Abstract
The nonobese diabetic/severe combined immune-deficient (NOD/SCID) mouse xenotransplantation assay is the most commonly used surrogate assay for the study of human candidate stem cells. In contrast to large animal and human studies, however, it is limited by the short life span of the recipients, the limited proliferative demand placed on the transplanted cells, and the inability to support differentiation into all hematopoietic lineages. In the present study, we directly compared hematopoietic repopulation in NOD/SCID mice with autologous reconstitution in the baboon, a well-established preclinical large animal model for stem cell transplantation. Baboon CD34-enriched marrow cells were retrovirally marked and infused into the irradiated baboon and the NOD/SCID mice. Although the percentage of gene-marked cells was high and remained stable in NOD/SCID mice up to 12 weeks and in those that underwent secondary transplantation, we observed a considerable decline and overall a significantly (10-fold) lower percentage of gene-marked cells in the baboons. In addition, clonal integration site analysis revealed common proviral vector integrants in NOD/SCID repopulating cells and in the baboon at 6 weeks but not at 6 months after transplantation. These results suggest that distinct hematopoietic stem/progenitor cells are responsible for hematopoietic reconstitution in NOD/SCID mice compared with nonhuman primates. (Blood. 2003;102:4329-4335)
Introduction
A significant obstacle in hematopoietic stem cell research has been the difficulty in identifying and characterizing human hematopoietic stem cells in vitro. Thus, a number of surrogate assays have been developed to define and study stem cells. Most definitions require that candidate stem cells provide stable multilineage repopulation and differentiation into all blood cell types after transplantation. For the study of human hematopoietic stem cells, the NOD/LtSz scid/scid (NOD/SCID) mouse model has been the most commonly used surrogate assay.1,2 The nonobese diabetic/severe combined immune-deficient (NOD/SCID) model has been shown to assay a defined population of primitive cells distinct from most of the more differentiated progenitors that are detected using short- and long-term in vitro culture assays.3 Although the existence of pluripotent SCID repopulating cells (SRCs) has been demonstrated,4,5 the NOD/SCID model is limited by the short life span of the recipients relative to the native host, the limited proliferative demand placed on the transplanted cells, and the inability to support differentiation into all hematopoietic lineages. Additionally, a significant contribution of lineage-restricted progenitor cells to engraftment in this model cannot be ruled out.
The NOD/SCID model has also been used extensively to study gene transfer into hematopoietic repopulating cells. SRCs can be transduced efficiently with retroviral and lentiviral vectors,6-9 in contrast to lower gene-marking levels in large animal studies and human gene therapy trials. This discrepancy has not been explained, and one question that remains is which aspects of human stem cell biology can be accurately predicted by the NOD/SCID xenotransplantation model. In this study, we therefore used retrovirally marked, CD34-enriched baboon bone marrow cells to directly compare their ability to repopulate irradiated NOD/SCID mice.
Materials and methods
Retrovirus vectors
The MNDEGFPSN and MNDEYFPSN vectors encode the enhanced green fluorescence protein (EGFP) and its yellow variant, EYFP, under the control of the 5′ modified viral long terminal repeat sequence (LTR) and the bacterial neomycin phosphotransferase (neo) gene conveying G418 resistance under the control of the SV40 promoter. Both vectors are identical with the exception of the 9 nucleotides distinguishing EGFP from EYFP. Individual clones from PG13 or Phoenix gibbon ape leukemia virus (GALV) retrovirus-producing packaging cells with similar titers were selected for the MNDEGFPSN/MNDEYFPSN comparisons.
Baboons
Healthy juvenile baboons were housed at the University of Washington Regional Primate Research Center under conditions approved by the American Association for the Accreditation of Laboratory Animal Care. Studies were conducted under protocols approved by the Institutional Review Board and Animal Care and Use Committees. Autologous baboon transplantations and all other procedures were performed as previously described.10-12
Gene transfer into baboon CD34-enriched cells
CD34+ cells were enriched from baboon marrow leukocytes using immunoglobulin M (IgM) monoclonal antibody (mAb) 12-8 with an immunomagnetic column technique (Miltenyi Biotec, Auburn, CA) as described.10-12 Equal numbers of CD34-enriched cells were prestimulated for 48 hours in tissue-culture-treated 75-cm2 canted-neck flasks (Corning, Corning, NY) in Iscove medium containing 10% fetal bovine serum (FBS; Hyclone, Logan, UT) in the presence of stem cell factor (SCF), granulocyte-colony-stimulating factor (G-CSF), and megakaryocyte growth and development factor (MGDF) at 100 ng/mL each and then transferred into non-tissue culture (TC)-treated 75-cm2 canted-neck flasks (Falcon, Franklin Lakes, NJ) that had been coated with CH-296 (RetroNectin; Takara Shuzo, Otsu Japan) at 2 μg/cm2 and preloaded twice with virally conditioned media (VCM).13 Cells were exposed to pure VCM for 4 hours in the presence of the same cytokines, then collected and resuspended in fresh media with cytokines; the next day, another 4-hour exposure to vector was performed before the cells were harvested. An aliquot of each transduction culture was removed for injection into NOD/SCID mice before the cells were pooled and reinfused into the irradiated baboon (1020 cGy total body irradiation [TBI]). A more detailed description of the baboon transplantation procedure and follow-up is described in Horn et al.12
Transplantation of NOD/SCID mice with baboon CD34+ cells
Cell transplantation into NOD/LtSz-scid/scid (NOD/SCID) mice was similar to a published standard protocol.1 All mice were offspring of breeders purchased from The Jackson Laboratory (Bar Harbor, ME). All animals were handled under sterile conditions and maintained in microisolators. Before transplantation, 6- to 8-week-old mice underwent TBI with 375 cGy at 20 cGy/min from a linear accelerator source. Within 24 hours, 2 × 106 CD34 enriched baboon cells were transplanted by tail-vein injection after retroviral transduction, after mock transduction, or uncultured, as noted.
Linear amplification-mediated polymerase chain reaction
Linear amplification-mediated (LAM)-polymerase chain reaction (PCR) analysis was performed as described by Schmidt et al.14,15 Insertion-site sequence data were aligned to the human genome, using NCBI Blast2 (http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html) and NCBI BlastN (http://www.ncbi.nlm.nih.gov:80/BLAST).
Secondary NOD/SCID transplantations
The total marrow content from both tibias and femurs of primary recipients was harvested. After red cell lysis and 2 washes, the cells were resuspended in phosphate-buffered saline (PBS) with 2% FBS. A 10% aliquot was removed for fluorescence-activated cell sorter (FACS) analysis, and the remaining cells (90%) were injected into secondary irradiated recipients. The total number of baboon cells in the bone marrow was estimated by multiplying the percentage of CD11a+ cells by the number of viable cells harvested, then by 4 (equivalent of 2 femurs plus 2 tibias = 25% of the equivalent of the total bone marrow of a mouse).16 This calculation is likely an underestimation because of the loss of cells during injection and during cell preparation.
Flow cytometric analysis of murine bone marrow
Six or 12 weeks after transplantation, mice were humanely killed and bone marrow was harvested from femura and tibiae. Red blood cells were lysed by adding 2.5 mL lysis buffer (0.155 M NH4Cl + 0.01 M KHCO3 + 104 M EDTA [ethylenediaminetetraacetic acid]), and the remaining white cells were washed in PBS containing 2% fetal calf serum (FCS). To assess overall engraftment and marking, aliquots containing at least 5 × 105 cells were stained with antihuman CD11a mAb conjugated to phycoerythrin (PE) (clone G43-25B; Becton Dickinson, San Jose, CA) as a pan-leukocytic marker for baboon hematopoietic cells. Immediately before flow cytometric analysis, 2 μg/mL propidium iodide (PI) was added to the cells on a FACSCalibur (Becton Dickinson). Subset analysis of engrafted baboon cells was performed by gating on CD11a+ cells and using PE-cyanine 5 (Cy5)-conjugated lineage-specific antibodies. Antibodies used were CD34 mAb (clone 563 or biotinylated 12-8 mAb in combination with streptavidin-PE-Cy5; both Becton Dickinson), CD20 mAb (clone B9E9; Immunotech, Westbrook, ME), or CD13 mAb (clone TÜK1; Caltag, Burlingame, CA). 4′,6-Diamidino-2-phenylindole dihydrochloride hydrate (DAPI) (Sigma, St Louis, MO) was used as a live/dead marker, and 4-color flow cytometric analysis was performed on an LSR (Becton Dickinson). For each mouse analyzed, an aliquot of cells labeled with conjugated isotype control mAbs was used as a control. In addition, bone marrow cells from NOD/SCID mice that did not undergo transplantation were stained with the same mAbs.
Analysis of EGFP/EYFP expression in a colony-forming unit assay
CD11a-expressing cells were sorted using a FACS Vantage (BD Biosciences, San Jose, CA) and were cultured (1000 cells per 35-mm plate) in triplicate in a double-layer agar culture system as described.11 Briefly, isolated cells were cultured in α minimal essential medium supplemented with 15% FBS (Hyclone), 0.1% bovine serum albumin (BSA; fraction V; Sigma), 0.3% (wt/vol) agar (BioWhittaker, Rockland, ME) overlaid on medium with 0.5% agar (wt/vol) containing 100 ng/mL SCF, interleukin-3 (IL-3), IL-6, granulocyte macrophage colony-stimulating factor (GM-CSF), G-CSF, and 4 U/mL erythropoietin (EPO; provided by G. Molineux, Amgen, Thousand Oaks, CA). Cultures were incubated at 37°C in a 5% CO2 humidified incubator. Colonies were evaluated for EGFP/EYFP expression at day 14 of culture using an inverted fluorescence microscope.
Wright and myeloperoxidase stain of baboon SRCs
Cytocentrifuge preparations of CD11a-sorted cells were subjected to Wright staining using the standard methodology. Similar cytocentrifuge preparations were also stained for myeloperoxidase using a brief formaldehyde fixation, followed by incubation with hydrogen peroxide and benzidine dihydrochloride for 30 seconds, and then counterstained with Giemsa.
Results
Engraftment of gene-marked CD34+ cells after autologous transplantation in the baboon
We have recently reported the development of an improved gibbon ape leukemia virus (GALV)-pseudotype retroviral vector packaging cell line based on human 293T cells.12 In this study we used GALV-pseudotype oncoretroviral vectors, which were produced by either the murine PG13/MNDEGFPSN or the human Phoenix-GALV/MNDEYFPSN producer cell line to transduce CD34-enriched marrow cells.10-12 In 3 separate experiments, transduced cells were infused into lethally irradiated baboons. The highest gene marking in baboons was observed in granulocytes at early time points after transplantation (up to 39% EYFP-expressing cells at 2 weeks after transplantation). The percentage of gene-marked cells then decreased over 2 to 3 months before reaching a stable plateau. The average marking level in the 3 baboons at 2 months after transplantation was 6.5% EYFP-expressing cells (Phoenix-GALV) compared with 2.6% EGFP-expressing cells (PG13). Marking was detected in all hematopoietic lineages, and at all time points vectors produced by the human producer cell line resulted in higher gene transfer into baboon repopulating cells than the murine-derived producer cell line.12
Engraftment of gene-marked baboon CD34+ cells in NOD/SCID mice
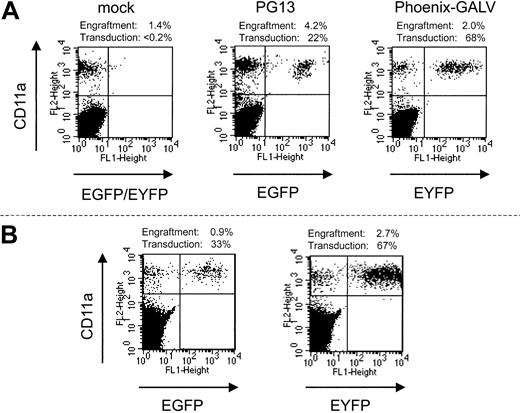
In each of the 3 experiments, an aliquot of the transduced CD34-enriched cells that was infused into the baboon was also infused into sublethally irradiated (375 cGy) NOD/SCID mice. To simplify flow cytometric analysis in the mice, EGFP- and EYFP-transduced cells were not pooled before infusion but, rather, infused into separate groups of mice. Thus, these mice received cells identical to those of the baboons, but the 2 experimental fractions were assayed independently (Figure 1). Thirty-two NOD/SCID mice underwent transplantation with transduced baboon CD34 cells in parallel with the 3 baboons. After 6 weeks, bone marrow was harvested and evaluated by flow cytometry for engraftment of baboon cells and transgene expression in the engrafted cells (Figure 2A). We were able to detect the engraftment of baboon cells in 30 of 32 mice, with a mean total baboon cell engraftment level of 4.5%. Table 1 summarizes the results from all 3 of these parallel transplantations. The mean percentage of engrafted baboon cells expressing EYFP (Phoenix GALV) and EGFP (PG13) in NOD/SCID mice from 3 separate experiments was approximately 60% and approximately 45%, respectively. Although the difference in gene transfer efficiency between the 2 vectors detected in the autologous nonhuman primate model was also reflected in the NOD/SCID mouse model, the actual percentage of gene-marked cells within the baboon SCID repopulating cells was significantly higher than the percentage in the blood of the baboons.
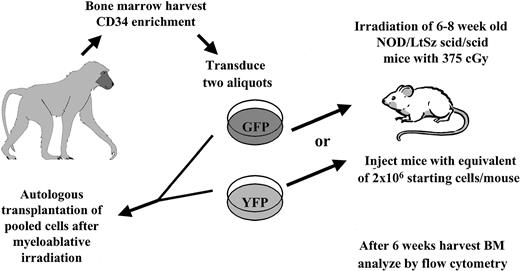
Experimental setup. Autologous transplantation of 3 baboons was performed as described in “Materials and methods.” An aliquot of cells was taken and transplanted into irradiated NOD/SCID mice, which received treated cells identical to those given to recipients of the autologous transplants.
Experimental setup. Autologous transplantation of 3 baboons was performed as described in “Materials and methods.” An aliquot of cells was taken and transplanted into irradiated NOD/SCID mice, which received treated cells identical to those given to recipients of the autologous transplants.
Engraftment of transduced baboon marrow cells in NOD/SCID mice. Displayed are examples from the 3 groups of mice that underwent autologous transplantation in parallel with baboons. (Left) mock-transduced cells; (middle) PG13; (right) Phoenix-GALV-transduced cells. (B) Engraftment with baboon SCRs is stable over 12 weeks. Two mice were observed for 12 weeks before bone marrow was harvested and analyzed. Neither the percentage of engrafted baboon cells nor transgene expression is markedly different than at 6 weeks after transplantation.
Engraftment of transduced baboon marrow cells in NOD/SCID mice. Displayed are examples from the 3 groups of mice that underwent autologous transplantation in parallel with baboons. (Left) mock-transduced cells; (middle) PG13; (right) Phoenix-GALV-transduced cells. (B) Engraftment with baboon SCRs is stable over 12 weeks. Two mice were observed for 12 weeks before bone marrow was harvested and analyzed. Neither the percentage of engrafted baboon cells nor transgene expression is markedly different than at 6 weeks after transplantation.
Oncoretroviral marking is higher in baboon SRCs than in autologous repopulating cells
. | Marking in baboons . | . | . | Mean SRC engraftment, % . | Marking in SRCs, % . | ||
|---|---|---|---|---|---|---|---|
| Animal . | 2 wk, % . | 6 wk, % . | 18 mo, % . | . | . | ||
| F99070 | 4.9 | ||||||
| PhoenixG | 33.8 | 7.3 | 4.8 | 54 | |||
| PG13 | 13.7 | 3.8 | 2.9 | 29 | |||
| 00021 | 2.6 | ||||||
| PhoenixG | 16.0 | 7.7 | 6.4 | 60 | |||
| PG13 | 4.3 | 2.6 | 3.7 | 45 | |||
| A00066 | 4.8 | ||||||
| PhoenixG | 29.1 | 4.4 | NA | 61 | |||
| PG13 | 16.0 | 2.4 | NA | 45 | |||
. | Marking in baboons . | . | . | Mean SRC engraftment, % . | Marking in SRCs, % . | ||
|---|---|---|---|---|---|---|---|
| Animal . | 2 wk, % . | 6 wk, % . | 18 mo, % . | . | . | ||
| F99070 | 4.9 | ||||||
| PhoenixG | 33.8 | 7.3 | 4.8 | 54 | |||
| PG13 | 13.7 | 3.8 | 2.9 | 29 | |||
| 00021 | 2.6 | ||||||
| PhoenixG | 16.0 | 7.7 | 6.4 | 60 | |||
| PG13 | 4.3 | 2.6 | 3.7 | 45 | |||
| A00066 | 4.8 | ||||||
| PhoenixG | 29.1 | 4.4 | NA | 61 | |||
| PG13 | 16.0 | 2.4 | NA | 45 | |||
Displayed are, for each baboon, the percentage of transgene-expressing cells of both experimental arms in white blood cells at approximately 2 weeks, 6 weeks, and 18 months after transplantation, the mean engraftment of baboon NOD/SCID repopulating cells (SRCs) after 6 weeks, and the percentage of transgene-expressing baboon SRCs.
NA indicates not applicable: the animal was transferred to a different study and humanely killed 6 months after transplantation.
To assess whether engraftment and transgene expression of baboon SRCs are stable over time, we monitored 2 mice from a separate experiment for 12 weeks (Figure 2B). Neither the percentage of engrafted baboon cells nor the percentage of transduced cells changed significantly between 6 weeks and 12 weeks, suggesting that engraftment and marking of baboon SRCs are stable over at least 12 weeks. This pattern was in contrast to marking levels in the respective baboons. In all 3 baboons the percentage of marked cells decreased within the first 6 weeks to levels that were significantly (10-fold) lower than in the NOD/SCID mice. Interestingly, the percentage of marked cells very early after transplantation was only approximately 2- to 3-fold lower in the baboons, suggesting that NOD/SCID repopulating cells more closely resemble short-term repopulating cells than long-term repopulating cells in the baboon.
Multilineage engraftment of baboon cells in NOD/SCID mice
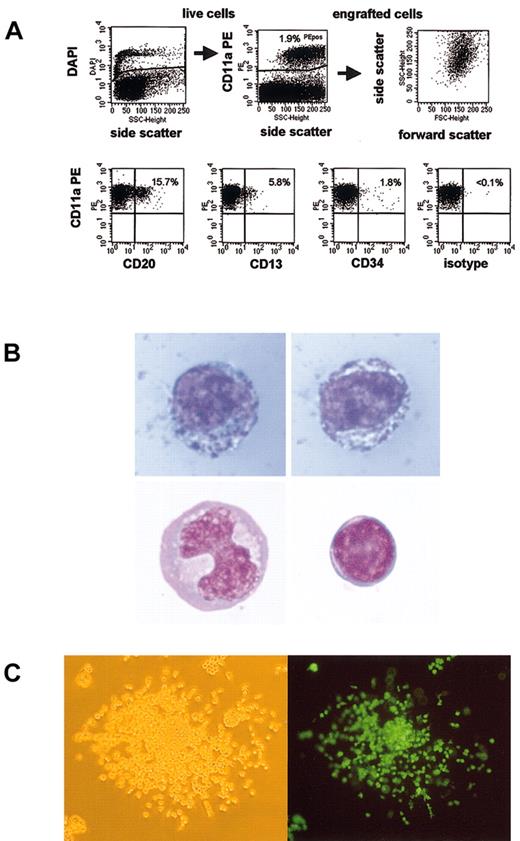
One potential explanation for the difference in marking in the 2 models is that the baboon CD34 cells did not truly engraft in these NOD/SCID mice but merely survived for these periods of time. Because multilineage differentiation of these cells has been used as an indicator of engraftment, we performed subset analysis of baboon cells recovered from mouse bone marrow. Figure 3A shows a subset analysis of baboon SRCs. We were able to detect CD13+ myeloid cells, CD20+ B-lymphoid cells, and CD34+ precursors. Gene-marked cells were detected in all subsets. Thus, multilineage engraftment of baboon SRCs was achieved. However, most cells (typically 75% to more than 95%) did not stain with any of these subset markers. This finding is consistent with a recent report by Norol et al.17
Baboon SRCs are capable of multilineage engraftment. (A) Displayed is the gating strategy and a representative subset analysis of engrafted baboon SRCs. Overall engraftment of baboon cells (CD11a+) was 1.9%. CD13 was expressed in 5.8%, CD20 in 15.7%, and CD34 in 1.8% of engrafted cells. (B) Myeloid precursors make up most engrafted baboon SRCs. Baboon cells from an engrafted NOD/SCID mouse were sorted, and cytospins were performed and stained. Although most engrafted SRCs appeared to be myeloid precursors (top panels, myeloperoxidase stain), mature monocytes (lower left panel) and lymphocytes (lower right panel) were also observed. Lower panels, Wright stain. Original magnification × 1000. (C) Baboon SRCs form colonies in agar. Baboon cells from engrafted NOD/SCID mice were sorted and plated into semisolid media. The percentage EGFP/EYFP-expressing colonies was similar to the percentage of transgene-expressing SRCs, as assessed by flow cytometry. (Left) White light microscopy; (right) fluorescence microscopy. Original magnification × 100.
Baboon SRCs are capable of multilineage engraftment. (A) Displayed is the gating strategy and a representative subset analysis of engrafted baboon SRCs. Overall engraftment of baboon cells (CD11a+) was 1.9%. CD13 was expressed in 5.8%, CD20 in 15.7%, and CD34 in 1.8% of engrafted cells. (B) Myeloid precursors make up most engrafted baboon SRCs. Baboon cells from an engrafted NOD/SCID mouse were sorted, and cytospins were performed and stained. Although most engrafted SRCs appeared to be myeloid precursors (top panels, myeloperoxidase stain), mature monocytes (lower left panel) and lymphocytes (lower right panel) were also observed. Lower panels, Wright stain. Original magnification × 1000. (C) Baboon SRCs form colonies in agar. Baboon cells from engrafted NOD/SCID mice were sorted and plated into semisolid media. The percentage EGFP/EYFP-expressing colonies was similar to the percentage of transgene-expressing SRCs, as assessed by flow cytometry. (Left) White light microscopy; (right) fluorescence microscopy. Original magnification × 100.
To further characterize these cells, baboon cells were sorted and stained either with a Wright stain for morphology or with a myeloperoxidase stain as an early myeloid marker. Figure 3B shows stained, engrafted baboon NOD/SCID repopulating cells. The blue granula in the cells displayed in the upper panel are consistent with myeloperoxidase activity, indicating that these cells are myeloid precursors. These cells made up most of the sorted cells, but mature monocytes (lower left panel), lymphocytes (lower right panel), and granulocytes (not shown) were also detected. Thus, most engrafted baboon SRCs had a different phenotype than human SRCs, which have been reported to differentiate predominantly into B-lymphoid cells.18 When these baboon cells were sorted and plated into semisolid media containing appropriate growth factors, colony-forming units (CFUs) grew (Figure 3C). Consistent with the high level of gene marking in SRCs, most colonies expressed the transgene, confirming that they were indeed derived from transduced baboon cells.
Common proviral integration sites between baboon and NOD/SCID repopulating cells
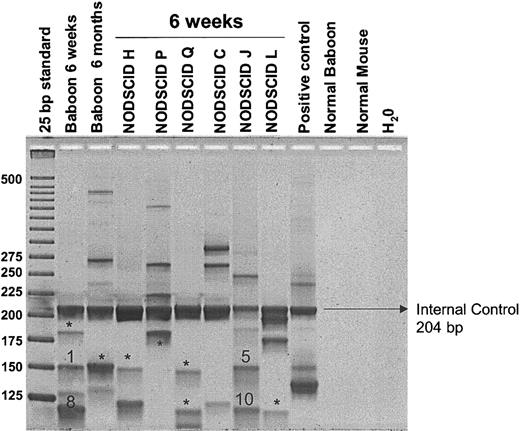
We used LAM-PCR to determine integration sites of transduced cells in baboon A00066 and the corresponding NOD/SCID mice. Multiple proviral integration sites were identified in the baboon and the corresponding NOD/SCID mice (Figure 4). No obvious difference in the number of integration sites was detected between 6 weeks and 6 months after transplantation in the baboon. Eleven bands, each representing a proviral integration site, were sequenced. No common integration sites were found between the early and late baboon samples or between the NOD/SCID mice and the late baboon sample. However, 2 common integration sites were determined for the early baboon peripheral blood sample and one of the corresponding NOD/SCID mice (Figure 4). Details of these common integration sites are given in Table 2. These results demonstrate that (1) multiple hematopoietic repopulating clones engraft in the NOD/SCID mice, (2) these clones are capable of contributing to short-term hematopoiesis in the baboon, and (3) the common vector integrants were only observed early after transplantation, thus supporting the conclusions from the marking results that the NOD/SCID repopulating cells are more reflective of short-term repopulating cells than long-term repopulating cells.
Detection of retrovirally marked hematopoietic clones contributing to engraftment in NOD/SCID mice compared with autologous engraftment. LAM-PCR was used to identify individual clones contributing to hematopoiesis after transplantation. Peripheral blood samples of baboon A00066 at 6 weeks and at 6 months after transplantation and NOD/SCID marrow samples were analyzed. Note that engraftment is oligoclonal in all NOD/SCID mice. *Bands that were subjected to insertion site sequencing and had unique proviral integration sites. Bands 1, 5, 8, and 10 were sequenced and showed common integration sites between baboon and NOD/SCID repopulating cells.
Detection of retrovirally marked hematopoietic clones contributing to engraftment in NOD/SCID mice compared with autologous engraftment. LAM-PCR was used to identify individual clones contributing to hematopoiesis after transplantation. Peripheral blood samples of baboon A00066 at 6 weeks and at 6 months after transplantation and NOD/SCID marrow samples were analyzed. Note that engraftment is oligoclonal in all NOD/SCID mice. *Bands that were subjected to insertion site sequencing and had unique proviral integration sites. Bands 1, 5, 8, and 10 were sequenced and showed common integration sites between baboon and NOD/SCID repopulating cells.
Integration site analysis*
Fragment . | Sample . | Size . | Chromosome . | Insertion site . | Insertion site details . | Insertion sequence . |
|---|---|---|---|---|---|---|
| 1 | Baboon 6 wk | |||||
| 73 bp | 12 | 101007-101074 | Homo sapiens BAC clone RP11-87C12 | ACTTCAGGTCAGGAGTTCGAGACCAATCTGGCCAAC | ||
| ATGGTGAAACCCCGCCTCTACTAAAAATACAAAAATT | ||||||
| 5 | NOD/SCID J | |||||
| 8 | Baboon 6 wk | Homo sapiens BAC clone RP11-203116 contains the gene for KIAA0970 protein, COX7CP1 (cytochrome c oxidase subunit VIIc pseudogene 1), a novel pseudogene, the GPR38 (G protein-coupled receptor 38) gene, ESTs, STSs, GSSs, and a CpG island, complete sequence | ||||
| 37 bp | 13 | 34070-34107 | TATAAAGACTTTCATGTGAAACTATTCGAAAACTTAA | |||
| 10 | NOD/SCID J |
Fragment . | Sample . | Size . | Chromosome . | Insertion site . | Insertion site details . | Insertion sequence . |
|---|---|---|---|---|---|---|
| 1 | Baboon 6 wk | |||||
| 73 bp | 12 | 101007-101074 | Homo sapiens BAC clone RP11-87C12 | ACTTCAGGTCAGGAGTTCGAGACCAATCTGGCCAAC | ||
| ATGGTGAAACCCCGCCTCTACTAAAAATACAAAAATT | ||||||
| 5 | NOD/SCID J | |||||
| 8 | Baboon 6 wk | Homo sapiens BAC clone RP11-203116 contains the gene for KIAA0970 protein, COX7CP1 (cytochrome c oxidase subunit VIIc pseudogene 1), a novel pseudogene, the GPR38 (G protein-coupled receptor 38) gene, ESTs, STSs, GSSs, and a CpG island, complete sequence | ||||
| 37 bp | 13 | 34070-34107 | TATAAAGACTTTCATGTGAAACTATTCGAAAACTTAA | |||
| 10 | NOD/SCID J |
Displays the 2 common integration sites between NOD/SCID and baboon repopulating cells at 6 weeks after transplantation. Wk refers to weeks after transplantation.
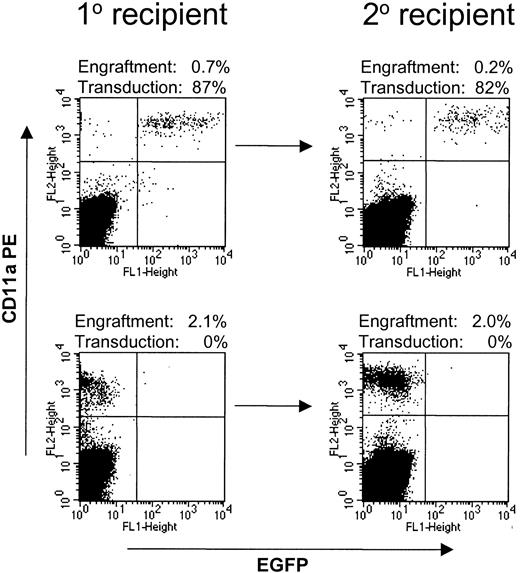
High-level engraftment in secondary NOD/SCID transplantations
Secondary NOD/SCID transplantations were performed with a separate group of 4 mice, 2 of which received untransduced baboon CD34-enriched cells and 2 of which received CD34-enriched cells transduced with Phoenix-GALV/MNDEYFPSN. After 6 weeks, bone marrow was harvested from femur and tibia bones of the mice; 90% of these cells were injected into irradiated secondary recipients, and 10% were used for analysis. All 4 secondary recipients engrafted at levels between 0.13% and 2% (Figure 5). The number of baboon-derived cells in the secondary recipients was on average 2.7 times higher than the number of injected cells, demonstrating an expansion of baboon cells in secondary recipients. The marking frequency in the primary NOD/SCID mouse recipients (53% and 87%) was conserved in the secondary recipients (54% and 82%, respectively).
Baboon SRCs are capable of engraftment into secondary recipients. Bone marrow from NOD/SCID mice transplanted with baboon CD34-enriched cells transduced with Phoenix-GALV/MNDEYFPSN (top row) or untransduced (bottom row) was harvested and injected into secondary recipients. All 4 secondary recipients engrafted at levels between 0.13% and 2%. Marking levels in the secondary recipient were similar to those in the primary recipient.
Baboon SRCs are capable of engraftment into secondary recipients. Bone marrow from NOD/SCID mice transplanted with baboon CD34-enriched cells transduced with Phoenix-GALV/MNDEYFPSN (top row) or untransduced (bottom row) was harvested and injected into secondary recipients. All 4 secondary recipients engrafted at levels between 0.13% and 2%. Marking levels in the secondary recipient were similar to those in the primary recipient.
NOD/SCID transplantation with marked CD34+ cells from a baboon 18 months after autologous transplantation
Based on the significant discrepancy between the level of marked cells in the NOD/SCID mice and in the baboons, we hypothesized that distinct hematopoietic cell populations were responsible for repopulating the NOD/SCID mice compared with the baboons. Given that the gene transfer efficiency after a transduction culture is generally higher in proliferating progenitor cells than in more quiescent stem cells, our data suggested that engraftment in the NOD/SCID mice was primarily the result of engraftment of progenitor cells rather than the type of stem cell assayed by autologous transplantation in the baboon. To further investigate this hypothesis we performed the following experiment: Bone marrow was harvested from a baboon that showed stable multilineage engraftment 18 months after autologous marrow transplantation with gene-modified cells. CD34+ cells were enriched and injected into 2 NOD/SCID mice. In this setting we would assume that the marking detected in progenitor cells or granulocytes was derived from the stem cell compartment and thus reflects the transduction level in “true” stem cells. Marking levels in NOD/SCID repopulating cells should therefore be similar to marking levels in the baboon. Our results showed that the percentage of transgene-expressing cells in CD34+ cells in the baboon marrow at that time was 13.5%. Transgene expression in baboon SRCs derived from these cells was 12.1% and 19.6% and thus not markedly different from the percentage of transduced cells injected. In addition, engraftment levels of uncultured CD34+ cells were similar to those of cultured, transduced cells (3.5% vs 4.5%), and the flow cytometric pattern was also similar. These results demonstrate that the NOD/SCID model does not per se have a skewed engraftment in favor of transduced cells. More important, it supports the hypothesis that a distinct, more mature, and more easily transducible cell population is responsible for repopulation of the NOD/SCID mouse.
Discussion
The present study describes a direct comparison between the hematopoietic repopulation of baboon CD34-enriched cells in the NOD/SCID xenotransplantation assay and autologous transplantation in the baboon. We used oncoretroviral marking to track hematopoietic repopulating cells in vivo and found that the overall percentage of gene-marked cells within the baboon SCID repopulating cells was significantly higher than the percentage of gene-marked, long-term repopulating cells in the baboon. The percentage of marked cells observed in the NOD/SCID model was more similar to the percentage of gene-marked cells in the very early engraftment period in baboons. In addition, clonal integration site analysis showed common vector integrants in the NOD/SCID mice and early after transplantation in the baboon, suggesting that NOD/SCID repopulating cells are more similar to short-term repopulating cells than long-term repopulating cells.
Retroviral marking of hematopoietic stem cells has been used for almost 20 years to study hematopoiesis after transplantation. Although retroviral gene transfer efficiency has been high in in vitro studies and studies in the mouse, only low marking efficiency has been obtained in human or large animal studies. More recently, a similar discrepancy in gene marking levels has been noted between SRCs and studies in large animals, prompting us to ask whether the NOD/SCID assay measures true hematopoietic stem cells or long-term repopulating cells, which are defined by their ability to provide long-term multilineage engraftment in a myeloablated host. Overall marking efficiency in the NOD/SCID mice in the present study was comparable with that of other studies, yet, after transplanting the same cells into the baboon, long-term marking efficiency was approximately 10-fold lower. Although marking in the NOD/SCID mice was remarkably stable between 6 and 12 weeks, there was a considerable decline in marking levels in the baboon during this time. Early after transplantation (at 2 weeks), marking in the baboons was more similar to marking levels in SRCs than they were 6 or more weeks after transplantation, suggesting that a distinct hematopoietic stem/progenitor cell population contributed to repopulation in NOD/SCID mice compared with the baboons. Because mature stem/progenitor cells are usually more easily transducible than immature long-term repopulating cells, our data suggest that engraftment in NOD/SCID mice was attributed to mature stem/progenitor cells than those responsible for long-term repopulation in the baboons.
To further characterize the cells that contribute to repopulation in the NOD/SCID compared with the large animal model, we performed clonal integration site analysis in NOD/SCID repopulating cells and in baboon repopulating cells early and later after transplantation. We were able to demonstrate 2 common retroviral integration sites between NOD/SCID and baboon repopulating cells. Each integration site was only detected early after transplantation in the baboon, supporting our findings from the marking studies that mature stem/progenitor cells are probably responsible for most NOD/SCID repopulation. This is the first direct evidence that the NOD/SCID xenotransplantation system assays a cell population relevant to the transplantation of higher animal species such as nonhuman primates and most likely also humans. However, in the autologous transplantation of the baboon, none of the clones contributing to long-term engraftment were detectable early after transplantation or in any NOD/SCID mouse. This finding is consistent with a report by Schmidt et al14 describing that long-term repopulating clones were not detectable within the first 6 weeks after autologous transplantation of nonhuman primates, and it supports the notion that short-term and long-term engraftment are provided by distinct cell populations.
Is it possible that our culture conditions could have skewed the engraftment kinetics in NOD/SCID mice and led to preferential engraftment of transduced cells? To test this possibility we transplanted into NOD/SCID mice the uncultured CD34+ cells from a baboon that had stable marking more than 1 year after autologous transplantation. Engraftment levels were similar to those after a 3-day transduction culture. In addition, the cells that engrafted in the NOD/SCID mice had a similar flow cytometric pattern. In this experiment, we observed similar marking in the NOD/SCID marrow cells and in baboon marrow cells, consistent with the interpretation that the NOD/SCID mouse model is an assay that primarily detects more committed cells than those capable of long-term engraftment after autologous transplantation in a large animal.
It is unlikely that the discrepancy between baboons and NOD/SCID mice resulted from an immune response against gene-modified cells in the baboons. In all 3 animals we observed persistent marking and gene expression. In addition, in 2 animals we performed immune assays and were unable to detect any immune responses against gene-modified cells.
In contrast to NOD/SCID transplantation with human progenitor cells, we detected mainly myeloid cells in the NOD/SCID mice that received transplanted baboon cells. It is possible that the culture conditions used for the transduction of these cells were responsible for a significant shift toward myeloid cells. A shift in phenotype toward more myeloid cells induced by ex vivo culture of cells before transplantation into NOD/SCID mice has been described previously for human SRCs.19,20 Another difference between our study and most NOD/SCID experiments with human cells described in the literature is the source of stem cells. Although most studies are performed using highly purified CD34+/CD38- cells from cord blood or mobilized peripheral blood, we used CD34-enriched cells from bone marrow of animals primed for 5 days with G-CSF and SCF.
This is the first time that engraftment of gene-marked cells has been directly compared between the NOD/SCID assay and autologous transplantation. We were able to demonstrate stable engraftment, multilineage differentiation, and proliferation of oncoretrovirally transduced baboon bone marrow CD34-enriched cells in NOD/SCID mice. Our data show that the difference in transduction efficiency between different vectors detected in an autologous, nonhuman primate transplantation model was also present in the NOD/SCID mouse model. However, gene-marking levels in baboon SRCs were markedly higher than in long-term repopulating cells in the baboons. Furthermore, we demonstrate that NOD/SCID repopulating clones are able to contribute to short-term repopulation in a large animal model. However, none of the NOD/SCID repopulating clones appeared to contribute to hematopoiesis at 6 months after transplantation, suggesting that a distinct, more committed, and more easily transducible cell population is responsible for NOD/SCID mice repopulation compared with autologous long-term reconstitution in primates. These findings emphasize that data from surrogate assays must be interpreted with caution with regard to their ability to predict human stem cell behavior.
Prepublished online as Blood First Edition Paper, June 19, 2003; DOI 10.1182/blood-2003-01-0082.
Supported by National Institutes of Health grants HL54881, HL53750, DK47754, DK56465, and RR00166. P.A.H. was supported by the German Krebshilfe. H.-P.K. is a Markey Molecular Medicine Investigator.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Laura J. Peterson and Jennifer Potter for technical assistance, Mike Gough and the staff of the University of Washington National Primate Research Center for assistance with the animals, Drs Beverly Torok-Storb and Irwin Bernstein for reviewing the manuscript, and Drs Christof von Kalle and Cynthia Dunbar for assistance with the LAM-PCR. We also thank Bonnie Larson and Helen Crawford for their help in preparing the manuscript.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal