Abstract
Short-term hematopoietic reconstituting cells have been identified in mice, nonhuman primates, and among human cells that engraft xenogeneic hosts. We now present clonal marking data demonstrating a rapid but unsustained contribution of cultured human autografts to the initial phase of hematologic recovery in myeloablated patients. Three patients received transplants of granulocyte colony-stimulating factor–mobilized autologous peripheral blood (PB) cells, of which a portion (8%-25% of the CD34+ cells) had been incubated in vitro with growth factors (5 days) and clinical grade LN retrovirus (3-5 days). More than 9% of the clonogenic and long-term culture-initiating cells harvested were transduced. Semiquantitative and linear amplification-mediated polymerase chain reaction analyses of serial PB samples showed that marked white blood cells appeared in all 3 patients within 11 days and transiently constituted up to 0.1% to 1% of those produced in the first month. However, within another 2 to 9 months, marked cells had permanently decreased to very low levels. Analysis of more than 50 vector insertion sites showed none of the clones detected in the first month were active later. Eighty percent of inserts were located within or near genes, 2 near CXCR4. These findings provide direct evidence of cells with rapid but transient repopulating activity in patients and demonstrate their efficient transduction in vitro.
Introduction
The existence of human hematopoietic stem cells able to permanently repopulate the hematopoietic system has been well established from clinical studies of allogeneic transplants,1 clonality analyses,2 and gene marking.3 Evidence of short-term repopulating human cells (STRCs) has come from comparative studies of the pattern of reconstitution obtained from different prospectively isolated subsets of human cells transplanted into preimmune fetal sheep4 or genetically immunodeficient mice.5-7 Results obtained using these xenograft models have identified 4 types of human cells with different phenotypes and biologically distinct differentiation and self-renewal properties: CD34+CD38+ myeloid-restricted short-term (3-week) repopulating cells (STRC-Ms), CD34+CD38– lympho-myeloid short term (6- to 12-week) repopulating cells (STRC-MLs), CD34+CD38– lympho-myeloid cells with long-term repopulating activities (CD34+ LTRCs) and CD34–CD38– lymphomyeloid cells with long-term repopulating activities (CD34– LTRCs). However, as yet there have been no studies to determine whether the human STRCs identified in these experimental models play a similar role in patients. To address this question we examined the appearance, persistence, clonality, and sites of proviral integration in retrovirally marked peripheral blood (PB) cells that were regenerated in 3 patients who received transplants with partially transduced autografts of mobilized PB cells.
Patients, materials, and methods
Clinical study
Between March 1998 and July 1999, 3 patients with chronic myelogenous leukemia (CML; no. 1 and no. 3 in chronic phase and no. 2 in early blast crisis) received transplants of partially transduced autologous PB cells at the University Hospital in Freiburg, Germany, according to a protocol approved by the Institutional Review Board, State and National Review Commission. Informed consent was provided according to the Declaration of Helsinki. All 3 patients were given 24 mg/m2 idarubicin, 450 mg/m2 etoposide phosphate, and 1600 mg/m2 cytosine-arabinoside followed by subcutaneous administration of granulocyte colony-stimulating factor (G-CSF). PB cells were then collected when the white blood cells (WBCs) reached 0.5 × 109/L (500/μL) or the CD34+ cells exceeded 1% of the mononuclear cells. From 2.9 × 106 to 6.2 × 106 CD34+ cells/kg body weight were isolated from a portion of the collection using the CellPro device (CellPro, Seattle, WA), and remaining cells were frozen. Patients were conditioned with 16 mg/kg body weight busulfan and received subcutaneous injections of 300 μg/d G-CSF starting 7, 9, or 13 days after transplantation until the neutrophils were greater than 1.5 × 109/L or the WBCs were greater than 5 × 109/L for 2 consecutive days.
Retroviral transduction
Supernatants of LN retrovirus–producing PG13 cells (A.D. Miller, Fred Hutchinson Cancer Research Center, Seattle, WA) containing 1.6 × 105 infectious units/mL were collected under good manufacturing practice (GMP) guidelines in a GMP facility (Magenta, Stirling, Great Britain) using a clinical grade serum-free medium (CellGro SCGM; Cell Genix, Freiburg, Germany). Transductions were also carried out in a GMP clean room facility as described8 with minor modifications. Briefly, cells from patients no. 1 and no. 2 were prestimulated for 48 hours in 850 cm2 roller bottles (Costar, Cambridge, MA) at 2 × 105 cells/mL in serum-free medium (CellGro) containing 100 ng/mL flt3-ligand (FL), 100 ng/mL stem cell factor (SCF), and 20 ng/mL each of interleukin (IL)–3, IL-6, and G-CSF (all from Genzyme, Cambridge, MA). The cells were then harvested, resuspended in virus-containing medium (VCM) plus 10–6 M hydrocortisone, 4 μg/mL protamine, and the same growth factors and transferred into new roller bottles. Cells from patient no. 3 were suspended directly in VCM with 10–6 M hydrocortisone, 4 μg/mL protamine, 300 ng/mL FL, 100 ng/mL SCF, and 30 ng/mL IL-3. Half of the VCM was replaced with fresh VCM each day. After a total of 5 days in culture, an aliquot (∼ 5% of the cells) was removed to perform helper virus assays (S+/L–), sterility testing, and other safety and gene transfer efficiency tests. The remaining cells were frozen. The fraction of colony-forming cells (CFCs) and 5-week long-term culture-initiating cells (LTC-ICs)9 transduced was inferred from colony yields in the presence and absence of 1.5 mg/mL G418 (Sigma Chemicals, St Louis, MO).
