Abstract
The number of bone marrow hematopoietic stem and progenitor cells as defined by the lineage-, Sca1++, c-kit+ (LSK) phenotype and their proliferative capacity in vitro are subject to quantitative genetic variation, and several quantitative trait loci (QTL) have been identified in young mice. Because some traits affecting hematopoiesis also change with age in a mouse strain-dependent fashion, we performed quantitative trait analysis in aged BXD recombinant inbred (RI) mice for the number and frequency of LSK cells, and for their proliferative capacity in vitro. Several novel QTL were identified. The number and frequency of LSK cells in old mice correlated inversely with lifespan. Furthermore, 4 of 7 lifespan QTL overlap with QTL contributing to the number, frequency, or proliferative capacity of LSK cells in young or old mice. Taken together, these data establish a close genetic, and perhaps functional, link between genetic variation in lifespan and characteristics of stem and progenitor cells. (Blood. 2004;104:374-379)
Introduction
Hematopoietic stem cells can self-renew and give rise to mature blood cells.1 Repopulating stem cells in mice are enriched in a population that does not express lineage-specific markers (ie, they are lin-) but does express high levels of the Sca1 (Ly6A/E) antigen and c-kit (lin-Sca1++kit+ or LSK cells).1-5 Several characteristics of the hematopoietic stem and progenitor cell compartment display mouse strain-dependent variation, and multiple quantitative trait loci (QTL) have been identified.6-15 One of these quantitative traits is the number of stem and progenitor cells as defined by phenotype.13,14 Using the lin-Sca1++Thy1lowkit+ phenotype, Morrison et al mapped a quantitative trait (QT) locus to chromosome (chr) 17 in an intercross mapping approach in AKR and C57BL/6Ka-Thy1.1 mice.13 We have identified 3 QTL contributing to the number and frequency of lin-Sca1++ and of LSK cells on chromosomes 2, 4, and 7 using recombinant inbred (RI) mouse strains with C57BL/6 and DBA/2 as progenitors (BXD RI strains). The QT locus on chr 2 was confirmed in congenic mice.14 The proliferation of purified progenitor and stem cells in response to the early-acting cytokines kit ligand (KL), flt3 ligand (Flt3L), and thrombopoietin (TPO) is also subject to quantitative genetic variation, and QTL were identified on chromosomes 2 and X.14 Many traits affecting hematopoiesis, such as the number of primitive day-35 cobblestone area-forming cells (CAFCd35), also change with age in a mouse strain-dependent fashion.16-21 Furthermore, several traits have been linked to longevity, either because overlapping QTL were identified (eg, the age-related change in CAFCd35 frequency on chr 2),19,21 or because of a correlation between trait value and lifespan together with overlapping QTL for both traits (eg, the cycling activity of CAFCd7).15 In a follow-up to our analysis in young mice,14 we performed linkage analysis for the same traits in aged BXD RI mice using the new web-based database and software for linkage analysis, webQTL.22 Several novel QTL were found. Furthermore, of the 7 putative QTL contributing to lifespan identified using the webQTL database, at least 4 overlap with QTL for traits affecting the LSK compartment in young or old mice. These data strongly suggest a link between longevity and the hematopoietic stem cell compartment, or between longevity and somatic stem and progenitor cells in a broader sense.
Materials and methods
Mice
The 8-week-old C57BL/6J, DBA/2J, and BXD RI mice were purchased from Jackson Laboratories (Bar Harbor, ME). The 18-month-old DBA/2 and C57BL/6 mice were either obtained from the National Institute on Aging or were aged in our animal facility in specific pathogen-free conditions. BXD RI mice were aged to 18 months in our facility. Experiments and animal care were performed in accordance with the Mount Sinai Institutional Animal Care and Use Committee (IACUC).
Antibodies and cytokines
Unconjugated CD2, CD3, CD8, CD4, B220, Ly6G/Gr1, Mac1, phycoerythrin (PE)-conjugated Sca1, Cychrome-conjugated streptavidin, and fluorescein isothiocyanate (FITC)-conjugated goat antirat antibodies (GARs) were purchased from Southern Biotechnologies (Birmingham, AL). Unconjugated Ter119 and biotinylated anti-c-kit were purchased from Pharmingen (San Diego, CA). Recombinant mouse Flt3 ligand (Flt3L), kit ligand (KL), and thrombopoietin (TPO) were purchased from R&D Systems (Minneapolis, MN).
Isolation of hematopoietic progenitor and stem cells from bone marrow
Femurs and tibias were flushed with Iscove modified Dulbecco medium (IMDM; Gibco, Grand Island, NY) supplemented with 5% fetal calf serum (FCS). Low-density bone marrow cells, obtained after density centrifugation, were stained with Ter119, CD2, CD3, CD8, CD4, B220, Mac1, and Gr1 for 20 minutes at 4°C, washed, and stained with GAR for 20 minutes at 4°C. After washing, the cells were stained for 20 minutes at 4°C with PE-conjugated Sca1 and biotin-conjugated CD117 (c-kit), washed with phosphate-buffered saline, and stained with streptavidin-Cychrome. The cells were sorted on a MoFlo (Cytomation, Fort Collins, CO) flow cytometer at 30 psi sheath pressure and at a rate of 12 000 to 15 000 events/second. Lin-Sca1++kit+ (LSK) cells were sorted as cells with a low side scatter, a low to medium forward scatter, a green (lineage) fluorescence lower than the median fluorescence of cells stained with isotype-matched control antibodies, an orange (Sca1) fluorescence twice the intensity (in terms of channel numbers) of the brightest cells in control samples, and a positive red (c-kit) fluorescence.14 Bone marrow pooled from at least 2 mice was used in each experiment. Absolute LSK numbers were estimated by recording the number of sorted, nonaborted events on the counter of the flow cytometer.14
Culture of Lin-Sca1++kit+ (LSK) cells
Sorted LSK cells, pooled from at least 2 mice in each experiment, were cultured in triplicate at 20 to 40 cells per well in flat-bottom 96-well plates in IMDM supplemented with 10% fetal calf serum (FCS), 100 mg/mL penicillin/streptomycin, and 50 ng/mL each of Flt3L, TPO, and KL. At 3 hours after plating, the exact number of cells per well was determined by visually counting the cells at × 40 magnification. After 5 days of liquid culture at 37°C and 5% CO2, the cells were again counted to assess responsiveness to early-acting factors.
Statistical analysis
Student t test for unpaired samples was used, unless mentioned otherwise. All results are expressed as mean ± SEM. A P value less than .05 was considered indicative of a statistically significant difference.
Linkage analysis in BXD recombinant inbred strains
Linkage analysis was performed in BXD recombinant inbred (RI) strains. These are commercially available and were generated by repeated inbreeding of F2 mice derived from the inbred progenitor strains, C57BL/6 and DBA/2. The resulting genome of RI strains is composed of a patchwork of homozygous chromosome segments derived from either progenitor strain, with each of the RI lines having a unique combination of “patches” from the progenitors.23 Each RI strain of a set is thus inbred and genetically distinct from the other strains in the set, since they inherited different combinations of chromosomal regions from each progenitor. Therefore, if a trait is determined by multiple genes, then the trait value will show a continuous distribution across a given set of RI strains. The distribution of the trait value among the RI strains is called the strain-distribution pattern (SDP). Polymorphic markers, derived from either one of the progenitors will also show a strain-distribution pattern. Linkage analysis consists of determining with which polymorphic markers the phenotypic SDP shows a statistically significant correlation. This analysis is performed using webQTL (http://www.webqtl.org).22 This web-based software for complex trait analysis uses an updated, error-checked database and is based on the Mapmanager software developed by Manly and Olson.24 It statistically analyzes the linkage of a given trait with previously typed polymorphic loci in the RI strains, which are inherited from either parental strain. The association between a marker and a trait is indicated by the likelihood ratio statistic (LRS) and an associated P value for point-wise linkage. However, the association of a phenotypic SDP with the SDP of hundreds of polymorphic markers is measured. Therefore, a close or identical match in SDPs may occur by chance,25,26 and this probability is higher if more markers are tested. This would lead to a very high number of false-positive QTL. To establish the genome-wide, as opposed to the point-wise, significance level of the association between a phenotypic and a marker SDP, a correction for multiple testing has to be introduced. One way to do this is to calculate the genome-wide probability of obtaining the observed linkages by random chance corresponding to a given error threshold using the nonparametric permutation method developed by Churchill and Doerge.25 In permutation testing, the phenotypic data are permutated 1000 to 5000 times, and the frequency of obtaining linkage anywhere in the genome at a given LRS value is calculated. This allows the determination of minimum LRS values to consider linkage suggestive (P < .5) or significant (P < .05) on a genome-wide basis. This algorithm is automatically implemented in the webQTL software. Suggestive QTL are deemed worth reporting but require confirmation.26 A weak statistical correlation between genotype and phenotype for a QT locus that is not a false negative can be caused by the fact that the QT locus near the marker has a small effect, or by the fact that a QT locus of large effect is located relatively far away from flanking markers. A subroutine of webQTL, interval mapping, uses computationally efficient regression equations to determine the interval where a QT locus is most likely located. Here, the most likely genotypes in between 2 markers are estimated at 1-centimorgan (cM) intervals, and these inferred genotypes are compared with the observed phenotype.27 Examples of the data obtained using interval mapping are shown in Figure 4. Finally, webQTL also allows for composite interval mapping, where linkage analysis is performed after removing variation caused by other linked QTL in the genome.
