Abstract
Here we report a new fusion gene, STRN3-RARA, in acute promyelocytic leukemia (APL). It cooperates with UTX deficiency to drive full-blown APL in mice. Although STRN3-RARA leukemia quickly relapses after all-trans retinoic acid treatment, it can be restrained by cepharanthine.
TO THE EDITOR:
Acute promyelocytic leukemia (APL) is a distinct subtype of acute myeloid leukemia that is generally driven by PML-RARA, resulting from a balanced chromosomal translocation t(15;17) (q24.1;q21.2).1,2 However, approximately 2% of patients with APL carry other variants of RARA fusions, which pose challenges for diagnosis and treatment.3,4 Here, we report a novel t(14;17) translocation in a patient with APL that gave rise to a new fusion gene, STRN3-RARA. STRN3 forms the striatin-interacting phosphatase and kinase complex, which couples kinases to protein phosphatase 2A and regulates their activities.5 Like other patients with APL with RARA variants, this patient quickly relapsed after the initial response to all-trans retinoic acid (ATRA) treatment.6,7 Mechanistically, we demonstrated that STRN3-RARA, in cooperation with UTX deficiency (which was mutated in the patient), drove APL formation in mice. Additionally, we found that STRN3-RARA cells were specifically susceptible to the tumor necrosis factor α (TNFα) pathway. Inhibition of this pathway by cepharanthine, a Food and Drug Administration–approved drug, effectively suppressed the growth of U937 cells with STRN3-RARA expression, as well as relapsed APL cells from the patient. Taken together, our study identified a new APL fusion gene, STRN3-RARA, and validated it as a functional APL driver, providing insight into the diagnosis and treatment of patients with nonclassic APL.
Informed consent was obtained from the patient’s family. All mouse experiments were approved by the Institutional Animal Care and Use Committee of West China Hospital of Sichuan University.
A 50-year-old man (WCH01 [patient 01 with STRN3-RARA fusion in West China Hospital]) was admitted to the hospital with leukopenia and coagulopathy. In the bone marrow (BM), 91.5% of cells were hypergranular promyelocytes (Figure 1A), combined with immunophenotype and other clinicopathological features; this WCH01 patient was diagnosed with APL (Figure 1B and supplemental Table 1, available on the Blood website). Further transcriptomic analysis of WCH01 showed enrichment of gene signatures of APL_PML_RARA8 (Figure 1C). However, the classic chromosome translocation t(15;17) was not detected using the fluorescence in situ hybridization test with PML and RARA probes (supplemental Figure 1A). Instead, a new translocation between chromosomes 14 and 17 was found in the karyotyping result, which was further confirmed by RNA sequencing (RNA-seq) and Sanger sequencing (Figure 1D-F). This fusion happened between STRN3 exon 3 and RARA exon 3, resulting in a new STRN3-RARA fusion. Additionally, several gene mutations, including UTX frameshift mutation, were detected by the RNA-seq of WCH01 APL cells (Figure 1G and supplemental Figure 1B).
Initially, the WCH01 patient achieved complete remission with standard ATRA and arsenic trioxide (ATO) combination treatment, followed by consolidation with anthracycline-based therapy and maintenance with ATRA and arsenic trioxide. But after 10 months, he quickly relapsed and became resistant to ATRA and ATO treatment (Figure 1H and supplemental Figure 1C-E). A novel RARA missense mutation (S232F) was detected in the relapse sample (supplemental Figure 1F). The WCH01 patient then received a combination of ATRA and venetoclax. But unfortunately, no response was seen. He quickly developed severe pneumonia and eventually died from respiratory failure after 14 days of treatment with ATRA and venetoclax (Figure 1H).
To investigate the role of the STRN3-RARA (SR) fusion, we cloned Flag-tagged SR complementary DNA into a green fluorescent protein (GFP)-expressing retrovirus-based construct (pMSCV-Flag-SR-IRES-GFP) and transduced it into cell lines. Like PML-RARA and other reported RARA variant fusion proteins, the SR protein was predominantly located in the nucleus and was sensitive to ATRA treatment9,10 (Figure 2A-B and supplemental Figure 2A). Transcriptomics analysis of U937 cells with SR showed that APL_PML_RARA-upregulated genes were significantly positively enriched in SR cells (Figure 2C), which was consistent with the significant overlap of the differentially expressed genes in PML-RARA and those in STRN3-RARA (supplemental Figure 2B-C). We further conducted CUT&Tag sequencing of Flag-SR U937 cells to discover the SR-binding sites (Figure 2D) and found that these SR-binding sites significantly overlapped with PML-RARA-binding genes11 (Figure 2E). Combined with RNA-seq data, we found that SR protein directly bound to genes and upregulated many APL-related genes or downregulated multiple differentiation-related genes (supplemental Figure 2B-E). These results suggest that, like PML-RARA, the STRN3-RARA oncogenic protein exerts both repressive and activating functions through direct binding.11
To investigate the role of SR in APL genesis, we transduced the hematopoietic stem and progenitor cells with SR and followed by transplantation into sublethal irradiated recipient mice. We found that SR alone was insufficient to induce a full-blown APL in mice over a 5-month period, which is consistent with the previous observations in the PML-RARA mouse model12 (supplemental Figure 3). Considering that the frameshift mutation of a known tumor suppressor gene UTX was detected13-16 (Figure 1G), we investigated the role of SR in murine APL formation with Utx single guide RNA (sgUtx). The results showed that all sgUtx;SR cohort mice had developed leukemia by 175 days after transplantation and none of the sgUtx;Vector control mice were sick (Figure 2F and supplemental Figure 4A-H). The peripheral blood smear results displayed blast cells with abnormal promyelocytic morphology (Figure 2G). Pathological examinations of the BM, spleen, and liver also revealed the infiltration of leukemic cells (supplemental Figure 4I-K). Besides, recipient mice transplanted with sgUtx;SR APL cells received ATRA or vehicle treatment, resulting in a reduction of GFP + mCherry + APL cells in the peripheral blood. The induction of differentiation was observed through blood smear and RNA-seq analysis (Figure 2H-I and supplemental Figure 5). Furthermore, the treatment significantly extended the survival of the recipient mice (Figure 2J). These results indicate that SR fusion with Utx deficiency drives APL.
To elucidate the molecular mechanisms underlying the cooperation between SR fusion and UTX loss, we studied the gene expression upregulated or downregulated in the WCH01 compared with normal human BM cells or in U937 cells with sgUTX;SR compared with that with empty vector and found that 454 upregulated genes (common_up_454) and 587 downregulated genes (common_down_587) were significantly overlapped17,18 (supplemental Figure 6A-C). According to these gene expression patterns, we divided the common_up_454 genes (or common_down_587) into 3 groups and labeled them with different colors. Genes in the green group were likely regulated mainly by SR, genes in the orange group were regulated mainly by UTX, and genes in the blue group seem to be under the control of both UTX and SR (supplemental Figure 6D-E).
To find a new inhibitor for sgUTX;SR, we searched for candidate drug-targetable genes that were specifically upregulated by sgUTX;SR but not PML-RARA. TNF gene and pathway were at the top of the list (Figure 2K-L). Therefore, we examined the impact of TNFα inhibitor cepharanthine, a Food and Drug Administration–approved drug, on the growth of U937 cells with vector, sgUTX, SR, or sgUTX;SR.19 Our results demonstrated that U937 cells with SR or sgUTX;SR were more sensitive to cepharanthine than control cells (Figure 2M). Furthermore, although clinical SR APL cells collected from WCH01 patient who relapsed showed no response to ATRA treatment, their viability was reduced by cepharanthine treatment (Figure 2N). These results suggest that cepharanthine could serve as a potential therapeutic method for patients with APL with STRN3-RARA fusion.
In conclusion, we have identified a novel fusion gene, STRN3-RARA, in a patient with APL. This SR APL displayed similar clinical and pathological characteristics as patients with classical APL carrying PML-RARA. Additionally, we have demonstrated that SR, with the Utx loss, drove APL genesis in mice, revealing the oncogenic role of this fusion gene in leukemogenesis. Furthermore, we found that SR APL cells collected from the WCH01 patient who relapsed were sensitive to the inhibitor of TNFα signaling pathway. Taken together, our findings not only advance the understanding of APL disease but also provide valuable insights for the diagnosis and treatment of patients with APL variants.
Acknowledgments
The authors thank the WCH01 patient and his family; all the members of the Chen and Liu laboratory for their technical support and suggestions; Yuquan Wei for his generous support; Zhihong Xu and Ying Lin at Sichuan Hua Xi Kindstar Medical Diagnostic Centre for their valuable assistance with karyotype and fluorescence in situ hybridization analysis; Qiaorong Huang at the Laboratory of Stem Cell Biology, West China Hospital, Sichuan University for assistance with flow cytometry analysis; and the Core Facilities of West China Hospital for technical support.
This work was supported by the National Natural Science Foundation of China (grants 82130007 [Y.L.]; 82300185 [Q.Z.]; 81670182 [Y.L.]; 82170171 [C.C.]; 81770157 [C.C.]), the China Postdoctoral Science Foundation (grants 2023M732465 [Q.Z.]; 2023M732434 [X.C.]), the Sichuan Science and Technology Program (grants 2020YFQ0059 [C.C.]; 2018JZ0077 [Y.L.]; 2020ZYD002 [Y.L.]; and 2017TJPT0005 [C.C.]), the 1.3.5. Project for Disciplines of Excellence, West China Hospital, Sichuan University (grants ZYGD22012 [Y.L.]; ZYJC21009 [Y.L.]; 2022HXFH040 [H.M.]; and ZYYC20004 [C.C.]), the Post-Doctor Research Project, West China Hospital, Sichuan University (grants 2023HXBH094 [Q.Z.]; and 2023HXBH033 [X.C.]), and the Sichuan University Postdoctoral Interdisciplinary Innovation Fund (grant JCXK2210 [X.C.]).
Authorship
Contribution: Y.L. and H.M. designed and supervised this study; T.L., C.C., and W.M. were involved in the study design; H.M., H.L., Y.W., and H.C. recruited and treated the patient; Q.Z., F.G., Lanxin Zhang, and T.C. designed and performed experiments; X.C. and Lu Zhang performed the bioinformatic analyses; Q.C. provided human cells; and Y.L., H.M., C.C., Q.Z., and X.C. analyzed the data and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hongbing Ma, Department of Hematology and Institute of Hematology, West China Hospital, Sichuan University. NO. 37 Guo Xue Xiang St, Chengdu 610041, China; e-mail: hongbingma@foxmail.com; and Yu Liu, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, 3-17 Renming Rd, Chengdu 610041, China; e-mail: yuliuscu@scu.edu.cn.
References
Author notes
∗Q.Z., H.L., and X.C. contributed equally to this work.
The RNA-seq and CUT&TAG data in this study are deposited in NCBI GEO: GSE224816.
Data are available upon request to the corresponding authors (hongbingma@foxmail.com and yuliuscu@scu.edu.cn).
The online version of this article contains a data supplement.


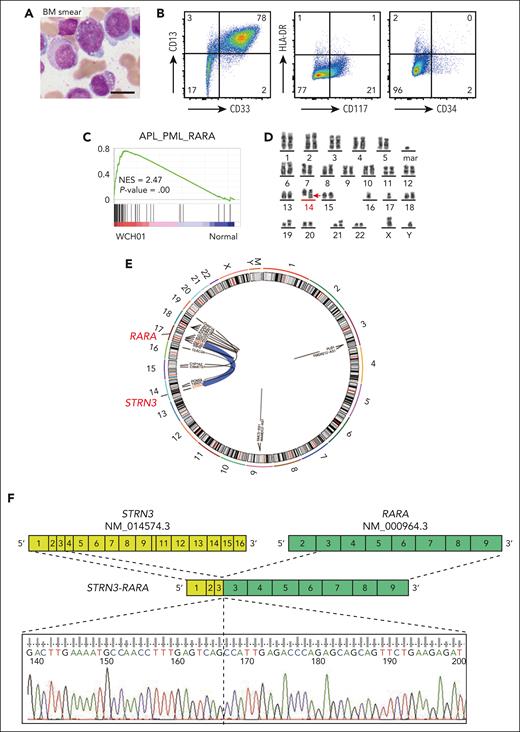
![Identifying an APL with STRN3-RARA translocation. (A) BM smear of the WCH01 patient before treatment. Scale bar, 10 μm. (B) Flow cytometry analysis of the WCH01 patient’s BM cells before treatment. The expression of CD13, CD33, CD117, CD34, and HLA-DR is shown. The numbers indicate the percentage of different cell populations. (C) Gene Set Enrichment Analysis (GSEA) showing the positive enrichment of the APL_PML_RARA gene set in the WCH01 patient, compared with normal gene sets (NES = 2.47; P = .00). (D) Cytogenetics analysis of the WCH01 patient’s BM cells before treatment. The karyotype analysis of the WCH01 patient reveals the following abnormalities: 45, XY, add(6)(q13), der(14)t(14,17)(q12;q21) dup(17)(q21q25), −16, −17, add(18)(p11), der(19)t(15;19)(q22;p13.3), add(21)(p11), +mar, inc[20]. The red arrow highlights the t(14,17) chromosome translocation. (E) The circular plot showing an overview of the top 10 fusion events between locations in chromosomes. (F) Schematic representation and Sanger sequencing of the STRN3-RARA fusion. The yellow blocks represent exons of the STRN3 gene (NM_014574.3), and the green blocks represent exons of the RARA gene (NM_000964.3). Numbers in the blocks indicate the exon number. The Sanger sequencing at the breakpoint site is shown at the bottom. (G) Sanger sequencing of the UTX locus obtained from the polymerase chain reaction product of the WCH01 patient. (H) Percentage of blast cells in the WCH01 patient’s bone marrow during treatment. The blocks indicate the drug treatments administered during the period. DA, daunorubicin and cytarabine; HA, homoharringtonine and cytarabine; NES, normalized enrichment score; Ven, venetoclax.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/142/17/10.1182_blood.2023020619/2/m_blood_bld-2023-020619-gr1b.jpeg?Expires=1766035784&Signature=EVjYAlo9GWesiVzcn3MU2jXD8h0L~F0nRYn71ioSKBm3lvFKauhezVT~EKYcOMtA-0O5K2-FtOtwHPtBgAsAoJZqy2ZKmHzaE36yIhjgOHMUJM-MbLin~dJsgZEYFIIC0-fLuj8JXoy52vr02mdkrsJkR0fl4Ra~PbI1RcGY0Cv2hUkDRf0g8C4wilTLOmavN47D89bsTln9IqbAZVtjL25LX~Aq9IUwQbYXEGi6pj5v5WKmz2KsULfCavd9Fsu0vZCxfAvvLeoFIw07FVI7kwfT90zpv5kBYVkL2TrBc02USUu5YYfg4RMZ1cebvjIqNwHA2wcfG~aKqD~nRHGZHQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
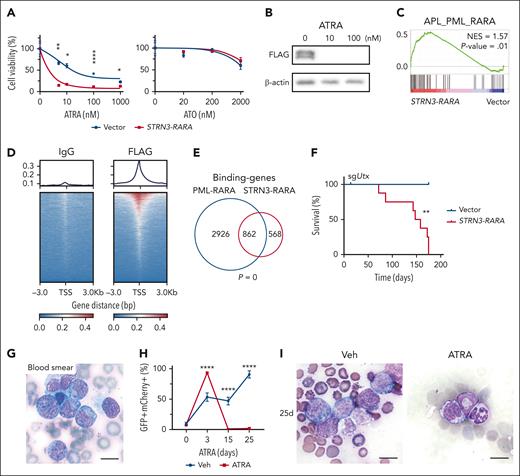
![The oncogenic role of STRN3-RARA fusion gene in leukemogenesis. (A) Treatment curve of U937 cells with ATRA and ATO. Cells were transduced with vector or STRN3-RARA. Means ± standard deviation (SD), n = 3; ∗P < .05, ∗∗P < .01, ∗∗∗∗P < .0001 (2-way analysis of variance [ANOVA]). (B) Western blot analysis of ATRA-treated 293T cells harvested 2 days after treatment. 293T cells were transduced with Flag-STRN3-RARA. (C) GSEA showing positive enrichment of the APL_PML_RARA gene set in U937 cell with STRN3-RARA overexpression, compared with vector (NES = 1.57; P = .01). (D) Levels of STRN3-RARA bound at the transcription start site (TSS) in U937 cells transduced with STRN3-RARA, measured by the CUT&Tag analysis. (E) The Venn diagram showing the overlap of binding-genes in STRN3-RARA and PML-RARA (hypergeometric test). (F) Kaplan-Meier tumor-free survival curves of recipient mice in vector and STRN3-RARA group with Utx knockout, n = 6 (vector), n = 8 (STRN3-RARA); ∗∗P < .01 (log-rank test). (G) Blood smear of sgUtx;SR recipient mouse before sacrificed. Scale bar, 10 μm. (H) The percentage of sgUtx;SR cells (GFP + mCherry + cells) along with vehicle and ATRA treatment. The x-axis indicates the number of days after treatment. Means ± SD, vehicle, n = 5, 4, 4, 2; ATRA, n = 5, 4, 4, 4; ∗∗∗∗P < .0001 (2-way ANOVA). (I) Blood smears from sgUtx;SR mice following a 25-day treatment with vehicle or ATRA. Scale bar, 10 μm. (J) Survival curve of sgUtx;SR mice following a 25-day ATRA or vehicle treatment. n = 5; ∗P < .05 (log-rank test). (K) Heat map showing the 140 drug targetable genes that were specifically upregulated in U937 cells with sgUTX;SR compared with that with empty vectors and not overlapped with the upregulated genes due to PML-RARA. (L) GSEA showing the positive enrichment of the HALLMARK_TNFA_SIGNALING_VIA_NFKB gene set in sgUTX;SR U937 cells, compared with vector (NES = 1.67; P = .00). (M) Relative viability of U937 cells treated with 1 μM cepharanthine. Vector, sgUTX, SR, and sgUTX;SR groups are shown. Means ± SD, n = 3; ∗P < .05, ∗∗P < .01 (1-way ANOVA). (N) Drug treatment survival curve of WCH01 relapsed cells. ATRA and cepharanthine were treated respectively. Means ± SD, n = 3; ∗∗P < .01, ∗∗∗P < .001 (2-way ANOVA). IgG, immunoglobulin G; Veh, vehicle.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/142/17/10.1182_blood.2023020619/2/m_blood_bld-2023-020619-gr2b.jpeg?Expires=1766035784&Signature=we2IIVkxDIJmZFSKN5sYlT8ewldXW0uWIRCK7MuHq07ScHwG1SOvs03VS38bio2qAmGIqAO1Z4APPqO79p3TKy0W~XI8~Q7z~ZoqKsAcxn4-LGZ5HW0rQOmjZg5Wuc9SQyTp1rrSt~BOTbEGHnnZ3hvZSzlMXS~FxsED8i-DQ28ZXlQl0Q8RZKe9J5rfm7~Putctfw3alXUwLfRAVW7UuKX2sFEM6oCx-3BPmqZ37WxT7j7sqVWEVtAXPZ5o2BEZXO-nqZhDXNRR4pp9ZiEYxOmuoWWdg39wxWC8ncFLLrR41cziygH3S8UjZputvMgp0QWFsArRErPz-dBo3gegcg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)