Key Points
Ruxolitinib caused DNA repair defects and sensitized MPN stem and progenitor cells to PARP inhibitors.
Quiescent and proliferating MPN cells were eliminated by ruxolitinib and olaparib plus or minus hydroxyurea.
Abstract
Myeloproliferative neoplasms (MPNs) often carry JAK2(V617F), MPL(W515L), or CALR(del52) mutations. Current treatment options for MPNs include cytoreduction by hydroxyurea and JAK1/2 inhibition by ruxolitinib, both of which are not curative. We show here that cell lines expressing JAK2(V617F), MPL(W515L), or CALR(del52) accumulated reactive oxygen species–induced DNA double-strand breaks (DSBs) and were modestly sensitive to poly-ADP-ribose polymerase (PARP) inhibitors olaparib and BMN673. At the same time, primary MPN cell samples from individual patients displayed a high degree of variability in sensitivity to these drugs. Ruxolitinib inhibited 2 major DSB repair mechanisms, BRCA-mediated homologous recombination and DNA-dependent protein kinase–mediated nonhomologous end-joining, and, when combined with olaparib, caused abundant accumulation of toxic DSBs resulting in enhanced elimination of MPN primary cells, including the disease-initiating cells from the majority of patients. Moreover, the combination of BMN673, ruxolitinib, and hydroxyurea was highly effective in vivo against JAK2(V617F)+ murine MPN-like disease and also against JAK2(V617F)+, CALR(del52)+, and MPL(W515L)+ primary MPN xenografts. In conclusion, we postulate that ruxolitinib-induced deficiencies in DSB repair pathways sensitized MPN cells to synthetic lethality triggered by PARP inhibitors.
Introduction
Philadelphia chromosome–negative (Ph−) myeloproliferative neoplasms (MPNs) include polycythemia vera (PV), essential thrombocythemia (ET), and primary myelofibrosis (PMF), which are associated with mutations in JAK2, CALR, and MPL genes.1,2 Current treatment options for Ph− MPNs include cytoreductive therapy with hydroxyurea, and the JAK1/2 inhibitor (JAK1/2i) ruxolitinib, which produce durable reductions in splenomegaly and improvement of symptoms and probably of survival, but do not eliminate the disease-initiating cell population.3,4 MPNs usually present in chronic phase, but they may eventually accelerate and transform into secondary acute myeloid leukemia, which carries a dismal prognosis and is always fatal.5 Therefore, it is imperative to generate new therapies, which alone or in combination with conventional treatments induce long-term remission, even in patients who have progressed to the acute leukemia stage. The combination of agents that target different mechanisms promises to provide a successful rational future strategy.6
MPN cells contain elevated levels of reactive oxygen species (ROS) and stalled replication forks, resulting in accumulation of high numbers of toxic DNA double-strand breaks (DSBs).7-12 Therefore, we reasoned that MPN cell survival may depend on DSB repair mechanisms.13-21 DSBs are repaired by 2 major mechanisms: BRCA1/2-mediated homologous recombination repair (HRR) and DNA-dependent protein kinase, catalytic subunit (DNA-PKcs)-mediated nonhomologous end-joining (D-NHEJ).22 In addition, poly-ADP-ribose polymerase 1 (PARP1) plays a central role in preventing/repairing lethal DSBs by activation of base excision repair/single-stranded DNA break repair, by stimulation of fork repair/restart, and by mediating the back-up NHEJ (B-NHEJ) repair.23-26
Accumulation of potentially lethal DSBs in MPN cells could create an opportunity to eliminate these cells by targeting DNA repair mechanisms. Here, we tested the hypothesis that the combination of ruxolitinib-mediated inhibition of DSB repair with a PARP inhibitor (PARPi) and/or hydroxyurea causes accumulation of lethal DSBs beyond reparable thresholds, resulting in enhanced elimination of MPN cells.
Materials and methods
Primary cells
Peripheral blood and bone marrow samples from patients with newly diagnosed MPNs (supplemental Table 1, available on the Blood Web site) were obtained from: (1) Department of Biomedicine, Basel University, Basel, Switzerland, (2) Department of Internal Medicine, Hematology and Oncology, Medical University, Aachen, Germany, (3) Department of Haematology, University of Cambridge, Cambridge, United Kingdom, and (4) Myeloproliferative Disorders Clinic, Huntsman Cancer Hospital, Salt Lake City, UT. Samples of normal hematopoietic cells were purchased from Cambrex Bio Science (Walkersville, MD). Lin−CD34+ cells were obtained from mononuclear fractions by magnetic sorting using the EasySep negative selection human progenitor cell enrichment cocktail followed by the human CD34+ selection cocktail (StemCell Technologies) as described previously.27
Cell lines
BaF3-JAK2(V617F)+EpoR, 32Dcl3-MPL(W515L), 32Dcl3-CALR(del52)+MPL(wt) cell lines, and their BaF3-EpoR and 32Dcl3-MPL(wt) parental counterparts were described previously.28-30 BaF3-HR2 and Jak2(V617F)+ BaF3-HR2 cells carrying the genome-integrated homologous recombination (HR)–enhanced green fluorescent protein (EGFP) cassette were generously provided by W. Vainchenker (INSERM UMR 1170, Gustave Roussy, Villejuif, France).31 They were cultivated in Iscove modified Dulbecco medium supplemented with 10% fetal bovine serum (FBS), interleukin-3 (IL-3) plus erythropoietin (Epo), and antibiotic cocktail.
Inhibitors/drugs
The following compounds were used: JAK1/2i ruxolitinib (Selleckchem), PARPi BMN673 and PARPi olaparib (Selleckchem), mutT homolog 1 (MTH1) inhibitor SCH51344 (Tocris), ROS scavenger vitamin E (Sigma), and ribonucleoside diphosphate reductase inhibitor hydroxyurea (Selleckchem).
Western analyses
Nuclear cell lysates and total cell lysates were obtained and resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis as previously described.27 Protein expressions were analyzed using primary antibodies detecting: BRCA1 (R&D Systems), BRCA2 (R&D Systems), RAD51 (Santa Cruz Biotechnology), DNA-PKcs (Bethyl Laboratories), Ku70 (Bethyl Laboratories), Ku80 (Thermo Fisher Scientific), PARP1 (Santa Cruz Biotechnology), PALB2 (Bethyl Laboratories), Lig3 (GeneTex), Lig4 (Abcam), STAT5 (Santa Cruz Biotechnology), phospho-STAT5A (Ser780; Santa Cruz Biotechnology), cleaved caspase-3 (Cell Signaling Technology), lamin B (Santa Cruz Biotechnology), and β-actin (Sigma).
Examination of DSB repairs
Cells were cultured in IMDM supplemented with 10% FBS, IL-3, and Epo in the presence or absence of 0.15 µM ruxolitinib. HRR events were measured as described previously with modification.32 Five million JAK2(V617F)+ Ba/F3-HR2 cells carrying the HR-EGFP cassette were nucleofected with 5 µg of pCBASCE1 and 2.5 µg of pDsRED-Mito plasmids using Nucleofector (program U-008, Human CD34 Cell Nucleofector kit; Lonza). Expression of I-SceI causes a DSB in the specific restriction site included in the HR-EGFP cassette, and pDsRed1-Mito encodes red fluorescent protein with a mitochondrial localization signal to control the efficiency of transfection. An HRR event restores functional EGFP expression, which is readily detected by fluorescent microscope 48 hours after transfection with I-SceI. After 72 hours, cells were analyzed by flow cytometry for the percentage of GFP+ cells to assess HRR activity. D-NHEJ was measured in cell-free extracts as described previously with modification.32 Briefly, 200 ng of the substrate plasmid (pBluescript KS+ linear plasmids digested XhoI + XbaI to generate noncompatible 5′ overhangs) was added to the reaction mix containing 10 μg of nuclear lysate and incubated for 1 hour at 37°C. Products of D-NHEJ reaction were resolved in 0.5% agarose gel containing 0.5 μg/mL ethidium bromide, scanned with Adobe Photoshop, and analyzed by ImageQuant TL (Amersham Biosciences).
In vitro treatment
Cells were cultivated in IMDM supplemented with 10% FBS and growth factors (100 ng/mL stem cell factor [SCF]; 10 ng/mL Flt3 ligand; 20 ng/mL IL-3, IL-6, granulocyte colony-stimulating factor, and granulocyte-macrophage colony-stimulating factor [GM-CSF]; 12 U/mL Epo; 2.5 ng/mL thrombopoietin. Ruxolitinib, hydroxyurea, vitamin E, SCH51344, olaparib, and/or BMN673 were added for 3 to 5 days followed by trypan blue exclusion counting and/or plating in methylcellulose in the presence of growth factors. Colonies were counted after 7 to 10 days. For quiescent/proliferating cells, Lin− cells were stained with cell trace violet (CTV; eBioscience) and incubated for 5 days in StemSpan SFEM medium (Stem Cell Technologies) supplemented with the cocktail of growth factors (see first sentence of this paragraph) and inhibitors when indicated. Quiescent (CTVmax) and proliferating (CTVlow) leukemia cells were detected by flow cytometry using fluorochrome-conjugated anti-Lin, anti-CD34, and anti-CD38 antibodies (all from BD Biosciences) as described previously.33
GFP+JAK2(V671F) murine MPN-like disease
C57BL/6 recipient mice (The Jackson Laboratory) were subjected to 900 Gy total body irradiation followed by IV injection of a 1:1 mixture of 106 GFP+JAK2(V671F) and 106 wild-type bone marrow cells as described previously.3 Five weeks later, mice were treated with vehicle, hydroxyurea (30 mg/kg twice daily intraperitoneally [IP]), ruxolitinib (30 mg/kg twice daily by oral gavage), BMN673 (0.33 mg/kg IV), and combinations of these drugs for 3 weeks. GFP+JAK2(V617F) cells were examined among total bone marrow cells, splenocytes, and peripheral blood leukocytes at the end of treatment; in addition, a fraction of GFP+JAK2(V617F) Lin−Sca1+c-Kit+ cells was assessed in the bone marrow population.
Primary MPN xenografts
NOD.Rag1−/−;γcnull mice expressing human IL-3, GM-CSF, and SCF (NRGS mice34 ; The Jackson Laboratory) were sublethally irradiated (600 Gy) and injected with 1 × 106 Lin−CD34+ primary MPN cells expressing JAK2(V617F), CALR(del52), or MPL(W515L). Three weeks later, mice were treated as described above for murine MPN-like disease with vehicle, hydroxyurea plus ruxolitinib, BMN673, and hydroxyurea plus ruxolitinib plus BMN673 for 3 weeks. Human CD45+ (hCD45+) cells, hCD45+Lin−CD34+ MPN progenitors, and stem cell–enriched hCD45+Lin−CD34+CD38− MPN cells were detected in bone marrow cells, splenocytes, and/or peripheral blood leukocytes at the end of treatment as previously described.33
Statistical analyses
Data are presented as mean plus or minus standard deviation (SD) from 3 independent experiments and were compared using the unpaired 2-tailed Student t test; values of P < .05 were considered significant. The response additivity approach was used to study the synergistic effects.35 This approach shows a positive drug combination effect when the observed combination effect is greater than the expected additive effect by the sum of the individual effects. The P value for the possible synergistic effect is given by the significance of the interaction effect in a factorial analysis of variance of the individual and combination effects.
Study approval
Studies involving human samples were approved by the Temple University Institutional Review Board and met all requirements of the Declaration of Helsinki. Animal studies were approved by the Temple University Institutional Animal Care and Use Committee.
Results
Wide-range sensitivity of MPN cells to PARPi
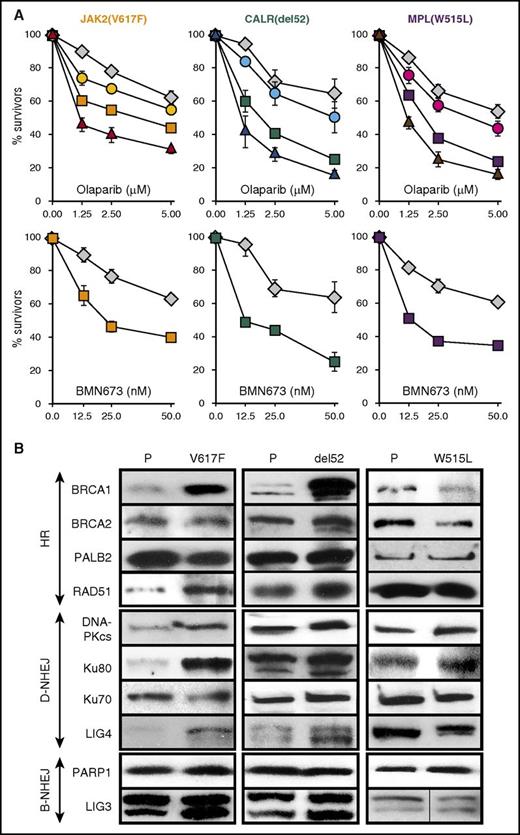
Murine cell lines expressing JAK2(V617F)+EpoR, MPL(W515L), and CALR(del52)+MPL(wt) displayed modestly increased (by 20%-40%) but statistically significant (P ≤ .002) sensitivity to PARPi olaparib and PARPi BMN673 when compared with nontransformed counterparts (Figure 1A). ROS scavenger vitamin E diminished, whereas MTH1 inhibitor SCH51344 (MTH1 sanitizes oxidized dNTP pools to prevent incorporation of damaged bases during DNA replication36 ) enhanced the toxic effect of olaparib in JAK2(V617F)+EpoR, MPL(W515L), and CALR(del52)+MPL(wt) cells.
Sensitivity of JAK2(V617F)+, CALR(del52)+, andMPL(ex10mut)+cells to PARPi. (A) Cell lines expressing JAK2(V617F)+EpoR, CALR(del52)+MPL(wt), or MPL(W515L) were incubated with olaparib alone (1.25, 2.5, 5.0 μM) (squares) or BMN673 alone (12.5, 25.0, 50.0 nM) (squares), olaparib plus 200 μM vitamin E (circles), or olaparib plus 2.5 μM SCH51344 (triangles) for 96 hours in the presence of IL-3 plus Epo. Parental cells (diamonds) were incubated with olaparib or BMN673 only. Living cells were counted in Trypan blue. Results represent mean plus or minus SD percentage of living cells in comparison with untreated control from 3 independent experiments. (B) Western analysis of the indicated proteins in parental cells (P) and in isogenic cells expressing JAK2(V617F)+EpoR, CALR(del52)+MPL(wt), and MPL(W515L).
Sensitivity of JAK2(V617F)+, CALR(del52)+, andMPL(ex10mut)+cells to PARPi. (A) Cell lines expressing JAK2(V617F)+EpoR, CALR(del52)+MPL(wt), or MPL(W515L) were incubated with olaparib alone (1.25, 2.5, 5.0 μM) (squares) or BMN673 alone (12.5, 25.0, 50.0 nM) (squares), olaparib plus 200 μM vitamin E (circles), or olaparib plus 2.5 μM SCH51344 (triangles) for 96 hours in the presence of IL-3 plus Epo. Parental cells (diamonds) were incubated with olaparib or BMN673 only. Living cells were counted in Trypan blue. Results represent mean plus or minus SD percentage of living cells in comparison with untreated control from 3 independent experiments. (B) Western analysis of the indicated proteins in parental cells (P) and in isogenic cells expressing JAK2(V617F)+EpoR, CALR(del52)+MPL(wt), and MPL(W515L).
Most of the key proteins regulating major DSB repair pathways, HR, D-NHEJ, and B-NHEJ were either not affected or upregulated in the presence of JAK2(V617F), MPL(W515L), and CALR(del52) (Figure 1B). However, it appears that expression of MPL(W515L) caused an approximately twofold reduction of the expression of BRCA1 and BRCA2 proteins. Because activation of MPL is associated with upregulation of D-NHEJ,37 it is plausible that BRCA1/2-mediated HR plays a secondary role in DSB repair in MPL(W515L)+ cells as reflected by downregulated BRCA proteins.
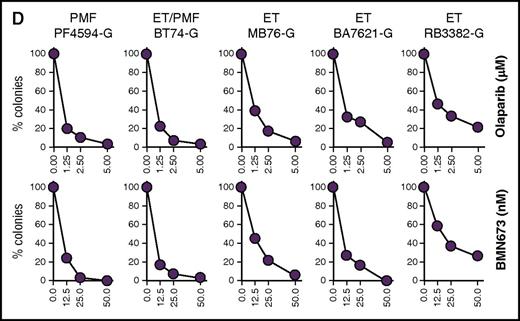
Next, the sensitivity to PARPi of primary Lin−CD34+ cells from healthy donors and MPN patients expressing JAK2(V617F), MPL(ex10mut), and CALR(del52) was tested in a clonogenic assay (Figure 2). Lin−CD34+ cells isolated from healthy donors were only partially sensitive to olaparib and BMN673 (Figure 2A). Eight JAK2(V617F) samples (6109-K, 013-S, 034-S, 8729-K, 338-S, 742-K, 4082-K, 1-P) were sensitive, whereas 3 samples (288-S, 4552-K, 10141-K) were only partially sensitive to olaparib and BMN673 (Figure 2B). On the other hand, CALR(del52) samples displayed the highest variability in sensitivity to PARPi, from sensitive (168-S and CV096-G), partially sensitive (055-S, 073-S, 215-S), to resistant (109-S) (Figure 2C). MPL(ex10mut) samples behaved similarly to JAK2(V617F) samples by being sensitive (PF4594-G, BT74-G, MB76-G, BA7621-G) or partially sensitive (RB3382-G) to PARPi (Figure 2D).
Sensitivity of individual MPN samples expressing JAK2(V617F), CALR(del52), and MPL(ex10mut) to PARPi. Lin−CD34+ primary cells from (A) healthy donors (n = 3) and from (B) JAK2(V617F)+, (C) CALR(del52)+, (D) MPL(ex10mut)+ MPN patients were incubated with olaparib (1.25, 2.5, 5.0 μM) or BMN673 (12.5, 25.0, 50.0 nM) for 96 hours in the presence of growth factors (100 ng/mL SCF; 10 ng/mL Flt3 ligand; 20 ng/mL IL-3, IL-6, granulocyte colony-stimulating factor, and GM-CSF; 12 U/mL Epo; 2.5 ng/mL thrombopoietin) followed by plating in methylcellulose. Colonies were counted after 7 to 10 days. Results represent the percentage of colonies in comparison with untreated control.
Sensitivity of individual MPN samples expressing JAK2(V617F), CALR(del52), and MPL(ex10mut) to PARPi. Lin−CD34+ primary cells from (A) healthy donors (n = 3) and from (B) JAK2(V617F)+, (C) CALR(del52)+, (D) MPL(ex10mut)+ MPN patients were incubated with olaparib (1.25, 2.5, 5.0 μM) or BMN673 (12.5, 25.0, 50.0 nM) for 96 hours in the presence of growth factors (100 ng/mL SCF; 10 ng/mL Flt3 ligand; 20 ng/mL IL-3, IL-6, granulocyte colony-stimulating factor, and GM-CSF; 12 U/mL Epo; 2.5 ng/mL thrombopoietin) followed by plating in methylcellulose. Colonies were counted after 7 to 10 days. Results represent the percentage of colonies in comparison with untreated control.
Ruxolitinib inhibited DSB repair and enhanced the sensitivity of MPN cells to PARPi
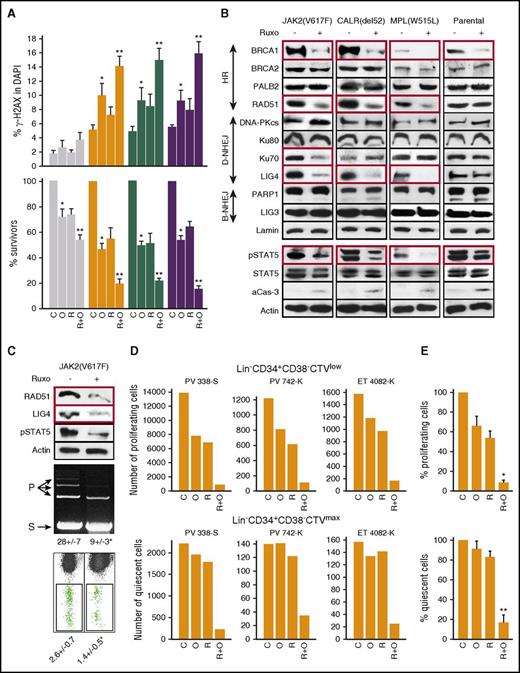
BaF3-JAK2(V617F)+EpoR, 32Dcl3-MPL(W515L), and 32Dcl3-CALR(del52)+MPL(wt) cells treated with olaparib and/or ruxolitinib accumulated elevated numbers of DSBs, especially in cells treated with ruxolitinib and olaparib (Figure 3A top panel). In addition, enhanced accumulation of DSBs in ruxolitinib plus olaparib–treated JAK2(V617F)+EpoR+, MPL(W515L)+, and CALR(del52)+MPL(wt)+ cells was associated with synergistic increase of cell death (Figure 3A bottom panel).
JAK2i ruxolitinib reduced HRR and D-NHEJ activity and enhanced the anti-MPN effect of PARPi olaparib. (Panel A) Parental cell lines and those expressing JAK2(V617F)+EpoR, CALR(del52)+MPL(wt), or MPL(W515L) were untreated (C) or treated with 5 μM olaparib (O) or 400 nM ruxolitinib (R) or ruxolitinib plus olaparib (R+O) in the presence of IL-3 plus Epo for 24 hours (γ-H2AX) and 96 hours (cell survival). DSBs were detected by γ-H2AX immunofluorescence overlapping with 4′,6-diamidino-2-phenylindole (DAPI) (top panel), and living cells were counted in Trypan blue (bottom panel; percentage of living cells in comparison with untreated control). Results represent means plus or minus SD from 3 independent experiments. *P < .05 in comparison with C using the Student t test; **P ≤ .001 in comparison with R and O groups using the response additivity approach. (Panel B) Western analysis of the indicated proteins in cells expressing JAK2(V617F)+EpoR, CALR(del52)+MPL and MPL(W515L), and in BaF3-EpoR cells (Parental) after 24-hour incubation with 400 nM ruxolitinib in the presence of IL-3 plus Epo. Proteins downregulated by ruxolitinib are in red boxes. (Panel C) HRR and D-NHEJ activities in JAK2(V617F)+ cells untreated (−) or treated for 24 hours with 400 nM ruxolitinib (+). Top panel, Western blots, Middle panel, D-NHEJ activity. S indicates linearized plasmid substrate; P indicates ligated plasmid products; results show the percentage of P. Bottom panel, HRR activity measured by restoration of EGFP expression. Results show the percentage of GFP+ cells; *P ≤ .01. (Panel D) Number of proliferating Lin−CD34+CD38−CTVlow and quiescent Lin−CD34+CD38−CTVmax cells from individual JAK2(V617F)+ MPN samples left untreated (C) or treated with ruxolitinib (R; 25 nM), olaparib (O; 1.25 μM), and ruxolitinib plus olaparib (R+O). (Panel E) Cumulative percentages from samples examined in panel D. *P < .001 in comparison with R or O groups using the Student t test; **P < .01 in comparison with R and O groups using the response additivity approach.
JAK2i ruxolitinib reduced HRR and D-NHEJ activity and enhanced the anti-MPN effect of PARPi olaparib. (Panel A) Parental cell lines and those expressing JAK2(V617F)+EpoR, CALR(del52)+MPL(wt), or MPL(W515L) were untreated (C) or treated with 5 μM olaparib (O) or 400 nM ruxolitinib (R) or ruxolitinib plus olaparib (R+O) in the presence of IL-3 plus Epo for 24 hours (γ-H2AX) and 96 hours (cell survival). DSBs were detected by γ-H2AX immunofluorescence overlapping with 4′,6-diamidino-2-phenylindole (DAPI) (top panel), and living cells were counted in Trypan blue (bottom panel; percentage of living cells in comparison with untreated control). Results represent means plus or minus SD from 3 independent experiments. *P < .05 in comparison with C using the Student t test; **P ≤ .001 in comparison with R and O groups using the response additivity approach. (Panel B) Western analysis of the indicated proteins in cells expressing JAK2(V617F)+EpoR, CALR(del52)+MPL and MPL(W515L), and in BaF3-EpoR cells (Parental) after 24-hour incubation with 400 nM ruxolitinib in the presence of IL-3 plus Epo. Proteins downregulated by ruxolitinib are in red boxes. (Panel C) HRR and D-NHEJ activities in JAK2(V617F)+ cells untreated (−) or treated for 24 hours with 400 nM ruxolitinib (+). Top panel, Western blots, Middle panel, D-NHEJ activity. S indicates linearized plasmid substrate; P indicates ligated plasmid products; results show the percentage of P. Bottom panel, HRR activity measured by restoration of EGFP expression. Results show the percentage of GFP+ cells; *P ≤ .01. (Panel D) Number of proliferating Lin−CD34+CD38−CTVlow and quiescent Lin−CD34+CD38−CTVmax cells from individual JAK2(V617F)+ MPN samples left untreated (C) or treated with ruxolitinib (R; 25 nM), olaparib (O; 1.25 μM), and ruxolitinib plus olaparib (R+O). (Panel E) Cumulative percentages from samples examined in panel D. *P < .001 in comparison with R or O groups using the Student t test; **P < .01 in comparison with R and O groups using the response additivity approach.
To determine whether ruxolitinib-mediated accumulation of olaparib-induced DSBs is associated with inhibition of DSB repair activity, we performed a western blot array to assess expression of key proteins in DSB repair pathways. JAK2(V617F)+EpoR+, MPL(W515L)+, and CALR(del52)+MPL(wt)+ cells and their parental counterparts were treated with ruxolitinib for 24 hours in the presence of IL3 plus Epo to inhibit JAK2 kinases as documented by downregulated phospho-STAT5A(Ser780) (Figure 3B). At the same time, ruxolitinib-treated cells were viable as assessed by Trypan blue exclusion, minimal caspase-3 activation, and uncleaved PARP1. Key proteins in HRR (BRCA1 and RAD51) and D-NHEJ (Lig4), but not B-NHEJ were downregulated in ruxolitinib-treated JAK2(V617F)+EpoR+, MPL(W515L)+, and CALR(del52)+MPL(wt)+ cells (Figure 3B).
Next, we examined whether ruxolitinib-induced downregulation of RAD51 and LIG4 proteins (Figure 3C top panel) caused reduction of HRR and D-NHEJ activities. D-NHEJ activity measured in vitro by nuclear cell lysate–mediated plasmid end-joining was inhibited by approximately threefold in ruxolitinib-treated JAK2(V617F)+ cells (Figure 3C middle panel). To measure HRR activity, an I-SceI endonuclease-mediated DSB was induced in Jak2(V617F)+ Ba/F3-HR2 cells carrying the HR-EGFP recombination reporter cassette integrated in their genome. HRR restores the expression of GFP detected by flow cytometry. Ruxolitinib-treated Ba/F3-HR2 cells displayed approximately twofold reduction in HRR activity (Figure 3C bottom panel).
We have previously reported that D-NHEJ–deficient quiescent and HRR/D-NHEJ–deficient proliferating tumor cells were sensitive to dual cellular synthetic lethality exerted by PARPi.33 Therefore, we tested whether ruxolitinib-induced downregulation of D-NHEJ and HRR sensitizes JAK2(V617F)+ quiescent and proliferating cells, respectively, to PARPi-mediated synthetic lethality. Intriguingly, ruxolitinib enhanced the sensitivity of leukemia stem cell–enriched Lin−CD34+CD38−CTVlow proliferating patient cells to olaparib (Figure 3D-E). Moreover, even if individual drugs did not affect leukemia stem cell–enriched Lin−CD34+CD38−CTVmax quiescent cells, the combination exerted a synergistic inhibitory effect (Figure 3D-E).
Ruxolitinib enhanced the effect of PARPi and/or hydroxyurea in vitro in preselected MNP samples
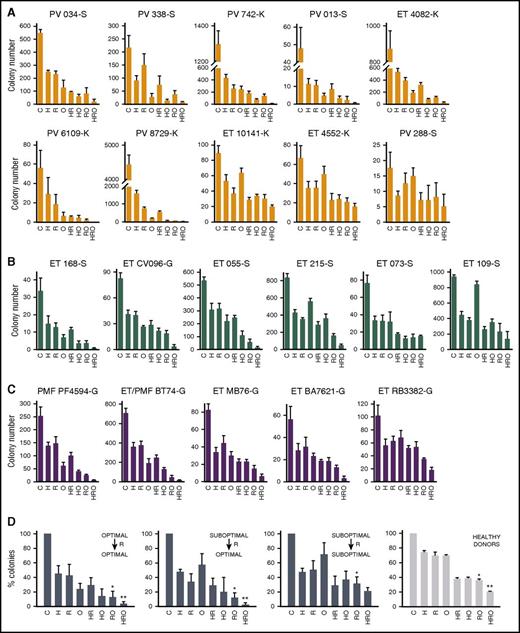
Clonogenic assay revealed that Lin−CD34+ primary MPN cells from a cohort of MPNs expressing JAK2(V617F), MPL(ex10mut), and CALR(del52), which were sensitive to PARPi (Figure 2B-D optimal response), responded favorably to the combination of ruxolitinib plus olaparib and ruxolitinib plus hydroxyurea plus olaparib (034-S, 338-S, 742-K, 013-S, 4082-K, 6109-K, 8729-K, 168-S, CV096-K, PF4594-G, BT74-G, MB76-G, BA7621-G in Figure 4A-C, and Figure 4D left panel). On the other hand, cells from samples displaying partial sensitivity or resistance to PARPi (suboptimal response) could be subdivided into 2 cohorts: those that responded favorably (055-S, 215-S in Figure 4A-C, and Figure 4D middle left panel) or unfavorably (10141-K, 5442-K, 288-S, 073-S, 109-S, RB3382-G in Figure 4A-C, and Figure 4D middle right panel) to ruxolitinib plus olaparib and/or ruxolitinib plus hydroxyurea plus olaparib. Lin−CD34+ cells from 3 healthy donors displayed a homogenous response pattern to the drugs (Figure 4D right panel), similar to that of the unfavorable MPN cohort (Figure 4D middle right panel).
The effect of ruxolitinib on the sensitivity of JAK2(V617F), CALR(del52), and MPL(ex10mut) MPN cells to PARPi. Lin−CD34+ cells from PV, ET, and PMF patients carrying (A) JAK2(V617F), (B) CALR(del52), and (C) MPL(ex10mut) were incubated with olaparib (O; 1.25 μM), hydroxyurea (H; 10 μM), and/or ruxolitinib (R; 25 nM) for 72 hours in the presence of growth factors (see Figure 2) and plated in methylcellulose. Colonies were counted after 7 to 10 days. Results represent mean number of colonies plus or minus SD from triplicates. (D) Ruxolitinib (R)-treated Lin−CD34+ cells from cohorts of MPN samples (steel blue bars) and healthy donors (gray bars) displayed heterogenic sensitivity to PARPi. *P < .05 in comparison with O, and **P < .05 in comparison with HO using the Student t test.
The effect of ruxolitinib on the sensitivity of JAK2(V617F), CALR(del52), and MPL(ex10mut) MPN cells to PARPi. Lin−CD34+ cells from PV, ET, and PMF patients carrying (A) JAK2(V617F), (B) CALR(del52), and (C) MPL(ex10mut) were incubated with olaparib (O; 1.25 μM), hydroxyurea (H; 10 μM), and/or ruxolitinib (R; 25 nM) for 72 hours in the presence of growth factors (see Figure 2) and plated in methylcellulose. Colonies were counted after 7 to 10 days. Results represent mean number of colonies plus or minus SD from triplicates. (D) Ruxolitinib (R)-treated Lin−CD34+ cells from cohorts of MPN samples (steel blue bars) and healthy donors (gray bars) displayed heterogenic sensitivity to PARPi. *P < .05 in comparison with O, and **P < .05 in comparison with HO using the Student t test.
Ruxolitinib enhanced the effect of PARPi and/or hydroxyurea in a retroviral murine model of JAK2(V617F)+ MPN
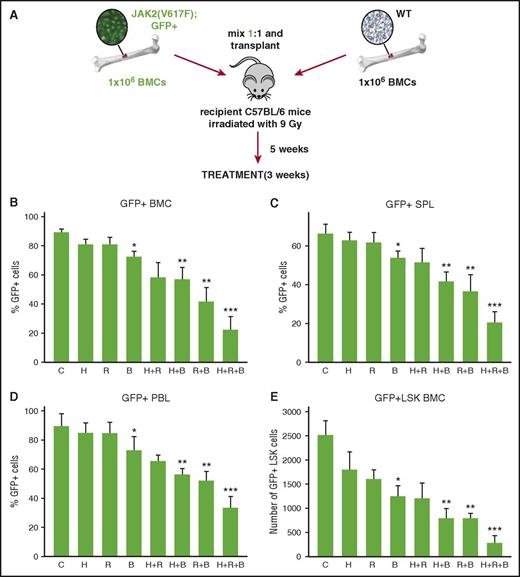
To test whether ruxolitinib enhances the effect of PARPi plus or minus the standard cytotoxic drug hydroxyurea, we applied a murine model of GFP+JAK2(V617F)+ PV (Figure 5A).3 BMN673 was used here because it displays better pharmacokinetic parameters in mice than olaparib.38 As expected, ruxolitinib and hydroxyurea when used individually did not reduce the percentage of GFP+JAK2(V617F)+ cells in peripheral blood, spleen, and bone marrow, and BMN673 exerted only a very moderate inhibitory effect (Figure 5B-D). However, ruxolitinib significantly enhanced the therapeutic effect of BMN673 and of BMN673 plus hydroxyurea. In addition, the population of stem cell–enriched GFP+Lin−Sca-1+c-Kit+ JAK2(V617F) cells was significantly reduced in mice treated with ruxolitinib combined with BMN673 or BMN673 plus hydroxyurea when compared with BMN673 plus or minus hydroxyurea (Figure 5E).
BMN673 exerted an anti-MPN effect in vivo. (Panel A) Experimental model. Lethally irradiated C57BL/6 recipient mice were injected with a 1:1 mixture of 106 GFP+JAK2(V671F) and 106 wild-type bone marrow cells. Five weeks later, mice were treated with vehicle (C), hydroxyurea (H; 30 mg/kg twice daily IP), ruxolitinib (R; 30 mg/kg twice daily oral gavage), BMN673 (B; 0.33 mg/kg IV), H+R, H+B, R+B, and H+R+B for 3 weeks. Percentage of GFP+JAK2(V617F) was measured in (panel B) bone marrow cells, (panel C) splenocytes, and (panel D) peripheral blood leukocytes; (panel E) number of GFP+JAK2(V617F) Lin−Sca1+c-Kit+ (LSK) cells per 106 bone marrow cells was calculated, too. *P < .05, **P < .05, and ***P < .05 when compared with control, single treatment, and double treatment, respectively, from 6 to 7 mice using the Student t test.
BMN673 exerted an anti-MPN effect in vivo. (Panel A) Experimental model. Lethally irradiated C57BL/6 recipient mice were injected with a 1:1 mixture of 106 GFP+JAK2(V671F) and 106 wild-type bone marrow cells. Five weeks later, mice were treated with vehicle (C), hydroxyurea (H; 30 mg/kg twice daily IP), ruxolitinib (R; 30 mg/kg twice daily oral gavage), BMN673 (B; 0.33 mg/kg IV), H+R, H+B, R+B, and H+R+B for 3 weeks. Percentage of GFP+JAK2(V617F) was measured in (panel B) bone marrow cells, (panel C) splenocytes, and (panel D) peripheral blood leukocytes; (panel E) number of GFP+JAK2(V617F) Lin−Sca1+c-Kit+ (LSK) cells per 106 bone marrow cells was calculated, too. *P < .05, **P < .05, and ***P < .05 when compared with control, single treatment, and double treatment, respectively, from 6 to 7 mice using the Student t test.
Major organs such as heart, lungs, liver, kidneys, bone marrow, and spleen in mice treated with the combination of ruxolitinib, hydroxyurea, and BMN67 showed normal morphologic features with no evidence of ischemia or drug toxicity (supplemental Figure 1). More detailed analysis of the hematopoietic system revealed only transient moderate toxicity in peripheral blood and bone marrow (supplemental Table 2).
In vivo PARPi treatment enhanced the effect of ruxolitinib plus hydroxyurea against preselected primary MPN xenografts in immunodeficient mice
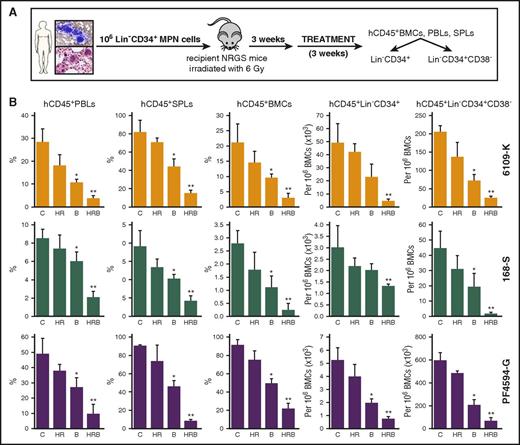
Primary MPN samples [JAK2(V617F)+ 6109-K, MPL(ex10mut)+ PF4594-G, and CALR(del52)+ 168-S] were preselected based on their favorable response to PARPi plus or minus ruxolitinib and hydroxyurea (Figures 2B-D and 4A-C). Primary Lin−CD34+ MPN cells from these patients also engrafted in NRGS mice (>5% hCD45+ cells in peripheral blood and splenomegaly after 3 weeks). NRSG mice bearing these MPN xenografts were treated with vehicle (control), hydroxyurea plus ruxolitinib (HR), BMN673, or HR plus BMN673 (Figure 6A). The therapeutic effect was measured by detection of hCD45+ cells in peripheral blood, spleen, and bone marrow, and of hCD45+Lin−CD34+ and hCD45+Lin−CD34+CD38− cells in bone marrow.
BMN673 exerted an anti-MPN xenograft effect in vivo. (Panel A) Experimental model. Sublethally irradiated NRGS recipient mice were injected with 106 primary Lin− MPN cells from individual MPN patients expressing JAK2(V617F), CALR(del52), or MPL(W515L). One week later, mice were treated with vehicle (C), hydroxyurea (H; 30 mg/kg twice daily IP) plus ruxolitinib (R; 30 mg/kg twice daily oral gavage) (HR), BMN673 (B; 0.33 mg/kg IV), and HR+B (HRB) for 3 weeks (3-5 mice per group). Indicated cells were detected by immunofluorescence. (Panel B) Percentage of hCD45+ cells was measured in peripheral blood leukocytes (PBLs), splenocytes (SPLs), and bone marrow cells (BMCs). Number of hCD45+ BMCs expressing Lin−CD34+ and Lin−CD34+CD38− per 106 cells was determined. *P < .05 and **P < .05 in comparison with C and all other groups, respectively, using the Student t test.
BMN673 exerted an anti-MPN xenograft effect in vivo. (Panel A) Experimental model. Sublethally irradiated NRGS recipient mice were injected with 106 primary Lin− MPN cells from individual MPN patients expressing JAK2(V617F), CALR(del52), or MPL(W515L). One week later, mice were treated with vehicle (C), hydroxyurea (H; 30 mg/kg twice daily IP) plus ruxolitinib (R; 30 mg/kg twice daily oral gavage) (HR), BMN673 (B; 0.33 mg/kg IV), and HR+B (HRB) for 3 weeks (3-5 mice per group). Indicated cells were detected by immunofluorescence. (Panel B) Percentage of hCD45+ cells was measured in peripheral blood leukocytes (PBLs), splenocytes (SPLs), and bone marrow cells (BMCs). Number of hCD45+ BMCs expressing Lin−CD34+ and Lin−CD34+CD38− per 106 cells was determined. *P < .05 and **P < .05 in comparison with C and all other groups, respectively, using the Student t test.
Ruxolitinib plus hydroxyurea did not consistently reduce the percentage and number of MPN xenograft cells (Figure 6B). On the other hand, BMN673 reduced the percentage of hCD45+ cells in peripheral blood, spleen, and bone marrow, and the number of hCD45+Lin−CD34+ and hCD45+Lin−CD34+CD38− cells in bone marrow of mice bearing JAK2(V617F)+, MPL(ex10mut)+, and CALR(del52)+ MPN xenografts. Importantly, the combination of BMN673 plus ruxolitinib plus hydroxyurea exerted the strongest anti-MPN effect when compared with BMN673 and ruxolitinib plus hydroxyurea.
Discussion
Our data and other reports indicated that MPN cells contain elevated levels of ROS and stalled replication forks, resulting in accumulation of potentially lethal DSBs.8,12 However, MPN cells are able to repair numerous DSBs because 2 major DSB repair pathways, HRR and D-NHEJ, are activated.31,37 PARPi and/or hydroxyurea generate additional DSBs that may overwhelm DSB repair activity in some MPN cells to cause cell death,39,40 but numerous cells can survive the treatment.
We have shown here that JAK1/2i ruxolitinib caused downregulation of key members of HRR (BRCA1, RAD51) and D-NHEJ (LIG4) in JAK2(V617F)+, MPL(ex10mut)+, and CALR(del52)+ cell lines, resulting in reduced HRR and D-NHEJ activities. This effect was associated with PARPi-induced accumulation of DSBs and enhanced elimination of MPN cells from numerous patient samples. Because defects in DNA repair sensitized tumor cells to PARPi, we postulate that ruxolitinib-induced HRR and D-NHEJ deficiencies triggered PARPi-mediated synthetic lethality.41
All 3 “driver” mutations [JAK2(V617F), CALR(del52), and MPL(W515L)] have been detected not only in mature MPN cells, but also in MPN stem cells, and therefore these cells must be eliminated to eradicate the disease.42-45 Because JAK1/2i did not eliminate the disease-initiating population, novel therapeutic approaches were needed.4
Ruxolitinib treatment inhibits proliferation of JAK2(V617)+, CALR(del52)+, and MPL(W515L)+ cells, but induces minimal degrees of apoptosis (Mazzacurati et al46 and supplemental Figure 2), and growth-arrested cells usually show poor sensitivity to cytotoxic drugs. We observed that ruxolitinib reduced the activity of DSB repair pathways playing a key role in proliferating (HRR/D-NHEJ) and quiescent (D-NHEJ) cells. We postulate that ruxolitinib-mediated inhibition of HR and D-NHEJ creates a unique opportunity to trigger PARPi-mediated dual synthetic lethality in HRR and D-NHEJ–deficient proliferating cells and in D-NHEJ–deficient G1/G0 cells expressing the “driver” mutations.
This statement is supported by 5 observations: (1) quiescent and proliferating Lin−CD34+CD38− human MPN-initiating cells42 were eliminated in vitro by ruxolitinib plus olaparib, (2) Lin−Sca-1+c-Kit+ murine MPN-initiating cells4,43 were eliminated by ruxolitinib plus BMN673 plus or minus hydroxyurea in syngeneic mice bearing JAK2(V617F)+ MPN-like disease, (3) Lin−CD34+CD38− human MPN-initiating cells were eliminated by ruxolitinib plus BMN673 plus or minus hydroxyurea in NRGS mice bearing primary MPN xenografts, (4) disease-initiating cells capable of engrafting secondary recipient mice were eliminated by ruxolitinib plus BM3673 plus hydroxyurea in NRGS mice bearing primary MPN xenograft (supplemental Figure 3), and (5) we reported that PARPi eliminated HRR/D-NHEJ–deficient proliferating and D-NHEJ–deficient quiescent acute and chronic leukemia cells.33 It has been reported that interferon α, which to date shows the highest degree of molecular remissions among the conventional drugs used to treat MPN patients, induced proliferation of JAK2(V617F) disease-initiating cells and promoted a predetermined erythroid differentiation.47 Our approach directly eliminates both proliferating and quiescent MPN-initiating cells, thus significantly expanding the MPN cells that can be targeted to the most primitive cell population.
Individual MPN samples displayed a high level of variability in responding to PARPi. Another report supported this observation and suggested that sensitivity to PARPi was associated with impaired HRR.48 Ruxolitinib induced HRR and D-NHEJ deficiency and enhanced sensitivity to PARPi in numerous patient samples that displayed optimal and suboptimal response to PARPi used alone. However, a cohort of MPN samples with suboptimal response to PARPi remained partially resistant to the inhibitor even when combined with ruxolitinib. There are several possible explanations for the heterogeneous response of individual MPNs to PARPi or ruxolitinib plus PARPi.
First, additional genetic/epigenetic factors inherently characteristic for individual MPNs (eg, mutations in TET2, ASXL1, DNMT3A, EZH2, IDH12 ) may regulate sensitivity to PARPi plus or minus ruxolitinib. This is supported by data suggesting that TET2 affects the response of JAK2(V617F)+ murine bone marrow cells to olaparib plus or minus ruxolitinib (supplemental Figure 4), that mutations in DNMT3a and IDH1 altered DNA repair activity and sensitivity to PARPi and anthracycyline,49-51 and that EZH2 downregulates the expression of BRCA1 and RAD51.52-55 In addition, deletion of Asxl1 and/or Tet2 deregulated expression of DNA repair genes including Rad51 in Lin−Sca-1+c-Kit+ murine bone marrow cells.56 The hypothesis that accompanying mutations may modulate sensitivity of MPN cells to PARPi plus or minus JAK1/2i is further supported by our data from primary cells indicating that TET2mut, EZH2mut, and ASXL1mut may enhance, whereas DNMT3Amut alone and RUNX1mut may diminish, sensitivity to PARPi used as single agents and also combined with ruxolitinib plus or minus hydroxyurea (supplemental Figure 5). In addition, mutations in the BRCA1-BRCA2–containing complex 3 (BRCC3) gene implicated in DNA repair are frequently concomitant with JAK2 and MPL mutations and may modulate the sensitivity to PARPi.57
Although heterozygosity/homozygosity of the mutated “driver” allele does not appear to regulate sensitivity to olaparib (supplemental Figure 6A), it may affect the response to the combination of ruxolitinib plus hydroxyurea plus olaparib (supplemental Figure 6B). The mutant allele burden did not appear to influence PARPi efficacy (supplemental Figure 7; Figure 2; supplemental Table 1).
In conclusion, we demonstrated that JAK1/2i-induced DNA repair deficiencies may be clinically explored in preselected MPN patients treated with a combination of ruxolitinib and, as an innovative therapeutic approach, PARPi plus or minus hydroxyurea to enhance elimination of MPN-initiating and progenitor cell populations. All of these drugs have been approved as therapeutic agents in oncology, thus facilitating such a clinical trial. Moreover, a similar therapeutic approach could also be undertaken in other hematological malignancies displaying constitutive activation of JAKs either by direct mutation [eg, JAK2(R683S) in pediatric acute lymphoblastic leukemia58 and JAK3(A572V) in acute megakaryocytic leukemia59 ] or by activating mutation upstream of JAKs [eg, CSF3R(T618I) in chronic neutrophilic leukemia60 ] because cell lines transformed with these mutants were sensitive to the combination of ruxolitinib and olaparib (supplemental Figure 8).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Ann Mullally (Division of Hematology, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA) for providing murine Tet2−/−, Tet2+/+, Jak2(V617F),Tet2−/−, and Jak2(V617F),Tet2+/+ cells, Charles Mullighan (St. Jude Children’s Research Hospital, Memphis, TN) for providing BaF3-EpoR cells expressing JAK2(R683S), Jeffrey Taub (Children’s Hospital of Michigan, Detroit, MI) for providing CMK cells carrying JAK3(A572V), and Jeffrey Tyner (Oregon Health and Science University, Portland, OR) for providing BaF3 cells expressing CSF3R(T618I).
This work was supported by the Leukemia & Lymphoma Society Quest-for-Cure grant 0860-15 and National Institutes of Health National Cancer Institute grant CA134458 (T.S.). S.F. was supported by Narodowe Centrum Nauki grant 2013/11/B/NZ7/02248. B.V.L. was supported by the European Union’s Horizon 2020 research and innovation program under Marie Sklodowska–Curie grant agreement 665735 and by funding from the Polish Ministry of Science and Higher Education for the implementation of international projects. Work in the T.R.G. laboratory was supported by the Cambridge National Institute for Health Research Biomedical Research Centre and by the Wellcome Trust-Medical Research Council Cambridge Stem Cell Institute.
Authorship
Contribution: M.N.-S. performed in vitro studies with patient samples and ran Western analyses; S.M. tested DNA repair activity and analyzed mice bearing primary MPN xenografts; Y.D. and K.S. analyzed mice bearing MPNs; S.F. determined sample sensitivity in the context of allele burden; B.V.L. analyzed heterozygous/homozygous cells; M.S. performed experiments with cell lines; E.A.B. performed histopathology analyses; L.K. provided murine cells for in vivo studies; M.N. examined sensitivity of cells expressing MPN-unrelated mutants; M.K. performed NGS analysis of MPN samples; H.Z. performed statistical analyses; J.T.P., S.K., and T.R.G. provided MPN primary cells; R.C.S. and A.R.M. provided MPN primary cells and murine cells; M.W. supervised E.A.B.; K.P. supervised B.V.L.; T.S. conceived the idea, supervised the project, and wrote the final version of the manuscript; and all authors read, corrected, and approved the final version of the manuscript.
Conflict-of-interest disclosure: S.K. received honoraria and travel support for conferences from Novartis, Incyte, and Bristol-Myers Squibb. The remaining authors declare no competing financial interests.
Correspondence: Tomasz Skorski, Department of Microbiology and Immunology and Fels Institute for Cancer Research and Molecular Biology, School of Medicine, Temple University, 3400 N. Broad St, MRB 548, Philadelphia, PA 19140; e-mail: tskorski@temple.edu.