Interval mapping of QTL for lifespan and for the regulation of LSK cells. Likelihood ratio statistic (LRS) values (y-axis) along chromosomes 2, 4, and 7 for lifespan and for traits related to the regulation of LSK cells in BXD RI mice, mentioned on top of each panel. The dashed horizontal lines in each panel represent the lower boundaries of LRS values indicating suggestive (lower line) and significant (upper line) genome-wide significance levels as determined by permutation analysis.25,26 The 2 different panels for lifespan along chr 4 were obtained by correcting for a different primary QT locus: chr 7 in the upper right, and chr 2 in the lower right panel.
Interval mapping of QTL for lifespan and for the regulation of LSK cells. Likelihood ratio statistic (LRS) values (y-axis) along chromosomes 2, 4, and 7 for lifespan and for traits related to the regulation of LSK cells in BXD RI mice, mentioned on top of each panel. The dashed horizontal lines in each panel represent the lower boundaries of LRS values indicating suggestive (lower line) and significant (upper line) genome-wide significance levels as determined by permutation analysis.25,26 The 2 different panels for lifespan along chr 4 were obtained by correcting for a different primary QT locus: chr 7 in the upper right, and chr 2 in the lower right panel.
Results
Age-related changes in LSK cells in C57BL/6 and DBA/2 mice
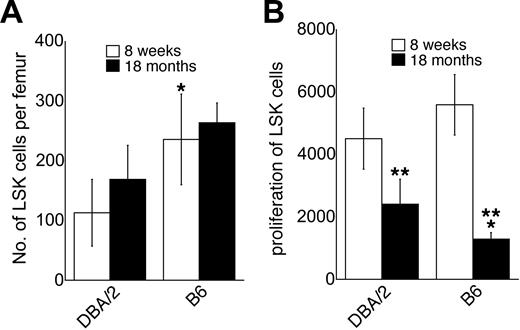
We have shown previously14 that C57BL/6 mice at the age of 8 weeks had approximately twice as many LSK cells as DBA/2 mice (Figure 1A). The number of LSK cells increased slightly, but not significantly, between the ages of 8 weeks and 18 months in both C57BL/6 and DBA/2 mice (Figure 1A). At 18 months of age, C57BL/6 mice still had approximately twice as many LSK cells as DBA/2 mice, as was the case at 8 weeks of age14 ; although in contrast to young mice, the difference between C57BL/6 and DBA/2 mice was not significant in old mice (Figure 1A). The proliferative response of LSK cells to the early-acting cytokines KL, Flt3L, and TPO in vitro was somewhat higher in young C57BL/6 mice than in young DBA/2 mice, but the difference did not reach statistical significance (Figure 1B). In both strains, however, the proliferative capacity of LSK cells decreased significantly with age (P < .05, Figure 1B). This finding is consistent with data obtained using human CD34+CD45RA+CD71low primitive hematopoietic progenitors.28 The age-related decrease in proliferative capacity of LSK cells in vitro tended to be more pronounced in C57BL/6 than in DBA/2 mice, so that in old mice, LSK cells from C57BL/6 were in fact significantly less responsive to early-acting factors than LSK cells from DBA/2 mice (Figure 1B). Genetic and age-related variation in the proliferative capacity of LSK cells in response to the early-acting cytokines KL, Flt3L, and TPO is likely biologically relevant, as mice with a homozygous deletion in the genes encoding these ligands or their receptors show moderate to severe defects in the hematopoietic stem and progenitor cell compartment.29-31
Number of LSK cells and their proliferative capacity in C57BL/6 and DBA/2 mice. (A) Absolute number of LSK cells in C57BL/6 (B6) and DBA/2 mice at the ages of 8 weeks and 18 months mice as determined by flow cytometric cell sorting (mean ± SEM; n = 6 independent experiments, where the bone marrow pooled from at least 2 mice was used). (B) Proliferative capacity of purified LSK cells from C57BL/6 and DBA/2 mice at the ages of 8 weeks and 18 months in liquid cultures supported by KL, Flt3L, and TPO. Results are expressed as the number of cells obtained after 5 days of culture per 50 cells at initiation of the cultures (n = 7). *Significantly different (P < .05 by paired analysis) from DBA/2 at the same age. **Significantly different from mice at the age of 8 weeks (P < .05).
Number of LSK cells and their proliferative capacity in C57BL/6 and DBA/2 mice. (A) Absolute number of LSK cells in C57BL/6 (B6) and DBA/2 mice at the ages of 8 weeks and 18 months mice as determined by flow cytometric cell sorting (mean ± SEM; n = 6 independent experiments, where the bone marrow pooled from at least 2 mice was used). (B) Proliferative capacity of purified LSK cells from C57BL/6 and DBA/2 mice at the ages of 8 weeks and 18 months in liquid cultures supported by KL, Flt3L, and TPO. Results are expressed as the number of cells obtained after 5 days of culture per 50 cells at initiation of the cultures (n = 7). *Significantly different (P < .05 by paired analysis) from DBA/2 at the same age. **Significantly different from mice at the age of 8 weeks (P < .05).
Linkage analysis in aged BXD RI mice
We first used webQTL to reanalyze our published data generated in young BXD RI mice,14 which were analyzed using the Mapmanager software developed by Manley and Olson24 and the genotypic database of polymorphic markers in BXD RI strains compiled by Williams et al.32 The QTL are shown in Table 1. We had previously identified QTL on chromosomes 2, 4, and 7 for the number of LSK cells in young mice.14 Reanalyzing the same data using the updated, error-checked database of webQTL,22 the QTL on chromosomes 4 and 7 were confirmed. In addition, 2 other, weak QTL were identified (chr 14 and chr 16). The QT locus on chr 2 we reported previously14 was just short of a suggestive level of genome-wide significance using webQTL. This QT locus is nevertheless real, since it was confirmed in congenic mice by us.14 A QT locus on chr 2 for the proliferative capacity of LSK cells in young mice14 was also confirmed using webQTL (Table 1).
QTL identified in young and old BXD RI strains
Trait and Chr* . | Peak LRS interval . | cM, range . | LRS . | High allele . |
|---|---|---|---|---|
| No. LSK cells 8 wk | ||||
| 4 | D4Mit54-D4Mit226 | 66-78.5 | 13.8 | DBA/2 |
| 7 | D7Mit17-D7Mit238 | 51-53 | 13.9 | DBA/2 |
| 2† | D2Mit493-D2Mit411 | 72.1-77.2 | NA | C57BL/6 |
| 16 | D16Mit146-D6Mit58 | 16.9-23.1 | 10.9 | C57BL/6 |
| 14‡ | Broad | Broad | 10.6 | C57BL/6 |
| % LSK cells 8 wk | ||||
| 4 | D4Mit312-D4Mit226 | 69.8-78.5 | 11.4 | DBA/2 |
| 7 | D7Mit17-D7Mit238 | 51-53 | 10.2 | DBA/2 |
| Proliferation 8 wk | ||||
| 2 | D2Mit493-D2Mit411 | 72.1-77.2 | 13.5 | C57BL/6 |
| X§ | NA | NA | NA | DBA/2 |
| No. LSK cells 18 mo | ||||
| 12 | D12Mit112-S12Gnf051.120 | 22-24 | 10.1 | C57BL/6 |
| 4∥ | D4Mit80-D4Mit303 | 37.7-48.5 | 9.8 | C57BL/6 |
| % LSK cells 18 mo | ||||
| 12 | D12Mit112-S12Gnf051.120 | 22-24 | 10.1 | C57BL/6 |
| Proliferation 18 mo | ||||
| 7 | D7Mit78-D7Mit190 | 0.5-2.5 | 14.9 | DBA/2 |
| 2∥ | D2Mit396-D2Mit304 | 69-72 | 13.8 | DBA/2 |
| 9 | D9Mit227-D9Mit4 | 23-27 | 9.2 | DBA/2 |
| Lifespan | ||||
| 2 | D2Mit396-D2Mit304 | 69-72 | 12.2 | DBA/2 |
| 4∥ | D4Mit312-D4Mit344 | 69.8-81.7 | 13.9 | C57BL/6 |
| 4∥ | D4Mit193-D4Mit237 | 7.5-13.3 | 11.5 | DBA/2 |
| X∥ | DXMit10-DXMit223 | 63-73 | 11.3 | DBA/2 |
| 7 | D7Mit178-D7Mit75 | 0.5-1.7 | 14.5 | C57BL/6 |
| 4∥ | D4Mit80-D4Mit303 | 37.7-48.5 | 12.4 | DBA/2 |
| 12∥ | D12Mit136-D12Mit235 | 13-19 | 9.8 | DBA/2 |
Trait and Chr* . | Peak LRS interval . | cM, range . | LRS . | High allele . |
|---|---|---|---|---|
| No. LSK cells 8 wk | ||||
| 4 | D4Mit54-D4Mit226 | 66-78.5 | 13.8 | DBA/2 |
| 7 | D7Mit17-D7Mit238 | 51-53 | 13.9 | DBA/2 |
| 2† | D2Mit493-D2Mit411 | 72.1-77.2 | NA | C57BL/6 |
| 16 | D16Mit146-D6Mit58 | 16.9-23.1 | 10.9 | C57BL/6 |
| 14‡ | Broad | Broad | 10.6 | C57BL/6 |
| % LSK cells 8 wk | ||||
| 4 | D4Mit312-D4Mit226 | 69.8-78.5 | 11.4 | DBA/2 |
| 7 | D7Mit17-D7Mit238 | 51-53 | 10.2 | DBA/2 |
| Proliferation 8 wk | ||||
| 2 | D2Mit493-D2Mit411 | 72.1-77.2 | 13.5 | C57BL/6 |
| X§ | NA | NA | NA | DBA/2 |
| No. LSK cells 18 mo | ||||
| 12 | D12Mit112-S12Gnf051.120 | 22-24 | 10.1 | C57BL/6 |
| 4∥ | D4Mit80-D4Mit303 | 37.7-48.5 | 9.8 | C57BL/6 |
| % LSK cells 18 mo | ||||
| 12 | D12Mit112-S12Gnf051.120 | 22-24 | 10.1 | C57BL/6 |
| Proliferation 18 mo | ||||
| 7 | D7Mit78-D7Mit190 | 0.5-2.5 | 14.9 | DBA/2 |
| 2∥ | D2Mit396-D2Mit304 | 69-72 | 13.8 | DBA/2 |
| 9 | D9Mit227-D9Mit4 | 23-27 | 9.2 | DBA/2 |
| Lifespan | ||||
| 2 | D2Mit396-D2Mit304 | 69-72 | 12.2 | DBA/2 |
| 4∥ | D4Mit312-D4Mit344 | 69.8-81.7 | 13.9 | C57BL/6 |
| 4∥ | D4Mit193-D4Mit237 | 7.5-13.3 | 11.5 | DBA/2 |
| X∥ | DXMit10-DXMit223 | 63-73 | 11.3 | DBA/2 |
| 7 | D7Mit178-D7Mit75 | 0.5-1.7 | 14.5 | C57BL/6 |
| 4∥ | D4Mit80-D4Mit303 | 37.7-48.5 | 12.4 | DBA/2 |
| 12∥ | D12Mit136-D12Mit235 | 13-19 | 9.8 | DBA/2 |
NA indicates not applicable.
All QTL with a suggestive level of genome-wide significance according to permutation analysis are shown.
Significance level below suggestive using webQTL, but confirmed in congenic mice.14
Broad low-suggestive QTL spanning the distal two thirds of chr 14.
Identified in reciprocal D2B6/B6D2 F1 hybrids.
After correction for the preceding major QTL in the table.
We next measured the number and frequency of LSK cells and their proliferative capacity in liquid cultures supported by KL, Flt3L, and TPO in 18-month-old BXD RI mice. As in young mice,14 for each of the traits examined, the phenotypic variation among old BXD RI mice was much more pronounced than among the 2 progenitor strains, C57BL/6 and DBA/2 (Figure 2). This is typical of a multigenic trait, where some of the BXD strains may have accumulated most of the positively or negatively acting alleles for a given trait, thus giving rise to more extreme trait values than in either of the progenitors.23 In contrast to the experiments shown in Figure 1, in which old and young C57BL/6 and DBA/2 mice were analyzed in side-by-side experiments, analysis of the LSK compartment in young and old BXD RI mice was not done in side-by-side experiments. The experiments on aged BXD RI mice were performed 12 to 18 months later than the experiments on young BXD RI mice, using different batches of antibodies, cytokines, and FCS, as well as different lasers on the flow cytometer. It is therefore difficult to numerically compare the trait values obtained in old and young BXD mice in these experiments. Nevertheless, there was a very good correlation between the number of LSK cells in young and old BXD RI mice (P = .0047, not shown). However, there was no correlation between proliferative capacity of LSK cells in young and old BXD RI mice (not shown), suggesting complex age-related changes in this trait. This notion is supported by the more rapid decline in the responsiveness of LSK cells to early-acting cytokines in C57BL/6 mice than in DBA/2 mice (Figure 1B). Linkage analysis in old BXD RI mice yielded several novel QTL. The QTL locations for each of the traits are summarized in Table 1. All QTL were in the suggestive range of genome-wide significance as determined by permutation analysis,25,26 although one QT locus was just short of significant (proliferation of LSK cells in aged mice, chr 7, 0.5-2.5 cM).
Strain-distribution patterns in BXD mice. Strain-distribution patterns of the absolute number of LSK cells (A), the relative fraction of LSK cells (B), and the proliferative capacity in response to KL, Flt3L, and TPO (C) in 18-month-old BXD RI mice (mean ± SEM; n = 1 to 3 independent experiments, where the bone marrow pooled from at least 2 mice was used; missing data, such as strains 2, 5, and 22 in panel C, are due to technical failure).
Strain-distribution patterns in BXD mice. Strain-distribution patterns of the absolute number of LSK cells (A), the relative fraction of LSK cells (B), and the proliferative capacity in response to KL, Flt3L, and TPO (C) in 18-month-old BXD RI mice (mean ± SEM; n = 1 to 3 independent experiments, where the bone marrow pooled from at least 2 mice was used; missing data, such as strains 2, 5, and 22 in panel C, are due to technical failure).
Overlapping QTL contributing to the regulation of LSK cells and to mean lifespan
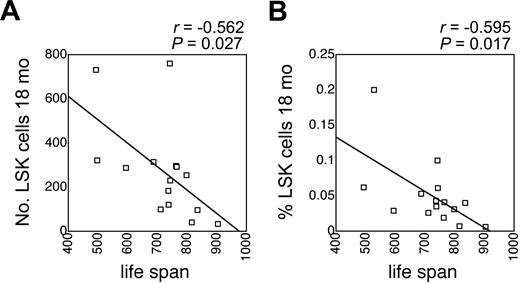
There is significant genetically determined variation in longevity among inbred mouse strains in general, and among BXD RI strains in particular. Mean lifespan of BXD RI mice was assessed by Gelman et al under carefully controlled housing conditions,33 and these data are publicly available from the database of BXD phenotypes in webQTL.22 Interestingly, the number and the frequency of LSK cells in old mice correlated significantly and inversely with mean lifespan (Figure 3). This correlation was less pronounced and not statistically significant in young mice (r = -0.387, P = .13 for the number of LSK cells; and r = -0.247, P = .34 for the fraction of LSK cells; not shown). These data suggested a close link between LSK cells and longevity.
Correlation between LSK cells and lifespan. Correlation between mean lifespan of BXD RI mice (data obtained from webQTL and published in Gelman et al33 ) and the absolute number (A) and the frequency (B) of LSK cells in 18-month-old BXD RI mice. P values and correlation coefficients are indicated on top of each panel.
Correlation between LSK cells and lifespan. Correlation between mean lifespan of BXD RI mice (data obtained from webQTL and published in Gelman et al33 ) and the absolute number (A) and the frequency (B) of LSK cells in 18-month-old BXD RI mice. P values and correlation coefficients are indicated on top of each panel.
The notion that lifespan and characteristics of LSK cells were genetically linked was further supported by quantitative trait analysis (Figure 4). A total of 7 suggestive QTL for lifespan were identified using webQTL (Table 1). Of these, 5 appear after correction for the effect of major QTL on chromosomes 2 and 7 (Table 1). Interestingly, at least 4 of the 7 QTL (chr 2, the telomeric region of chr 4, the middle portion of chr 4, and chr 7; see Table 1) overlap with QTL, contributing to the regulation of the LSK population in young (telomeric region of chr 4) or old (middle portion of chr 4) mice, or of the proliferative capacity of LSK cells in old mice (chr 2, chr 7) (Figure 4; Table 1). The patterns of LRS values and the positions of peak LRS values for lifespan and for traits related to LSK cells along each of the relevant chromosomes, obtained by interval mapping as implemented in webQTL, are shown in Figure 4. Overlapping QTL can occur by chance, however. The average size of the marker intervals to which a peak LRS value could be assigned was approximately 5 cM. Assuming a genome of 1600 cM and 7 QTL contributing to lifespan, the probability of finding a lifespan QTL and an unrelated QTL within the same 5-cM interval by chance is 0.022 (7/[1600/5]). Because we identified a total of 11 separate QTL contributing to the regulation of LSK cells (Table 1), the probability that 1 of these 11 QTL would overlap with a lifespan QT locus is 0.022 × 11 = 0.24. However, the probability that this would happen by chance for 4 QTL is (7 × 11)/(1600/5) × (7 × 10)/(1600/5 - 1) × (7 × 9)/(1600/5 - 2) × (7 × 8)/(1600/5 - 3) = 0.0018. For 3 of these QTL (chr 2, middle portion of chr 4, and chr 7), the peak LRS value was found within the same 2-cM interval (Figure 4). The probability that such close linkage would occur by chance is 0.00066. In addition, a fifth lifespan QT locus maps very near a QT locus on chr 12 determining the number of LSK cells in aged mice (Table 1). Thus, even though all the QTL in Table 1 are in the suggestive range of genome-wide significance and some of these might therefore be false, it is highly unlikely that the association between multiple QTL regulating lifespan and LSK cells occurred purely by chance, indicating that longevity and stem and progenitor cell number and kinetics are intimately linked.
Discussion
We demonstrated a remarkable association in BXD RI mouse strains between QTL contributing to the regulation of the number of LSK cells and their responsiveness to early-acting factors on one hand and QTL contributing to genetic variation in mean lifespan on the other hand. These data strongly suggest a genetic link between lifespan and hematopoietic stem and progenitor cells.
The QTL observed in 18-month-old mice were different from those we had previously identified14 in 8-week-old mice (Table 1). It is possible that changes occur in the LSK population upon aging, and that in aged mice, other genes become important in the regulation of the number, frequency, and proliferative capacity in vitro of LSK cells. Age-related changes have been demonstrated for several hematopoietic traits such as stem cell pool size as determined by the CAFCd35 assay,19 the number of stem and progenitor cells as determined by phenotype,34,35 and the proliferative capacity of purified stem and progenitor cells in response to early-acting cytokines (Figure 1). Although, in our hands, the increase in the number of LSK cells in old mice did not reach statistical significance, our data are probably consistent with those of Morrison et al who observed a significant increase in the fraction of lin-Sca1++Thy1lowkit+ cells in old C57BL/6Ka-Thy1.1 mice34 and of Sudo et al who reported a significant increase in the fraction of CD34-/lowkit+Sca1+lin- cells in old C57BL/6 mice.35 An additional explanation for the different QTL detected in young and in old mice is that in linkage analysis using BXD RI mice, many QTL are missed in each given series of experiments because of the relatively limited number of strains, and therefore informative recombinations, available.23,36 Thus, some of the QTL identified may in fact contribute to the regulation of LSK cells in both old and young mice. Finally, since all QTL were identified with a suggestive level of genome-wide significance, it is likely that some are in fact false.25,26
We have previously identified a potential mechanism contributing to the genetic variation in the number and function of LSK cells in young mice. One isoform of transforming growth factor-beta (TGF-β), TGF-β2, is a positive regulator of hematopoietic stem cell function in vivo.37 In vitro, TGF-β2 had a stimulatory effect on the proliferation of LSK cells at low concentrations37 that requires the presence of serum factors and shows mouse strain-dependent variation (E.H. et al, submitted manuscript). A QT locus for this effect was identified on the telomeric region of chr 4,37 where QTL for the number of LSK cells and for lifespan are located (Table 1). Furthermore, this effect of TGF-β2 on LSK cells in vitro increases with age in a genetically determined fashion and a QT locus was mapped to the middle portion of chr 2 (E.H. et al, submitted manuscript), near, but not overlapping with, the QTL on chr 2 contributing to lifespan. It will be of interest to investigate the effect of TGF-β2 deficiency and overexpression on lifespan. It is striking how many QTL related to the hematopoietic stem and progenitor cell compartment were identified by us and by other investigators on chr 2. These include the age-related change in the number of CAFCd35,19,21 the mobilization of progenitor cells,11 the proliferative capacity of LSK cells in young14 and in old mice (Table 1; Figure 4), the age-related change in the aforementioned effect of TGF-β2 on LSK cells (E.H. et al, submitted manuscript), and the number of LSK cells in young mice.14 All these QTL occur within a genetic distance of approximately 30 cM. Further research will be needed to clarify whether these are all separate QTL.
Data supporting the hypothesis that there is a close genetic and functional link between lifespan and the biology of stem and progenitor cells have been obtained independently by other investigators who performed linkage analysis for different hematopoietic traits. de Haan and Van Zant showed that cycling of progenitor cells and lifespan negatively correlated and mapped to overlapping QTL, on the centromeric region of chr 7 and on chr 11.15 The QT locus on chr 7 for the cycling activity of progenitor cells overlaps completely with the QT locus identified here for the proliferative capacity of LSK cells from aged mice. It is possible that these are identical QTL.
There are several explanations, which are not mutually exclusive, for the close association between QTL regulating hematopoiesis and lifespan. It is possible that our linkage analysis and that of other investigators relied on traits that change with age, and are therefore associated with biologic age and longevity, thus explaining the observation of overlapping QTL. While such a phenomenon may have contributed to our results, it is unlikely to be the sole explanation, because the QTL for the number of LSK cells on the telomeric region of chr 414 and the QTL for the cycling of progenitor cells on chr 715 were identified in young mice (6 to 8 weeks). Since several loci are involved, it is doubtful that each of the multiple genetic mechanisms that regulate the hematopoietic stem and progenitor cell compartment also independently affects lifespan, although this may be true for some QTL. A direct impact on longevity of the number and the kinetics of hematopoietic progenitor and stem cells, and possibly of somatic stem and progenitor cells in general, cannot be excluded, at least in mice with an allele pool derived from C57BL/6 and DBA/2.
It has been proposed that maintenance of stem cell function with aging or a larger stem pool might increase lifespan by enhancing tissue renewal.38 On the other hand, given the potential role of stem and progenitor cells in oncogenesis,39-41 a larger or more actively cycling stem and progenitor cell compartment could increase the risk of malignancy, and thus limit lifespan. This may explain the inverse correlation between LSK number and frequency on one hand and lifespan on the other hand in BXD RI strains. An apparent contribution of the hematopoietic stem and progenitor cell compartment to longevity could be explained in several ways. One explanation may be that the functional characteristics of hematopoietic stem cells (HSCs) are determined by molecular mechanisms that are shared by other organ-specific stem cells, and that the genetic variation in the HSC compartment is a reflection of genetic variation in somatic stem cells in general. There is evidence for common gene expression patterns in several types of adult stem cells (HSCs, neural stem cells) and embryonic stem cells, although the data from different laboratories for similar stem cell populations showed little overlap.42,43 Mouse strain-dependent quantitative variation has been demonstrated in neurogenesis in the hippocampus,44 although it is unclear how genetic variation in the hematopoietic and the neural stem cell compartments correlate. Another possibility is that HSCs show plasticity, so that genetic variation in the hematopoietic stem cell compartment may affect renewal or oncogenesis in multiple tissues. The concept of plasticity of HSCs is highly debated and debatable, however, and evidence for extensive plasticity that would affect organ function in the recipients is weak.45,46 A third hypothesis is that the LSK fraction in the bone marrow contains other types of stem cells that are critical for longevity. Here too, evidence is scant to absent. Although other types of stem cells reside in the bone marrow, such as multipotent adult progenitors47 and mesenchymal stem cells,48 they are not enriched in the LSK population. Finally, it is possible that the function and the dynamics of the hematopoietic system itself affect the health of aged mice through their effects on tumor immune surveillance, defense against infectious agents, tissue oxygenation, and the development of hematologic malignancies. Further research will be needed to test these intriguing hypotheses.
In summary, quantitative trait analysis shows that genetic mechanisms contributing to the regulation of early hematopoiesis and to longevity are closely linked at multiple loci in BXD RI mice. Although still correlative, these data constitute the strongest support available thus far for the hypothesis that stem and progenitor cells may play a critical role in the aging process and in longevity, at least in the mouse model.21,38
Prepublished online as Blood First Edition Paper, February 26, 2004; DOI 10.1182/blood-2003-12-4304.
Supported by National Institutes of Health (NIH) grant RO1 AG16327 to H.-W.S. E.H. was supported by a fellowship from the Belgian American Educational Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal