MIPI-c included as categorical variable did not show significant individual effect; data not shown). Surprisingly, MIPI high-risk and Ki67 ≥ 30% included as separate values did not show independent prognostic value (data not shown). All factors with P < .05 in univariate analyses were included in the multivariate analyses: TP53 and NOTCH1 mutations, TP53 and CDKN2A deletions, blastoid morphology, and MIPI-c high-risk. WHSC1 was only included for OS, as it had no significant effect for PFS and CIR in the univariate log-rank analyses (Table 2; supplemental Table 4).
Combining TP53 mutations and deletions in the multivariate analyses also showed significant prognostic value (OS: HR, 3.1 [P = .0004]; PFS: HR, 2.8 [P < .0001]; CIR: HR, 3.1 [P < .0001]). The different influence on outcome between TP53 mutations and deletions is illustrated in Figure 3D-F. Inclusion of the combined NOTCH1 and NOTCH2 mutations did not change the multivariate results (data not shown).
Characteristics of patients with MCL with TP53 mutations
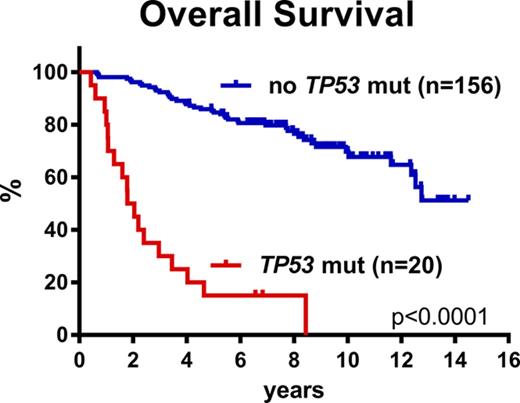
The above multivariate analyses clearly demonstrated that cases with TP53 mutations represent a unique subset of MCL. Stratification of all 176 patients according to TP53 mutational status revealed 2 cohorts of very different outcomes. The median OS for the TP53-mutated cases was 1.8 years, median PFS was 0.9 years, and median time to relapse was 1.0 years compared with a median OS not reached, median PFS of 10.2 years, and median time to relapse of 12.3 years for the TP53-unmutated cases (Figure 2D-F; supplemental Table 4). Comparably, the median OS, PFS, and time to relapse were 8.0, 3.1, and 3.1 years for the TP53-deleted cases, respectively (Figures 2A-C and 3D-F). To gain further insight into the specific characteristics of patients with and without TP53 mutations, we analyzed these cohorts separately.
Nineteen patients carried a single TP53 mutation, and 1 patient carried 2. Of the 21 detected mutations, 16 were missense mutations in the DNA binding domain (supplemental Table 2).
TP53-mutated cases displayed highly aggressive baseline characteristics and were highly associated with blastoid morphology, Ki67 ≥ 30%, MIPI high-risk, and MIPI-c high-risk, and significantly fewer TP53-mutated patients achieved CR after induction chemotherapy and ASCT, respectively (Table 3). Furthermore, MRD evaluation showed a significant association between PCR positivity and TP53 mutations post-ASCT and pre-ASCT (Table 3).
Comparing TP53 mutations with already available data on micro-RNA expression from the same cohort,25 we found a significant correlation of TP53 mutations and higher expression of miR-18b and miR-378d, but not miR-92a-3p (P = .001, P = .043, and P = .20, respectively) (supplemental Table 6).
We found no significant relation of TP53 mutations to IGHV mutational data (available for 93 patients, including 14 with TP53 mutations; data not shown).
One patient (no. 3098; supplemental Table 2) carried a TP53 mutation, p.Pro191Arg (c.572C>G), with variant allele frequency 48.9% despite no morphologic BM involvement. This mutation had not previously been reported in relation to hematological cancer in COSMIC, or as a rare SNP. Sanger sequencing of normal DNA from the patient confirmed its germ line origin. The patient was still in first remission after 7 years of follow-up and had never experienced any other cancer. This case was referred to the local department of clinical genetics, who interpreted the mutation as a rare variant of unknown significance, class 3. Hence, the mutation was excluded from our analyses.
All these features advocate that MCL with TP53 mutations represents a molecular and clinical distinct disease entity.
Patients with unmutated TP53
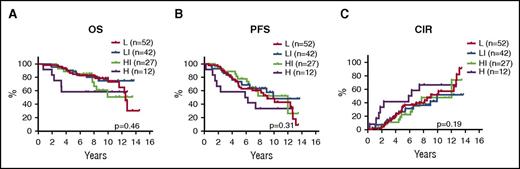
With TP53 mutations confidently assigning 11% of patients to a dismal outcome, we investigated the possibility of stratifying the remaining 89% of TP53-unmutated cases. Interestingly, among the 156 TP53-unmutated cases, no other biomarker showed any prognostic value (mutations in the remaining 7 genes, TP53 and CDKN2A deletions, MIPI, blastoid morphology, Ki67 > 30%, and MIPI-c; Figure 4). However, MIPI-c high-risk cases showed a significantly shorter time to relapse compared with MIPI-c non-high-risk cases (5.9 vs 12.0 years; P = .035), but did not have significant prognostic value for OS (not reached vs 12.8 years; P = .18) or PFS (5.9 years vs 10.2 years; P = .11).
Prognostic impact of MIPI-c index in TP53-unmutated cases. Kaplan-Meier estimates of OS (A), PFS (B), and CIR (C) stratified by the MIPI-c index into low (L), low-intermediate (LI), high-intermediate (HI), and high (H) risk groups.
Prognostic impact of MIPI-c index in TP53-unmutated cases. Kaplan-Meier estimates of OS (A), PFS (B), and CIR (C) stratified by the MIPI-c index into low (L), low-intermediate (LI), high-intermediate (HI), and high (H) risk groups.
Discussion
Here, we report the prognostic value of 8 recurrently mutated and 2 recurrently deleted genes on outcome for younger patients with MCL treated in the Nordic MCL2 and MCL3 trials, representing current standard-of-care regimens. Despite the high efficacy of these regimens, cases with TP53 mutations were resistant. Thus, our data show that TP53 mutations identify a unique MCL subtype associated with high-risk baseline characteristics, dismal response to standard treatment, and poor clinical outcome.
TP53 mutations have repeatedly been associated with a poor prognosis in previous MCL studies; however, these cohorts were heterogeneously treated and included patients of all ages.14,17,18 Interestingly, both Greiner et al18 and Halldórsdóttir et al17 reported a median OS of 1.1 years for the TP53-mutated cases (16 and 17 patients, respectively). Here, we demonstrate for the first time that TP53 mutations are associated with poor prognosis in a homogeneously treated cohort of younger, fit patients with MCL. The addition of rituximab and high-dose cytarabine to high-dose chemotherapy has improved responses significantly for younger patients with MCL in general (median OS, 12.7 years), but TP53-mutated MCL cells seem to escape eradication by these drugs, and patients with TP53-mutated MCL still have a dismal outcome with a median survival of 1.8 years.
Deletions of CDKN2A and TP53 have been thoroughly investigated in MCL. Although TP53 deletions have shown diverse results, CDKN2A deletions have consistently been associated with inferior clinical outcome.12,17,19-21 Importantly, 1 recent study reported CNVs in patients from the MCL Younger trial,22 of whom 72 of 134 had been treated with a rituximab- and cytarabine-containing induction regimen followed by ASCT; that is, a regimen similar to the cohort in our study. Using a panel of 8 gene regions, the authors showed that only deletions of TP53 (22%) and CDKN2A (25%) retained individual prognostic impact; hence, only these 2 genomic regions were analyzed for deletions in our studies. We detected slightly lower frequencies of TP53 and CDKN2A deletions (16% and 20%, respectively), which can possibly be explained by the unsorted selection of BM samples in our studies, whereas Delfau-Larue et al22 selected samples with higher tumor content. Importantly, we confirmed the prognostic effect of both deletions in the univariate setting; however, both were highly associated with TP53 mutations and, hence, lost effect in the multivariate analyses.
Our data show a clear difference in impact on all outcomes of TP53 deletions vs mutations (Figure 3D-E). This pattern has previously been reported in a more heterogeneous cohort of 119 Swedish patients with MCL, which showed prognostic significance of TP53 mutations, but not of TP53 deletions.17 Similar results have been reported in diffuse large B-cell lymphomas.26 The explanation for this difference may relate to the dominant negative effect of some TP53 mutations: although some mutated forms of p53 disrupt the function of the entire p53 protein-tetramer, deletions only reduce the amount of transcribed p53, and thus may affects the protein function to a lesser degree.27 Furthermore, an evaluation of the effect of homo- versus heterozygous deletion is needed.
In this report, we confirm the prognostic impact of blastoid morphology, Ki67 ≥ 30%, MIPI, and MIPI-c in the largest cohort of younger, optimally treated patients with MCL reported to date. Interestingly, all these factors were strongly associated with TP53 mutations in our study cohort, and only MIPI-c high-risk maintained independent prognostic impact in the multivariate analyses (Table 3). Moreover, in the cohort of TP53-unmutated patients (n = 156), none of these widely validated biomarkers showed any prognostic value; still, this remains to be validated in an independent cohort of patients stratified according to TP53 mutations. Indeed, for MIPI and MIPI-c, it needs to be stressed that these indices were trained for an unselected cohort of patients, rather than a TP53-stratified cohort.6,7
Mutations of NOTCH1 and NOTCH2 have previously been reported to have prognostic impact.14,15 In our study, NOTCH1 mutations (n = 7) were associated with a poor outcome in univariate analyses, but lost prognostic value in multivariate analyses, as they were strongly associated with TP53 mutations (5 of 7). We did not find any prognostic impact of NOTCH2 mutations.
For mutation detection, we used a targeted next-generation sequencing panel based on the most commonly mutated MCL genes according to previous whole-genome/exome sequencing studies.10,14 We found a similar distribution of mutations,10,14,28,29 but the frequencies observed in our study were generally lower. This difference most likely reflects differences in study cohorts, sample origin, processing of material, and method of sequencing. Our patient cohort consisted of fit younger, untreated patients with MCL, whereas other studies included both diagnostic and relapse samples from patients of various age. We consistently investigated BM samples, whereas other studies investigated DNA from a mixture of blood, lymph nodes, BM, and other sources. Because of the high coverage (>2900×) of our targeted approach, we decided to include patients regardless of known BM involvement from morphological assessment; however, we still detected significantly fewer mutations in patients with no morphological BM involvement, suggesting we miss identification of some mutated cases in patients with no or very little BM involvement of MCL.
TP53 aberrations have been widely examined in hematologic cancer and showed negative prognostic value in most.30 In chronic lymphocytic leukemia, TP53 aberrations (mutations and deletions) have consistently been shown to confer poor prognosis and treatment resistance, and recently, the novel small molecule inhibitors, ibrutinib, venetoclax, and idelalisib, have been approved for first-line treatment in TP53-disrupted chronic lymphocytic leukemia, as they seem to partly overcome this deleterious effect.31-33 Interestingly, ibrutinib and venetoclax have shown high response rates in relapsed MCLs,34,35 and hence, it is tempting to speculate whether a similar approach could be applied in MCL. At this time, novel trials, including these targeted drugs, are appearing in both the frontline and relapse setting, and it will be interesting to investigate the responses in light of the TP53 status.36 Our preliminary results from the Nordic relapse trial, MCL6 Philemon (ibrutinib, lenalidomide, and rituximab), indicate that the presence of a TP53 mutations may have less effect on outcome with this nonchemotherapeutic regimen; however, longer follow-up is needed to substantiate this finding.37 Interestingly, from our MCL2 and MCL3 cohort, 2 patients with TP53 mutations seemed to respond to salvage therapy. Both progressed during induction treatment, and both received bortezomib- and cytarabine-containing regimens for their relapses, the combination of which has previously been shown to have a synergistic effect.38 One patient remains alive in second remission after 6.5 years, and the other went on to allogeneic stem cell transplantation but died of complications after 4.5 years, still being in MCL remission.
In conclusion, we show that TP53-mutated MCL represents a phenotypically distinct and highly aggressive disease entity with poor or no response to the high-dose regimens including cytarabine, rituximab, and ASCT. Thus, we suggest stratification of patients with MCL according to TP53 mutational status, and inclusion in separate clinical trials exploring novel targeted agents.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the medical and nursing staffs of all contributing departments of the MCL2 and MCL3 trials, the blood banks for their contribution, and the patients for their willingness to participate.
This work was supported by grants from the Danish Council for Independent Research, the Novo Nordisk foundation, Rigshospitalet’s Research Foundation, Lundbeck Foundation, Danish Cancer Research Foundation, the Højmosegård Grant, Inge and Per Refshall’s Research Grant, Fabrikant Einar Willumsen’s Memorial Foundation, and the A.P. Møller Foundation for the Advancement of Medical Science.
Authorship
Contribution: C.W.E., C.D., J.W.H., M.W., C.F., S.E., M.K.A., P.G., M.J., and K.G. conceived and designed the experiments; C.W.E., C.D., J.W.H., L.B.P., C.P.M.-A., S.H., and A.P. performed the experiments; C.W.E., C.D., J.W.H., P.B., P.G., and K.G. interpreted the data; A.K., R.R., C.N., C.H.G., and M.J. conducted the trials, handled patient material, and collected clinical data; C.W.E., C.D., M.J., and K.G. wrote the manuscript; and all authors critically reviewed the final version of the manuscript.
Conflict-of-interest disclosure: K.G. is on Celgene and Janssen advisory boards. M.J. received research funding from Janssen, Celgene, AbbVie, and Gilead. C.H.G. is a member of the advisory boards for Janssen, Celgene, and Novartis. A.K. has received research funding from Merck and Roche and is a member of the scientific advisory board for Nordic Nanovector. C.N. received research support from AbbVie; has been involved with consultancy with Roche, Gilead, AbbVie, and Jannsen; and has received travel grants from Roche, Novartis, Gilead, and AbbVie. The remaining authors declare no competing financial interests.
Correspondence: Kirsten Grønbæk, Department of Hematology/ The Epi-/Genome Laboratory, Rigshospitalet Department 3733, Bartholin Institute, Copenhagen Biocenter, Building 2, 3rd Floor, Ole Maaløes Vej 5, 2200 Copenhagen N, Denmark; e-mail: kirstengroenbaek@regionh.dk.