Quantification of retrovirally marked hematopoietic cells
Semiquantitative polymerase chain reaction (PCR) was performed to measure the contribution of gene-marked cells in samples of PB cells obtained from all patients at regular intervals after the transplantation was performed. DNA was isolated using the blood and cell culture DNA kit (Qiagen, Valencia, CA). A 589-base pair (bp) fragment of the 5′ vector sequence was amplified from 1 μg of each sample and compared with dilutions of DNA isolated from HeLa cells containing a single copy of LN provirus. Taq polymerase (2.5 U; Qiagen) was used in 35 PCR cycles (denaturation at 95°C for 60 seconds, annealing at 60°C for 45 seconds, extension at 72°C for 60 seconds) after initial denaturation for 5 minutes and before final extension for 10 minutes with primers LTR1 (5′-GTGGTCTCGCTGTTCCTT-3′; Roth, Karlsruhe, Germany) and MISC (5′-CGCTCGACATCTTTCCAGT-3′; Roth). PCR products were separated on a 2% agarose gel and quantified by comparing the intensity of the test signal to a limited dilution set of HeLa cell controls.
5′-Long terminal repeat integration site analysis
Genomic proviral junction sequences were detected by linear amplification-mediated PCR (LAM-PCR).10,11 In brief, 1 μg DNA from PB leukocytes was used as template for 2 rounds of a 50-cycle linear PCR with a biotinylated vector-specific primer followed by immobilization on streptavidin-coupled paramagnetic beads and separation using a magnet. This step was followed by hexanucleotide random priming, restriction digestion with Sse9I or isoschizomers, ligation of the linker, and exponential PCR. Two percent of the first PCR product served as template (either directly or after an additional magnetic capture purification step) for a second (semi-) nested PCR that enabled the visualization of the LAM-PCR amplicons (Figure 1B). LAM-PCR amplicons derived from DNA isolated from patients 1 and 2 were separated on a high-resolution Spreadex gel (Elchrom Scientific, Cham, Switzerland). For patient 3, PCR products were separated on a 2% agarose gel, transferred to a nylon membrane (Hybond-N; Amersham, Piscataway, NJ) by pressure blot (Stratagene, La Jolla, CA), probed with a digoxigenin-labeled LTR/extended packaging signal probe, and documented by chemiluminescence exposure to x-ray film (Roche Diagnostics, Mannheim, Germany). LAM amplicons were either isolated, purified, and directly sequenced or shotgun cloned into the TOPO TA vector (Invitrogen, Carlsbad, CA) and sequenced. The length of 37 bp in the unprimed LTR was taken to indicate the specificity and correctness of the insertion sites sequenced. A representative analysis of 96 wells showed 14 contained no bacterial colonies, and in 9 the genomic-LTR junction could not be sequenced. As a result, from 96 wells, a total of 73 specific inserts were isolated.
Results
Retrovirally marked mobilized PB cells display transient repopulating activity in autografted patients
G-CSF–mobilized PB cells were harvested from 3 patients with CML (2 in chronic phase and 1 in early blast crisis), and a portion of the total CD34+ cells present (8%, 25%, and 25%, respectively; Table 1) were then isolated and cultured for 5 days in the presence of a potent growth factor cocktail (FL, SCF, and IL-3 ± G-CSF and IL-6). During the same 5 days, or only during the last 3 days, the cells were also exposed to a clinical grade preparation of a neo-encoding retrovirus, as described in “Materials and methods.” These cultured cells were then harvested and transplanted together with the unmanipulated cells into their respective autologous hosts who, in the interim, had been treated with a myeloablative dose of busulfan. The proportion of transduced (G418-resistant) CFCs was at least 13%, and the total number of transduced CFCs infused varied from 2.9 to 3.8 × 104 per kg body weight (Table 1). Corresponding values for the LTC-ICs harvested from the same cultures were greater than 9% marked LTC-ICs and a calculated 2 to 11 × 104 transduced LTC-ICs per kg body weight transplanted (Table 1). Hematologic recovery was timely in all 3 patients, with achievement of a sustained WBC count of greater than 109/L and a platelet count of greater than 2 × 1010/L within 13 to 18 days.
Numbers of transduced cells transplanted
Patient no. . | No. CD34+ cells infected × 106/kg bwt (% of total) . | Total no. cells harvested from transduction cultures and transplanted × 106/kg . | No. transduced CFCs transplanted/kg . | No. transduced LTC-ICs transplanted/kg . |
|---|---|---|---|---|
| 1 | 1.0 (25) | 4.0 | 29 000 | 4300 |
| 2 | 1.1 (25) | 8.8 | 38 000 | 1100 |
| 3 | 0.5 (8) | 1.1 | 35 000 | 2100 |
Patient no. . | No. CD34+ cells infected × 106/kg bwt (% of total) . | Total no. cells harvested from transduction cultures and transplanted × 106/kg . | No. transduced CFCs transplanted/kg . | No. transduced LTC-ICs transplanted/kg . |
|---|---|---|---|---|
| 1 | 1.0 (25) | 4.0 | 29 000 | 4300 |
| 2 | 1.1 (25) | 8.8 | 38 000 | 1100 |
| 3 | 0.5 (8) | 1.1 | 35 000 | 2100 |
The fraction of CFCs and 5-week LTC-ICs transduced was inferred from a comparison of the yields of colonies obtained in methyl cellulose cultures containing 1.5 mg/mL G418 or not. Bwt indicates body weight.
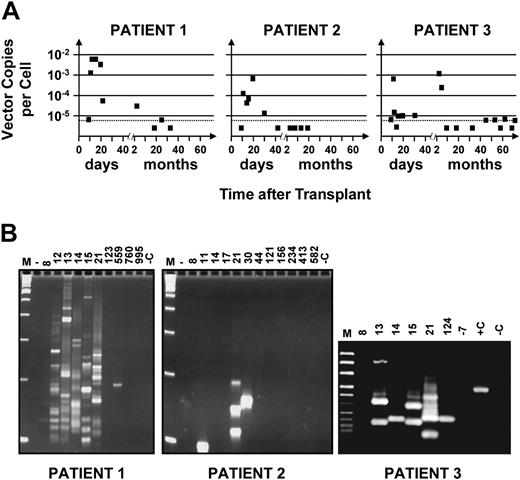
Figure 1 shows the results of semiquantitative PCR and highly sensitive LAM-PCR analyses of sequential PB samples obtained from all 3 patients from 10 days after transplantation until follow-up was terminated (18, 33, and 72 months later). Marked cells were first detected in all 3 patients on day 11 (Figure 1A). In one case (patient no. 2), such cells were detected only at that time among several samples taken before day 21 and then again on days 21 and 30 but not thereafter. In the other 2 patients (no. 1 and no. 3), evidence of a first peak of marked WBC production persisted throughout the period between days 11 and 21. Quantification of the amount of proviral DNA within the WBCs sampled during the first month showed that marked cells contributed approximately 0.1% to 1% of the total, assuming 1 copy of proviral DNA per transduced cell. Only approximately 10% of the CD34+ cells transplanted were exposed to the original retroviral preparation, and it is unlikely that the number of cells with STRC activity would have increased during the 5-day transduction culture.7,12 Thus, from the level of marking detected in these patients, a transduction efficiency of approximately 1% to 10% can be inferred for the cells infused that were able to generate an initial burst of WBC production. This would be consistent with the assumption of 1 copy of proviral DNA per transduced cell.
Kinetics of appearance and disappearance of marked cells in recipients of retrovirally transduced mobilized PB cell autografts. (A) Time course studies revealing an early contribution of marked cells to the WBCs produced in the first 3 weeks in all 3 patients studied with variable patterns of contribution at later times. DNA was extracted from sequentially obtained samples of PB cells from each patient, and proviral sequences were detected using primers designed to amplify a 589-bp 5′ vector sequence. Quantification of the proportion of WBCs that were marked was achieved by comparing the intensity of the signal obtained from each sample to control DNA from HeLa cells that contained a single copy of the provirus that was then serially diluted in DNA extracted from nontransduced HeLa cells. Analysis of 1 μg template DNA (∼ 1.5 × 105 test cells) at each time point allowed the limit of gene-marked cells detectable to be reduced to close to 0.001% (assuming 1 vector copy per cell, dashed horizontal line). (B) Detection of different proviral integration sites in PB cells using LAM-PCR. The data indicate that multiple clones contributed to the early output of retrovirally marked WBCs in all 3 patients. M indicates a 100-bp DNA ladder; +C, positive control (0.02 ng DNA from monoclonal HeLa cells transduced with the same vector); and –C (1 μg nontransduced HeLa DNA), as well as –7 (DNA extracted from the PB of this patient taken 7 days before the transplantation was performed), negative controls.
Kinetics of appearance and disappearance of marked cells in recipients of retrovirally transduced mobilized PB cell autografts. (A) Time course studies revealing an early contribution of marked cells to the WBCs produced in the first 3 weeks in all 3 patients studied with variable patterns of contribution at later times. DNA was extracted from sequentially obtained samples of PB cells from each patient, and proviral sequences were detected using primers designed to amplify a 589-bp 5′ vector sequence. Quantification of the proportion of WBCs that were marked was achieved by comparing the intensity of the signal obtained from each sample to control DNA from HeLa cells that contained a single copy of the provirus that was then serially diluted in DNA extracted from nontransduced HeLa cells. Analysis of 1 μg template DNA (∼ 1.5 × 105 test cells) at each time point allowed the limit of gene-marked cells detectable to be reduced to close to 0.001% (assuming 1 vector copy per cell, dashed horizontal line). (B) Detection of different proviral integration sites in PB cells using LAM-PCR. The data indicate that multiple clones contributed to the early output of retrovirally marked WBCs in all 3 patients. M indicates a 100-bp DNA ladder; +C, positive control (0.02 ng DNA from monoclonal HeLa cells transduced with the same vector); and –C (1 μg nontransduced HeLa DNA), as well as –7 (DNA extracted from the PB of this patient taken 7 days before the transplantation was performed), negative controls.
In patients no. 1 and no. 3, evidence of a second transient output of marked cells was seen 4 to 5 months after transplantation. However, thereafter, marked cells permanently decreased to undetectable or barely detectable levels in both.
Multiple transient clones contribute to the early phase of hematologic recovery in autografted patients
We also used LAM-PCR and sequence analysis of proviral integration sites to investigate the clonal composition of the marked PB cells identified at different times after transplantation (Figure 1B; Table 2 and Table S1, which is available on the Blood website; see the Supplemental Table link at the top of the online article). None of the insertion sites were repeated in sequential samples taken from patient no. 1 on days 12, 13, 14, 15, or 21 after transplantation or from patient no. 3 on days 13, 14, and 15 after transplantation. Of note, the clone identified in the second wave of marked WBC production that occurred 4 to 5 months after transplantation in patient no. 3 was different from those seen earlier or detected later. However, a repeat analysis performed on an early sample from patient no. 2 demonstrated some bands of identical size and one with a different size. Since we can estimate that the template DNA analyzed for LAM-PCR from these samples was derived from fewer than 150 marked cells, this finding supports the inference that low levels of marked cells reflect small numbers of transduced cells in the transplant. Taken together, these results confirm the transient nature of the populations of marked cells detected in this initial phase of hematologic recovery and, thus, support the concept of their origin from STRCs.
LAM-PCR analysis of provirus integrants in identified sites
Patient no. and Seq no. . | Time after transplantation, d . | Genomic length . | LAM amplicon length, bp . | Integrant-host-junction sequence . | Identity, % . | Chromosome . | Sequence orientation . | Integration locus . | Genomic locus/RefSeq gene . |
|---|---|---|---|---|---|---|---|---|---|
| 1 | |||||||||
| 80852 H02 | 12 | 54 | 147 | TCCATTTAATACAGTtgaaagaccc | 100.0 | 4 | Plus | 48114519 | 1777 bp upstream of TEC |
| 78933 B06 | 12 | 177 | 270 | GACCAGATGTGGGAAtgaaagaccc | 98.4 | 5 | Plus | 94448967 | 6025 bp upstream of MCTP1 |
| 78933 C06 | 12 | 139 | 232 | TGCTATAGTTACCGCtgaaagaccc | 100.0 | 10 | Minus | 121145584 | 187767 bp in intron 3 of GRK5 |
| 80852 F01 | 12 | 26 | 119 | ATAAAAGGGAAGCCTtgaaagaccc | 100.0 | 11 | Plus | 128037827 | 31372 bp upstream of FLI1 |
| 78933 B07 | 13 | 38 | 131 | CACACACACTCATAAtgaaagaccc | 100.0 | 1 | Minus | 155874598 | 14878 bp upstream of AIM2 |
| 78933 F07 | 13 | 77 | 170 | GACCTCAGGTGATCCtgaaagaccc | 100.0 | 1 | Plus | 164251126 | 3588 bp in intron 1 of CREG1 |
| 78933 D07 | 13 | 52 | 145 | TTGCTTACCCCACTGtgaaagaccc | 100.0 | 2 | Minus | 136728494 | 19044 bp upstream of CXCR4 |
| 79697 C08 | 13 | 130 | 223 | ATGTCACAGACAAAGtgaaagaccc | 100.0 | 2 | Plus | 17225602 | 41799 bp upstream of VSNL1 |
| 78933 H06 | 13 | 28 | 121 | AACAAATGAGGTTAGtgaaagaccc | 100.0 | 6 | Plus | 2935514 | 1582 bp in exon 1 of DKFZp6861 15217 |
| 78933 E07 | 13 | 125 | 218 | CTTGAAGGGTGGGTGtgaaagaccc | 100.0 | 7 | Minus | 94707759 | 1187 bp in intron 1 of PON2 |
| 80852 G03 | 13 | 289 | 382 | CTCCCTATATGATTGtgaaagaccc | 100.0 | 7 | Plus | 43647712 | 27533 bp downstream of BLVRA |
| 80852 C04 | 13 | 79 | 172 | CCAATAGCCTACCTTtgaaagaccc | 100.0 | 8 | Plus | 62757554 | 32002 bp in intron 2 of ASPH |
| 80852 G04 | 13 | 80 | 173 | ATCGCTTGAACCCGGtgaaagaccc | 98.8 | 9 | Minus | 129009529 | 8983 bp downstream of IER5L |
| 78933 F06 | 13 | 27 | 120 | TCTGTCTTCAAGTTCtgaaagaccc | 100.0 | 12 | Minus | 107639068 | 11307 bp upstream of CORO1C |
| 80852 E03 | 13 | 278 | 371 | TCCAGTAAGAGCTATtgaaagaccc | 99.7 | 12 | Minus | 92744212 | 170593 bp in intron 2 of CRADD |
| 80852 A04 | 13 | 26 | 119 | GCCGTGCAATACCTTtgaaagaccc | 100.0 | 12 | Plus | 15927381 | 768 bp in intron 1 of STRAP |
| 79697 C09 | 14 | 26 | 119 | GGAACTACTTTTTCCtgaaagaccc | 100.0 | 4 | Plus | 185981905 | 35854 bp in exon 9 of FLJ33167 |
| 80852 A06 | 14 | 60 | 153 | CATAGTTTCAAATAAtgaaaggccc | 100.0 | 5 | Minus | 59965457 | 66256 bp in intron 7 of DEPDC1B |
| 79697 E09 | 14 | 93 | 186 | GTGGGAGGATCGCTTtgaaagaccc | 100.0 | 6 | Minus | 109393783 | 117465 bp in intron 17 of ARMC2 |
| 78933 A09 | 14 | 49 | 142 | GAAAGTTATTTAGTTtgaaagaccc | 100.0 | 8 | Plus | 124166005 | 11904 bp in intron 4 of MGC21654 |
| 80852 G07 | 14 | 24 | 117 | GAACGGTCCTCTGACtgaaagaccc | 100.0 | 15 | Plus | 48215414 | 16703 bp upstream of ATP8B4 |
| 79697 F09 | 14 | 23 | 116 | CAGGATAGGATGCAGtgaaagaccc | 100.0 | 18 | Minus | 40593847 | 58380 bp in intron 1 of SETBP1 |
| 78933 H07 | 14 | 119 | 212 | TTAAGAGCTTGAGGCtgaaagaccc | 99.2 | 20 | Minus | 30763159 | 31877 bp in intron 3 of COMMD7 |
| 78933 A08 | 14 | 207 | 300 | ATACAGGTTCCAGGGtgaaagaccc | 99.1 | 20 | Plus | 237848 | 8887 bp downstream of ZCCHC3 |
| 80852 A08 | 15 | 76 | 169 | GTGTGGCCTAATAAtgaaagaccc | 98.7 | 2 | Plus | 36543852 | 48804 bp in intron 2 of CRIM1 |
| 80852 H08 | 15 | 159 | 252 | ATCTTAACAAACAGGtgaaagaccc | 100.0 | 3 | Plus | 15815709 | 2372 bp upstream of ANKRD28 |
| 80852 A09 | 15 | 89 | 182 | GTTAAGATATTTCATtgaaagaccc | 100.0 | 3 | Minus | 159553012 | 242418 bp in intron 5 of MGC12197 |
| 80852 G08 | 15 | 21 | 114 | TTTCATTCAAGAATCtgaaagaccc | 100.0 | 13 | Plus | 77171831 | 41659 bp upstream of FLJ30046 |
| 79697 H10 | 21 | 137 | 230 | AATATGACTGACATCtgaaagaccc | 100.0 | 5 | Minus | 10558333 | 40196 bp downstream of ROPN1L |
| 78933 E11 | 21 | 167 | 260 | TGCTTTGTGTCGCAGtgaaagaccc | 98.3 | 7 | Minus | 23111987 | 193 bp upstream of C7orf30 |
| 78933 E02 | 21 | 22 | 115 | AGGTGCCATTCCTTTtgaaagaccc | 100.0 | 12 | Minus | 121193424 | 16278 bp in Intron 1 of MGC35140 |
| 78933 H10 | 21 | 176 | 269 | CCCCCACCCACCTACtgaaagaccc | 99.5 | 17 | Minus | 7181218 | 612 bp in intron 1/exon 1 of CENTB1 |
| 78933 A11 | 21 | 86 | 179 | TGCAAAGATTGTTTCtgaaagaccc | 100.0 | 18 | Plus | 27468862 | 49764 bp in intron 5 of B4GALT6 |
| 78933 D10 | 21 | 115 | 208 | GTCTGGGGTTCAAACtgaaagaccc | 97.4 | 19 | Plus | 44428610 | 1159 bp upstream of IL28B |
| 80852 F12 | 21 | 101 | 194 | TCCCCTCAGTCCTCAtgaaagaccc | 100.0 | 20 | Plus | 5573746 | 34074 bp upstream of KIAA1434 |
| 2 | |||||||||
| 78933 A02 | 21 | 54 | 147 | CTGAATACTTACTATtgaaagaccc | 100.0 | 16 | Minus | 28531941 | 3791 bp upstream of SULT1A1 |
| 3 | |||||||||
| 9084 | 13 | 79 | 172 | GTCTTTGTTTCATAGtgaaagaccc | 100.0 | 2 | Minus | 136732882 | 23434 bp upstream of CXCR4 |
| 9086 | 14 | 110 | 203 | GAGCTGAGATCGCGCtgaaagaccc | 99.1 | 12 | Minus | 120560220 | 10060 bp in intron 1 of LOC283385 |
| 9088 | 15 | 89 | 182 | ACATTGGCACATCGCtgaaagaccc | 100.0 | 12 | Plus | 18867724 | 84336 bp upstream of CAPZA3 |
| 4065 | 152 | 212 | 305 | GGGGAAGGAAAAAACtgaaagaccc | 99.6 | 1 | Plus | 181568775 | 106526 bp in intron 5 of C1orf24 |
| 9706* | 335 | 93 | 186 | CAAGGAGAGTTTGATtgaaagaccc | 94.6 | 2 | Plus | 128781865 | 10617 bp in intron 1 of HS6ST1 |
Patient no. and Seq no. . | Time after transplantation, d . | Genomic length . | LAM amplicon length, bp . | Integrant-host-junction sequence . | Identity, % . | Chromosome . | Sequence orientation . | Integration locus . | Genomic locus/RefSeq gene . |
|---|---|---|---|---|---|---|---|---|---|
| 1 | |||||||||
| 80852 H02 | 12 | 54 | 147 | TCCATTTAATACAGTtgaaagaccc | 100.0 | 4 | Plus | 48114519 | 1777 bp upstream of TEC |
| 78933 B06 | 12 | 177 | 270 | GACCAGATGTGGGAAtgaaagaccc | 98.4 | 5 | Plus | 94448967 | 6025 bp upstream of MCTP1 |
| 78933 C06 | 12 | 139 | 232 | TGCTATAGTTACCGCtgaaagaccc | 100.0 | 10 | Minus | 121145584 | 187767 bp in intron 3 of GRK5 |
| 80852 F01 | 12 | 26 | 119 | ATAAAAGGGAAGCCTtgaaagaccc | 100.0 | 11 | Plus | 128037827 | 31372 bp upstream of FLI1 |
| 78933 B07 | 13 | 38 | 131 | CACACACACTCATAAtgaaagaccc | 100.0 | 1 | Minus | 155874598 | 14878 bp upstream of AIM2 |
| 78933 F07 | 13 | 77 | 170 | GACCTCAGGTGATCCtgaaagaccc | 100.0 | 1 | Plus | 164251126 | 3588 bp in intron 1 of CREG1 |
| 78933 D07 | 13 | 52 | 145 | TTGCTTACCCCACTGtgaaagaccc | 100.0 | 2 | Minus | 136728494 | 19044 bp upstream of CXCR4 |
| 79697 C08 | 13 | 130 | 223 | ATGTCACAGACAAAGtgaaagaccc | 100.0 | 2 | Plus | 17225602 | 41799 bp upstream of VSNL1 |
| 78933 H06 | 13 | 28 | 121 | AACAAATGAGGTTAGtgaaagaccc | 100.0 | 6 | Plus | 2935514 | 1582 bp in exon 1 of DKFZp6861 15217 |
| 78933 E07 | 13 | 125 | 218 | CTTGAAGGGTGGGTGtgaaagaccc | 100.0 | 7 | Minus | 94707759 | 1187 bp in intron 1 of PON2 |
| 80852 G03 | 13 | 289 | 382 | CTCCCTATATGATTGtgaaagaccc | 100.0 | 7 | Plus | 43647712 | 27533 bp downstream of BLVRA |
| 80852 C04 | 13 | 79 | 172 | CCAATAGCCTACCTTtgaaagaccc | 100.0 | 8 | Plus | 62757554 | 32002 bp in intron 2 of ASPH |
| 80852 G04 | 13 | 80 | 173 | ATCGCTTGAACCCGGtgaaagaccc | 98.8 | 9 | Minus | 129009529 | 8983 bp downstream of IER5L |
| 78933 F06 | 13 | 27 | 120 | TCTGTCTTCAAGTTCtgaaagaccc | 100.0 | 12 | Minus | 107639068 | 11307 bp upstream of CORO1C |
| 80852 E03 | 13 | 278 | 371 | TCCAGTAAGAGCTATtgaaagaccc | 99.7 | 12 | Minus | 92744212 | 170593 bp in intron 2 of CRADD |
| 80852 A04 | 13 | 26 | 119 | GCCGTGCAATACCTTtgaaagaccc | 100.0 | 12 | Plus | 15927381 | 768 bp in intron 1 of STRAP |
| 79697 C09 | 14 | 26 | 119 | GGAACTACTTTTTCCtgaaagaccc | 100.0 | 4 | Plus | 185981905 | 35854 bp in exon 9 of FLJ33167 |
| 80852 A06 | 14 | 60 | 153 | CATAGTTTCAAATAAtgaaaggccc | 100.0 | 5 | Minus | 59965457 | 66256 bp in intron 7 of DEPDC1B |
| 79697 E09 | 14 | 93 | 186 | GTGGGAGGATCGCTTtgaaagaccc | 100.0 | 6 | Minus | 109393783 | 117465 bp in intron 17 of ARMC2 |
| 78933 A09 | 14 | 49 | 142 | GAAAGTTATTTAGTTtgaaagaccc | 100.0 | 8 | Plus | 124166005 | 11904 bp in intron 4 of MGC21654 |
| 80852 G07 | 14 | 24 | 117 | GAACGGTCCTCTGACtgaaagaccc | 100.0 | 15 | Plus | 48215414 | 16703 bp upstream of ATP8B4 |
| 79697 F09 | 14 | 23 | 116 | CAGGATAGGATGCAGtgaaagaccc | 100.0 | 18 | Minus | 40593847 | 58380 bp in intron 1 of SETBP1 |
| 78933 H07 | 14 | 119 | 212 | TTAAGAGCTTGAGGCtgaaagaccc | 99.2 | 20 | Minus | 30763159 | 31877 bp in intron 3 of COMMD7 |
| 78933 A08 | 14 | 207 | 300 | ATACAGGTTCCAGGGtgaaagaccc | 99.1 | 20 | Plus | 237848 | 8887 bp downstream of ZCCHC3 |
| 80852 A08 | 15 | 76 | 169 | GTGTGGCCTAATAAtgaaagaccc | 98.7 | 2 | Plus | 36543852 | 48804 bp in intron 2 of CRIM1 |
| 80852 H08 | 15 | 159 | 252 | ATCTTAACAAACAGGtgaaagaccc | 100.0 | 3 | Plus | 15815709 | 2372 bp upstream of ANKRD28 |
| 80852 A09 | 15 | 89 | 182 | GTTAAGATATTTCATtgaaagaccc | 100.0 | 3 | Minus | 159553012 | 242418 bp in intron 5 of MGC12197 |
| 80852 G08 | 15 | 21 | 114 | TTTCATTCAAGAATCtgaaagaccc | 100.0 | 13 | Plus | 77171831 | 41659 bp upstream of FLJ30046 |
| 79697 H10 | 21 | 137 | 230 | AATATGACTGACATCtgaaagaccc | 100.0 | 5 | Minus | 10558333 | 40196 bp downstream of ROPN1L |
| 78933 E11 | 21 | 167 | 260 | TGCTTTGTGTCGCAGtgaaagaccc | 98.3 | 7 | Minus | 23111987 | 193 bp upstream of C7orf30 |
| 78933 E02 | 21 | 22 | 115 | AGGTGCCATTCCTTTtgaaagaccc | 100.0 | 12 | Minus | 121193424 | 16278 bp in Intron 1 of MGC35140 |
| 78933 H10 | 21 | 176 | 269 | CCCCCACCCACCTACtgaaagaccc | 99.5 | 17 | Minus | 7181218 | 612 bp in intron 1/exon 1 of CENTB1 |
| 78933 A11 | 21 | 86 | 179 | TGCAAAGATTGTTTCtgaaagaccc | 100.0 | 18 | Plus | 27468862 | 49764 bp in intron 5 of B4GALT6 |
| 78933 D10 | 21 | 115 | 208 | GTCTGGGGTTCAAACtgaaagaccc | 97.4 | 19 | Plus | 44428610 | 1159 bp upstream of IL28B |
| 80852 F12 | 21 | 101 | 194 | TCCCCTCAGTCCTCAtgaaagaccc | 100.0 | 20 | Plus | 5573746 | 34074 bp upstream of KIAA1434 |
| 2 | |||||||||
| 78933 A02 | 21 | 54 | 147 | CTGAATACTTACTATtgaaagaccc | 100.0 | 16 | Minus | 28531941 | 3791 bp upstream of SULT1A1 |
| 3 | |||||||||
| 9084 | 13 | 79 | 172 | GTCTTTGTTTCATAGtgaaagaccc | 100.0 | 2 | Minus | 136732882 | 23434 bp upstream of CXCR4 |
| 9086 | 14 | 110 | 203 | GAGCTGAGATCGCGCtgaaagaccc | 99.1 | 12 | Minus | 120560220 | 10060 bp in intron 1 of LOC283385 |
| 9088 | 15 | 89 | 182 | ACATTGGCACATCGCtgaaagaccc | 100.0 | 12 | Plus | 18867724 | 84336 bp upstream of CAPZA3 |
| 4065 | 152 | 212 | 305 | GGGGAAGGAAAAAACtgaaagaccc | 99.6 | 1 | Plus | 181568775 | 106526 bp in intron 5 of C1orf24 |
| 9706* | 335 | 93 | 186 | CAAGGAGAGTTTGATtgaaagaccc | 94.6 | 2 | Plus | 128781865 | 10617 bp in intron 1 of HS6ST1 |
All patient samples analyzed were derived from cloned and sequenced PB LAM products as shown in Figure 1. Each sequenced LAM-PCR amplicon was aligned to the human genome database, and the insertion site was identified using University of California, Santa Cruz BLAT search tools (http://genome.ucsc.edu/). Integrant-host-junction sequence present in the first 15 nucleotides of the genomic DNA flanking the 5′-LTR (10 nucleotides). Genomic length denotes the size of the LAM-PCR amplicon without linker and LTR sequences. A complete list of all integrants analyzed is provided in Table S1. All cells analyzed were WBCs, except as shown by the asterisk (*), where purified granulocytes were analyzed.
The numbers of unique integration sites detected in all 3 patients during the first month after transplantation (59 inserts detected in cells from patient no. 1, 3 in cells from patient no. 2, and 5 cells from patient no. 3) also demonstrates the multiplicity of clones active during this time, assuming 1 clone per insert. The greater number of clones (∼ 10-fold) of gene-marked cells identified in the WBCs produced within the first 3 weeks in patient no. 1 by comparison to patients 2 and 3 is consistent with the correspondingly greater representation of marked cells in the PB of patient no. 1 during this period (Figure 1A).
Sequencing of shotgun-cloned LAM-PCR products revealed 69 unique integration sites (Table 2 and Table S1). Fifty-one of these integration sites could be mapped unequivocally to previously identified sequences in the human genome (44 from patient no. 1, 1 from patient no. 2, and 6 from patient no. 3). Forty-one of the 51 mapped inserts (80%) were found near (± 100 kb) or within genes defined in RefSeq, and all but 2 of these were located within introns. Interestingly, 2 of the integration sites detected in WBCs produced early after transplantation in 2 different patients (no. 1 and no. 3) were 19 kb and 23 kb upstream of CXC chemokine receptor 4 (CXCR4), the gene encoding the receptor for stromal-derived factor-1 (SDF-1).
Discussion
In this study, we analyzed serial PB blood cell samples from 3 recipients of partially marked autografts. The results provide new evidence that human hematopoietic cells with predetermined short-term repopulating activity contribute to the initial but not later phases of hematopoietic reconstitution seen in such patients. This conclusion is based on the following findings. First, in all 3 patients, there was a well-defined early wave of marked WBCs that reached a peak between the 11th and 20th days after transplantation. Second, the kinetics and ultimate level of early and late repopulation in individual patients was found to be highly dissociated. Third, multiple (differently marked) clones were found to be uniquely active during the first 4 weeks after transplantation with different clones showing activity at later times. These studies thus extend to human autografts the concept of a hierarchy of cells with in vivo repopulating ability that vary in their durability of mature cell output.
It is noteworthy that the substantial, but transient, early burst of marked WBCs seen in the patients followed in this study mirrors closely in time and content the output of cells produced during the first 3 weeks after transplantation in immunodeficient nonobese diabetic (NOD)/SCID-β2microglobulin–/– mice that received transplants of CD34+CD38+ STRC-Ms.5-7 These results suggest that STRCs defined by their repopulating activities in such xenogeneic recipients display similar kinetics of growth and differentiation when transplanted into myeloablated human hosts, thus underscoring the promise of such xenotransplantation assays for predicting WBC recoveries in patients receiving transplants.13
Transplantation experiments to examine the properties of individual repopulating cells have further suggested that hematopoietic recovery in mice can be carried out by a very small number of clones when grafts of freshly isolated murine cells are transplanted.14-17 In humans, it seems much less likely that the neutrophils and platelets produced immediately after transplantation would represent clonal populations for a number of reasons. First, the cell numbers required to meet the daily needs for new mature PB cells in adult humans are much larger (∼ 100 times those needed in mice). Second, human cells also have longer cell cycle times (∼ 2 times longer than murine cells). Thus, it would not be anticipated that single human cells could achieve clonal dominance within 2 to 3 weeks in patients with a normalized WBC count by that time. Assuming an 80-kg adult human normally contains approximately 1010 WBCs, a single repopulating cell would have to produce a minimum of 108 progeny within 11 days to regenerate just 1% of these. This, in turn, would require a minimum of 26 divisions in the same time frame, assuming no lag time and no cell loss during this phase of cell expansion in vivo. The present evidence of oligo/polyclonal reconstitution of human recipients of marked autologous cells is consistent with these calculations. In fact, clonal repopulation of humans with transplanted cells has rarely been documented and then only after a minimum of 28 days.2,11 Similarly in nonhuman primates, early repopulation by marked cells has also been found to be highly polyclonal.10
We have recently shown that the leukemic clone in patients with chronic-phase CML contains cells with features of STRCs18 as well as more primitive leukemic cells with a more prolonged repopulating activity detectable in immunodeficient mice.19-21 It is, therefore, conceivable that some of the repopulating activity detected in the present study might have been attributable to BCR-ABL+ cells. Although the data available cannot completely exclude this possibility, we believe such cells did not play a significant role in the repopulation obtained in the autografting studies reported here for several reasons. First, in 2 cases (patients no. 2 and no. 3), fluorescence in situ hybridization analyses were performed on the CD34+ cells isolated from the freshly harvested leukapheresis products subsequently used as autografts, and these analyses showed the frequency of BCR-ABL+ cells to be either low (7% and 30% in the 2 collections obtained from patient no. 2) or undetectable (< 6% in the case of patient no. 3). In the case of patient no. 2, who received a transplant after showing symptoms of incipient blast crisis and who relapsed with terminal blast-phase disease 18 months after transplantation, no evidence of gene marking was detectable in the relapse PB or marrow cells using a sensitive nested (LTR) PCR procedure (data not shown). Finally, the chemotherapy used in this study has been demonstrated to mobilize predominantly normal hematopoietic progenitors in patients with chronic-phase CML, and we and others have previously shown that the number of primitive BCR-ABL+ cells (LTC-ICs) declines rapidly in vitro under the type of culture conditions used in our 5-day transduction protocol.22
Our results also add significantly to a growing body of data documenting a preferential integration of Moloney virus-based oncoretroviral vectors, and lentiviral vectors near or within genes.23-26 In this study we were able to identify a total of 51 integration sites by sequence alignment and found that 80% of these inserts were also located within or near genes (± 100 kb). The concept that such inserts occur preferentially in genes whose expression is relevant to the function of the cells transduced was reinforced here by the finding that 2 inserts in cells from different patients were sited within 25 kb upstream of the CXCR4 gene. CXCR4 is the receptor for SDF-1 and is known to be expressed on the surface of primitive human hematopoietic cells where it participates in the regulation of both their cell cycle arrest27,28 and chemotactic/homing responses to SDF-1.29 No untoward consequences of retroviral insertional mutagenesis were noted in any of the 3 recipients of marked autografts in the present study. Nevertheless, our findings point to the need for continuing vigilance of this possibility in future trials using transduced grafts, particularly given the demonstrated efficiency (∼ 10%) with which STRCs can be retrovirally transduced, even in the virtual absence of transduction of cells with long-term repopulating activity.
Prepublished online as Blood First Edition Paper, April 21, 2005; DOI 10.1182/blood-2004-07-2859.
Supported by grants from the Dr Mildred Scheel Stiftung fur Krebsforschung, Bonn, Germany, and the Verein zur Foerderung der Leukaemie und Tumor Forschung, Freiburg, Germany (H.G.); by the Deutsche Forschungsgemeinschaft (Ka 976/4-1) and the German Minister for Education and Research (D1 KV9527/7) (C.v.K); by the National Institutes of Health (P01-HL55435), the National Cancer Institute of Canada (013003; with funds from the Terry Fox Run), and the Stem Cell Network (C.J.E.).
H.G. and M.S. contributed equally to this study. C.E. and C.v.K. contributed equally to this study.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dagmar Wider and Eva Samek for technical assistance, CellGenix for gifts of reagents, and Adrienne Wanhill for assistance in manuscript preparation. We also thank Dr C. Peters and Dr R. Mertelsmann for helpful discussions and support.
Supplemental data
All patient samples analyzed were derived from cloned and sequenced PB LAM products as shown in Figure 1. Each sequenced LAM-PCR amplicon was aligned to the Human Genome Database and the insertion site identified using University of California–Santa Cruz BLAT search tools. Integrant-host-junction sequences were present in the first 15 nucleotides of the genomic DNA flanking the 5'LTR (10 nucleotides). Genomic length denotes the size of the LAM-PCR amplicon without linker and LTR sequences. Pat indicates patient number; WBC, white blood cells; and (G), purified granulocytes.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal