Abstract
Endomitosis is a unique form of cell cycle used by megakaryocytes, in which the latter stages of mitosis are bypassed so that the cell can increase its DNA content and size. Although several transcription factors, including GATA-1 and RUNX-1, have been implicated in this process, the link between transcription factors and polyploidization remains undefined. Here we show that GATA-1–deficient megakaryocytes, which display reduced size and polyploidization, express nearly 10-fold less cyclin D1 and 10-fold increased levels of p16 compared with their wild-type counterparts. We further demonstrate that cyclin D1 is a direct GATA-1 target in megakaryocytes, but not erythroid cells. Restoration of cyclin D1 expression, when accompanied by ectopic overexpression of its partner Cdk4, resulted in a dramatic increase in megakaryocyte size and DNA content. However, terminal differentiation was not rescued. Of note, polyploidization was only modestly reduced in cyclin D1–deficient mice, likely due to compensation by elevated cyclin D3 expression. Finally, consistent with an additional defect conferred by increased levels of p16, inhibition of cyclin D-Cdk4 complexes with a TAT-p16 fusion peptide significantly blocked polyploidization of wild-type megakaryocytes. Together, these data show that GATA-1 controls growth and polyploidization by regulating cyclin D-Cdk4 kinase activity.
Introduction
Cyclin D1 is one of a battery of cell-cycle regulatory proteins, including cyclins, cyclin-dependent kinases (Cdks), and Cdk inhibitors, that is required for proper regulation of cell cycle and cell differentiation.1 G1-specific cyclin-Cdk complexes, including the D-type cyclins and Cdk4/6, promote progression into S phase through the phosphorylation and inhibition of pocket proteins (pRb, p107, and p130).2 Hypophosphorylated pocket proteins bind and inhibit the E2F family of transcription factors that control genes involved in proliferation and DNA replication, including cyclin E.3 Phosphorylation of pocket proteins by cyclin D1–Cdk4/6 causes their dissociation from E2F proteins, leading to transcriptional activation and a commitment to cell-cycle progression.4 However, recent data have described a more intricate balance between different phosphorylation states and function of pRb. Ezhevsky et al have shown that cyclin D–Cdk4/6 hypophosphorylates pRb in early G1, activating pRb as a transcriptional repressor and allowing it to bind E2F and E1A proteins.5 In late G1, cyclin E–Cdk2 complexes hyperphosphorylate pRb, thereby inactivating it and allowing for progression into S phase. These findings lend credence to the model that cyclin D and cyclin E activities are not equivalent. Of note, cellular proliferation is not affected when individual D-type cyclins or Cdk4 or Cdk6 are disrupted in mice, although individual knockout lines display specific growth defects. For example, cyclin D1–deficient animals are smaller than their littermates and show hypoplasia of the retina and mammary epithelium.6–9 Most tissues express multiple D-type cyclins, however, which allows for compensation when one is knocked out. Notably, mice that are deficient for all 3 D-type cyclins show gross defects in multiple cell types, including hematopoietic progenitors.10
Cdk inhibitors offer another means of regulating cell-cycle progression. For example, p16ink4A (p16), a member of the INK4 family of cell-cycle inhibitors that is comprised of p15, p16, p18, and p19 proteins, acts by specifically binding monomeric Cdk4/6 to disrupt interactions with D-type cyclins and drive cells to arrest in G0.11 Similar to gene-targeting studies of cyclins and other Cdks, p16-deficient mice are viable and generally do not seem to have defects in cell-cycle progression.12 However, consistent with these proteins functioning as tumor suppressors, p16-deficient mice do display a subtle increase in susceptibility to developing tumors. Furthermore, overexpression of INK4 proteins leads to an arrest in the G1 phase of the cell cycle, indicating a necessity for cyclin D–Cdk4/6 to progress to S phase in proliferating cells.13,14
Progression through the cell cycle is not limited to dividing cells, as a small number of cell types undergo repeated rounds of DNA replication without cell division. Examples include vascular smooth muscle cells, trophoblast giant cells, Drosophila salivary glands, and platelet producing megakaryocytes.15 Committed megakaryocyte progenitors, including the BFU-Mks and CFU-Mks, burst-forming and colony-forming unit megakaryocytes, respectively, proliferate to a limited extent, giving rise to a colony of megakaryocytes. Within these colonies, individual cells undergo terminal differentiation and eventually shed platelets. Unlike all other differentiating hematopoietic cells, megakaryocytes progress through repeated rounds of DNA synthesis without cell division, in a process termed endomitosis. This phenomenon allows megakaryocytes to accumulate DNA up to 128N and greatly increase their size and protein production.16 These increases in cell size, DNA content and protein levels are associated with the development of long cytoplasmic extensions, termed proplatelet forms that eventually shed platelets.17
To elucidate factors that govern endomitosis, we investigated the cause of the defect in growth and polyploidization of megakaryocytes that lack the essential transcription factor GATA-1. Previous studies have shown that GATA-1 is required for both polyploidization and platelet production.18,19 Here we report that the growth and ploidy defect of GATA-1–deficient megakaryocytes are due, in part, to reduced expression of cyclin D1, which is a direct GATA-1 target gene. Ectopic expression of cyclin D1, when accompanied by Cdk4, led to a dramatic increase in size and DNA content of GATA-1–deficient megakaryocytes, but did not rescue other aspects of terminal maturation, such as proplatelet production. We also show that p16, whose expression is elevated in the absence of GATA-1, is a potent inhibitor of polyploidization. These results demonstrate that cyclin D-Cdk4 kinase activity is essential for this distinct mode of cell cycle progression.
Materials and methods
Expansion and purification of primary megakaryocytes
Primary mouse hematopoietic progenitor cells were isolated from fetal livers of E13.5 embryos using the EasySep negative selection mouse hematopoietic progenitor enrichment kit (Stem Cell Technologies, Vancouver, BC). Progenitors were expanded for 2 days in serum-free expansion media described previously.20 Cells were differentiated into megakaryocytes by culturing in differentiation media (RPMI medium 1640 supplemented with 10% FCS containing SCF 1/100 and thrombopoietin [TPO, 10 ng/mL; R&D Systems, Minneapolis, MN]) for 3 days. Following differentiation, megakaryocytes were isolated on a 1.5%/3.0% discontinuous bovine serum albumin (BSA) gradient.21 Purity of megakaryocytes was assessed using acetylcholinesterase (ACHE) staining following standard protocols.
Analysis of DNA content and cell size and flow cytometry
BSA gradient–purified or CD41+ megakaryocytes were fixed in 75% ice-cold ethanol for one hour and stained with 4′,6-diamidino-2-phenylindole (DAPI; Sigma; St Louis, MO)/Triton X-100 solution containing 0.1% Triton X-100 and 1 μg/mL DAPI (freshly prepared). Surface staining for mouse CD41 (glycoprotein IIb [GPIIb]; BD Pharmingen, San Diego, CA) and CD42b (GPIbα; Emfret Analytics, Wurzburg, Germany) was performed by incubation for 30 minutes with phycoerythrin (PE)–conjugated antibodies, in Ca2+-free, Mg2+-free phosphate-buffered saline (PBS). DNA content and cell size were determined using flow cytometry on an LSR II (BD Biosciences, Mountain View, CA). Data were analyzed using FlowJo software (Treestar, Ashland, OR).
Mice
GATA-1KD (ΔneoΔHS-strain) mice harbor a targeted deletion of a DNase I hypersensitive (HS) site between the 2 GATA-1 promoters, and as a consequence fail to express detectable GATA-1 in megakaryocytes but show normal levels of GATA-1 expression in the erythroid lineage.18,19 These animals are maintained in a mixed C57Bl/6-129 background. Control wild-type animals included littermates, C57Bl/6-129 F2 hybrids, and C57Bl/6 mice (Jackson Labs, Bar Harbor, ME). Cyclin D1 knockout mice were maintained in a mixed C57Bl/6-129 background. Animal studies were approved by the University of Chicago Institutional Animal Care and Use Committee.
Retroviral transduction
The murine cyclin D1 cDNA was subcloned with a hemagglutin (HA) tag into the MIGR1 retroviral vector that coexpresses enhanced green fluorescent protein (EGFP) through an internal ribosomal entry site (IRES) element. Murine Cdk4 was subcloned with an HA tag into the pCS retroviral vector, which coexpresses human CD25 through an IRES element. Primary murine fetal liver progenitors were infected using the spinoculation method as described previously22 on days 2 and 3 of expansion. Differentiation was initiated after the final spinoculation.
qRT-PCR
RNA was extracted from multiple outgrowths of BSA-purified wild-type and GATA-1 knockdown megakaryocytes using TRIzol reagent (Invitrogen, Carlsbad, CA). cDNA was generated from RNA using SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturer's recommendations. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed as described previously.20 All expression values were normalized to the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Primer sequences are provided in Figure S4, available on the Blood website; see the Supplemental Figures link at the top of the online article.
Luciferase assays
The cyclin D1 promoter (2 kb) and various deletions were amplified and subcloned, according to mapping of the murine cyclin D1 promoter,23 into the pGL3-basic vector. Primers used for amplification of the promoter fragments are listed in Figure S4. NIH-3T3 cells were transfected with pGL3 constructs and a β-galactosidase vector (for normalization) using Polyfect transfection reagent (Qiagen, Valencia, CA) and following the manufacturer's protocol. Cells were cultured in 6-well plates and transfected at 40% to 80% confluency with a maximum of 1.5 μg DNA. The total amount of DNA was kept constant by the inclusion of appropriate amounts of empty vectors. Cells were harvested 48 hours after transfection with Passive Lysis Buffer (Promega, Madison, WI). After one freeze-thaw cycle, the supernatant was collected by centrifugation for 1 minute. The supernatant was evaluated for luciferase activity using the Luciferase Reporter Assay System (Promega). β-Gal activity was measured using the Galacto-Star detection system (Applied Biosystems, Foster City, CA) following the manufacturer's protocol. Luciferase and β-gal activity was measured on a Monolight 2010 (Analytical Luminescence Laboratory, San Diego, CA). Luciferase activity was normalized by dividing luciferase units by β-gal units for a given sample.
Chromatin immunoprecipitations
Results
GATA-1–deficient megakaryocytes are smaller and less polyploid than their wild-type counterparts
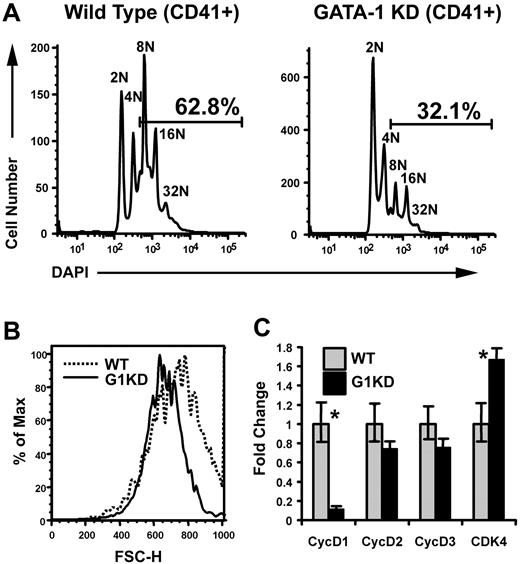
Previous studies have shown that megakaryocytes derived from GATA-1 knockdown mice (G1KD), which fail to express GATA-1 in the megakaryocyte lineage but show normal levels of GATA-1 expression in erythroid cells,18,19 undergo polyploidization to a lesser extent than wild-type cells.19 To confirm this defect, we cultured wild-type (WT) and G1KD lineage-depleted bone marrow cells with TPO and SCF for 2 to 3 days. The resulting megakaryocytes were stained with DAPI and the megakaryocyte marker CD41, and then analyzed by flow cytometry. As expected, fewer G1KD megakaryocytes reached a ploidy class of 8N or greater compared to those generated in wild-type cultures (Figure 1A). Of note, we failed to observe a ploidy defect in earlier experiments20 and attribute this to differences in culture conditions. We previously used a differentiation cocktail that included EPO, whereas these studies include TPO and SCF to promote megakaryocyte differentiation. We have noted more robust megakaryocyte differentiation with our current conditions, which likely allows for defects in polyploidization to be revealed. Next, we performed flow cytometry to analyze the size of the megakaryocytes derived from WT and G1KD cultures using forward scatter (FSC), which is an indicator of cell size. As a population, CD41+ G1KD cells were smaller than those generated from wild-type progenitors (data not shown). Since cell size is related to the degree of polyploidization, we also compared the sizes of a specific ploidy class: gating on the 16N population confirmed that the G1KD megakaryocytes were smaller than wild-type cells (Figure 1B). Taken together, these studies confirm that the absence of GATA-1 leads to defects in both cell growth and polyploidization of megakaryocytes.
Megakaryocytes lacking GATA-1 show defects in both polyploidization and accumulation of cell mass. (A) Ploidy analysis of wild-type and GATA-1 KD bone marrow progenitors differentiated into megakaryocytes for 3 days. The cells were stained with anti-CD41 and DAPI to detect DNA content. Gates indicate the percentage of cells that is 8N or greater. One representative of 3 experiments is shown. (B) Cell size analysis of wild-type and GATA-1 KD cells within the 16N ploidy class were compared in a forward scatter–height histogram. The dotted line indicates the wild-type cells and the solid line, the GATA-1 KD cells. Mean FSC-A for WT and G1KD megakaryocytes was 740 and 668, respectively; confidence in difference is 99.9% based on Kolmogorov-Smimov test. One representative of 3 experiments is shown. (C) mRNA levels were determined by real-time quantitative RT-PCR of RNA isolated from wild-type and GATA-1 KD, BSA gradient–purified megakaryocytes. GATA-1 KD mRNA levels are shown relative to wild type, which were arbitrarily set to one. The means ± 1 SD are shown. Asterisks indicate P values determined by Student t test to be .05 or less. One representative of 2 experiments, each in triplicate, is shown.
Megakaryocytes lacking GATA-1 show defects in both polyploidization and accumulation of cell mass. (A) Ploidy analysis of wild-type and GATA-1 KD bone marrow progenitors differentiated into megakaryocytes for 3 days. The cells were stained with anti-CD41 and DAPI to detect DNA content. Gates indicate the percentage of cells that is 8N or greater. One representative of 3 experiments is shown. (B) Cell size analysis of wild-type and GATA-1 KD cells within the 16N ploidy class were compared in a forward scatter–height histogram. The dotted line indicates the wild-type cells and the solid line, the GATA-1 KD cells. Mean FSC-A for WT and G1KD megakaryocytes was 740 and 668, respectively; confidence in difference is 99.9% based on Kolmogorov-Smimov test. One representative of 3 experiments is shown. (C) mRNA levels were determined by real-time quantitative RT-PCR of RNA isolated from wild-type and GATA-1 KD, BSA gradient–purified megakaryocytes. GATA-1 KD mRNA levels are shown relative to wild type, which were arbitrarily set to one. The means ± 1 SD are shown. Asterisks indicate P values determined by Student t test to be .05 or less. One representative of 2 experiments, each in triplicate, is shown.
We recently compared the gene expression profiles of wild-type and G1KD megakaryocytes by Affymetrix (Santa Clara, CA) microarray analysis.20 Among the cell-cycle–related genes, cyclin D1 expression was reduced 4-fold in GATA-1–deficient cells, while other cyclins remained relatively unchanged. To verify these microarray data, we compared the expression of cyclin D1 by quantitative real-time PCR in mRNA isolated from BSA gradient–purified wild-type and G1KD megakaryocytes and found that cyclin D1 mRNA was down-regulated nearly 10-fold in the G1KD megakaryocytes (Figure 1C). Notably, there were no significant differences in the expression of cyclins D2 or D3 in the absence of GATA-1. Furthermore, expression of the cyclin D–binding partner, Cdk4, was only modestly elevated in the absence of GATA-1 (1.6-fold, Figure 1C).
Cyclin D1 is a direct target of GATA-1 in megakaryocytes
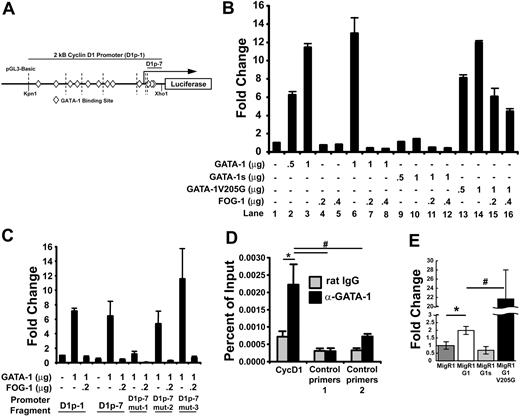
The observed decrease in cyclin D1 expression in GATA-1–deficient cells could be a reflection of differences in the maturation stage of the cells, or a direct consequence of the inability of GATA-1 to promote its transcription. To determine whether cyclin D1 is a direct target of GATA-1, we first cloned 2 kb of the cyclin D1 promoter upstream of the firefly luciferase gene (Figure 2A) and assayed the ability of GATA-1 to induce expression in transient reporter assays in NIH3T3 cells. As shown in Figure 2B, the cyclin D1 promoter displayed a dose-dependent induction in response to GATA-1 (lanes 1-3), whereas coexpression of FOG-1 had a potent repressive effect on luciferase expression (lanes 6-8). The V205G mutant of GATA-1, which is defective in its ability to interact with FOG-1,26 was also able to induce cyclin D1 promoter activity and was largely resistant to FOG-1 coexpression (lanes 13-16). Furthermore, expression of GATA-1s, the short isoform of GATA-1 that lacks the N-terminal activation domain and is associated with megakaryoblastic leukemia,27 was unable to activate the cyclin D1 promoter (lanes 9-12).
GATA-1 binds and activates the cyclin D1 promoter, while FOG-1 represses its transcription. (A) The pGL3-basic construct containing 2 kb of the cyclin D1 promoter upstream of the luciferase gene. Putative GATA-1–binding sites are indicated as diamonds, and dotted vertical lines indicate serial deletions made of the cyclin D1 promoter. D1p-1 is the full 2-kb fragment, and D1p-7 is the shortest 76-bp fragment. (B) Luciferase assay performed by transient transfection of NIH3T3 cells with the indicated micrograms of pcDNA3 vector containing GATA-1, GATA-1s, GATA-1 V205G, and FOG-1. Luciferase levels are shown as fold change relative to cells transfected with reporter construct alone. The total amount of DNA transfected was kept constant by transfection with empty pcDNA3 vector. One representative of 2 experiments, each in triplicate, is shown. (C) Luciferase assay performed as in panel B with luciferase vectors containing mutations in the 3 putative GATA-binding sites within the last 76 base pairs. Mutation of the first GATA site (D1p-7 mut-1) showed reduced response to GATA-1, whereas mutation of 2 other putative GATA sites (D1p-7 mut-2 and D1p-7 mut3) does not alter GATA-1 activation of the cyclin D1 promoter. D1p-1 and D1p-7 are described in panel A. One representative of 3 experiments, each in triplicate, is shown. (D) Chromatin immunoprecipitation assay performed in primary mouse fetal liver–derived megakaryocytes with primer sets directed toward the functional GATA-binding site identified in luciferase assays, as well as, downstream (control primers 1) and upstream (control primers 2) primer sets. Control rat IgG and anti–GATA-1 (N6) antibodies were used for immunoprecipitation and indicate GATA-1 enrichment at the functional GATA-1–binding site in the cyclin D1 promoter. P values were generated using the Student t test and are as follows: *P ≤ .04; #P ≤ .01. One representative of 2 experiments, each in triplicate, is shown. (E) Real-time quantitative PCR data showing expression levels of cyclin D1 mRNA following reconstitution of GATA-1 KD megakaryocytes with the empty MigR1 retroviral vector or containing wild-type GATA-1, GATA-1s, or GATA-1 V205G. Values are shown relative to empty MigR1-infected GATA-1 KD expression levels. P values were generated using the Student t test and are as follows: *P ≤ .006; #P ≤ .007. Changes between expression values of MigR1 and MigR1-G1s were not statistically significant. One representative of 3 experiments, each in triplicate, is shown. For each study in B-E, the means ± 1 SD is shown.
GATA-1 binds and activates the cyclin D1 promoter, while FOG-1 represses its transcription. (A) The pGL3-basic construct containing 2 kb of the cyclin D1 promoter upstream of the luciferase gene. Putative GATA-1–binding sites are indicated as diamonds, and dotted vertical lines indicate serial deletions made of the cyclin D1 promoter. D1p-1 is the full 2-kb fragment, and D1p-7 is the shortest 76-bp fragment. (B) Luciferase assay performed by transient transfection of NIH3T3 cells with the indicated micrograms of pcDNA3 vector containing GATA-1, GATA-1s, GATA-1 V205G, and FOG-1. Luciferase levels are shown as fold change relative to cells transfected with reporter construct alone. The total amount of DNA transfected was kept constant by transfection with empty pcDNA3 vector. One representative of 2 experiments, each in triplicate, is shown. (C) Luciferase assay performed as in panel B with luciferase vectors containing mutations in the 3 putative GATA-binding sites within the last 76 base pairs. Mutation of the first GATA site (D1p-7 mut-1) showed reduced response to GATA-1, whereas mutation of 2 other putative GATA sites (D1p-7 mut-2 and D1p-7 mut3) does not alter GATA-1 activation of the cyclin D1 promoter. D1p-1 and D1p-7 are described in panel A. One representative of 3 experiments, each in triplicate, is shown. (D) Chromatin immunoprecipitation assay performed in primary mouse fetal liver–derived megakaryocytes with primer sets directed toward the functional GATA-binding site identified in luciferase assays, as well as, downstream (control primers 1) and upstream (control primers 2) primer sets. Control rat IgG and anti–GATA-1 (N6) antibodies were used for immunoprecipitation and indicate GATA-1 enrichment at the functional GATA-1–binding site in the cyclin D1 promoter. P values were generated using the Student t test and are as follows: *P ≤ .04; #P ≤ .01. One representative of 2 experiments, each in triplicate, is shown. (E) Real-time quantitative PCR data showing expression levels of cyclin D1 mRNA following reconstitution of GATA-1 KD megakaryocytes with the empty MigR1 retroviral vector or containing wild-type GATA-1, GATA-1s, or GATA-1 V205G. Values are shown relative to empty MigR1-infected GATA-1 KD expression levels. P values were generated using the Student t test and are as follows: *P ≤ .006; #P ≤ .007. Changes between expression values of MigR1 and MigR1-G1s were not statistically significant. One representative of 3 experiments, each in triplicate, is shown. For each study in B-E, the means ± 1 SD is shown.
To narrow down the specific cis-acting sequences that are required for activation by GATA-1, a series of shorter promoter constructs were generated and analyzed in the same way. These studies revealed that the most proximal 76 bases, contained in the D1p-7 reporter, were sufficient for full activity (Figure 2C). As the D1p-7 reporter includes 3 putative GATA-1–binding sites (Figure 2A diamonds), we subsequently mutated each site independently in 3 constructs (mut-1, mut-2, or mut-3), and then assayed for luciferase activity. These experiments established that one of the GATA sites, at +118, was essential for transactivation, while mutations in the second or third GATA sites did not interfere with the ability of GATA-1 to activate the reporter (Figure 2C). Thus, a functional GATA-1–binding site likely resides at +118 relative to the predicted transcriptional start.
To demonstrate that GATA-1 is bound to the cyclin D1 promoter in vivo, chromatin immunoprecipitation (ChIP) assays were performed using chromatin prepared from primary fetal liver–derived megakaryocytes. Quantitative PCR with primers that amplified the proximal promoter (encompassing the predicted GATA-1–binding site identified by the luciferase assays in Figure 2C) revealed that GATA-1 was indeed enriched on the cyclin D1 promoter (Figure 2D). Although the enrichment on this promoter was noted to be about 10-fold less than that observed on the GPIIb promoter, binding of GATA-1 to this site was specific, as primer sets designed to amplify upstream or downstream sequences within the cyclin D1 gene did not reveal significant binding. GATA-1 was not associated with the cyclin D1 promoter in G1ER cells, an erythroid cell line that harbors an inducible GATA-1:ER fusion protein (Figure S1).28 This latter observation suggests that GATA-1 controls cyclin D1 expressing specifically in maturing megakaryocytes.
As further evidence that GATA-1 regulates cyclin D1 expression in vivo, we noted a strong correlation between the size of primary megakaryocytes expressing wild-type or mutant GATA-1 proteins and the extent of cyclin D1 expression. Cyclin D1 expression levels were compared between G1KD cells transduced with wild-type GATA-1, GATA-1s, or the V205G mutant. Consistent with the reporter assay data, G1KD cells reconstituted with wild-type GATA-1 displayed increased cyclin D1 expression, whereas cells expressing GATA-1s failed to up-regulate cyclin D1 expression. In contrast, cyclin D1 mRNA was markedly elevated in G1KD cells transduced with V205G GATA-1 (22-fold increase) (Figure 2E). This latter observation is consistent with the strong repression of the D1 promoter mediated by FOG-1 (Figure 2B). Of interest, we previously reported that megakaryocytes that express GATA-1 V205G in place of wild-type GATA-1 grew to a very large size were highly polyploid.20 Taken together, these results demonstrate that GATA-1 binds and activates the cyclin D1 promoter in megakaryocytes during endomitosis and that both the N-terminus and the FOG-interaction domain of GATA-1 are required for precise regulation of cyclin D1 expression.
Overexpression of cyclin D1–Cdk4 rescues growth and polyploidization of GATA-1–deficient megakaryocytes
Given the role that cyclin D1 plays in cell cycle progression, we predicted that it would also contribute to megakaryocyte growth and polyploidization and, further, that restoration of its expression would rescue these defects in G1KD cells. To test this hypothesis, hematopoietic progenitor cells were isolated from E13.5 fetal livers of G1KD mice and infected with the MigR1 retrovirus harboring cyclin D1 and EGFP (via an IRES element) or GFP alone. Following 3 days of culture, megakaryocytes were isolated on a discontinuous BSA gradient and the GFP+ population was analyzed by flow cytometry. Surprisingly, overexpression of cyclin D1 resulted in a decreased extent of polyploidization when compared to control cells expressing GFP alone (data not shown). Furthermore, cells overexpressing cyclin D1 were slightly smaller than control cells (data not shown).
Since cyclin D1 functions in complex with Cdk4, we next assayed whether overexpression of both cyclin D1 and Cdk4 affected polyploidization and/or cell growth. The full-length Cdk4 cDNA was cloned into the pCS retrovirus, which coexpresses CD25 (via an IRES element). Protein and mRNA expression were confirmed for single- and double-infected cells for HA–cyclin D1 and HA-Cdk4 in retroviral producer cells and infected primary fetal liver cells by Western blot and real-time PCR, respectively (Figure S2). Following infection of G1KD fetal liver progenitors and ex vivo differentiation, the resulting megakaryocytes were enriched on a BSA gradient and then analyzed by flow cytometry, gating specifically on the GFP and CD25 double-positive cells. First, we repeated the overexpression of cyclin D1 in the presence of the empty pCS vector and confirmed the observation from the previous paragraph of a slight reduction in polyploidization and cell size compared to cells infected with both empty vectors MigR1 and pCS (Figure 3A-B,E-F). Conversely, simultaneous overexpression of cyclin D1 and Cdk4 resulted in a striking increase in DNA content and cell size, compared to control cells or cells expressing only Cdk4 or cyclin D1 alone (Figure 3B-D): 66.3% of the cyclin D1-Cdk4–overexpressing cells reached a DNA content of 8N or greater compared to 43% in the control population. This dramatic shift in DNA content was accompanied by an increase in cell size: cyclin D1–Cdk4 cells showed a mean FSC-A of 1275 versus 737 for control cells (Figure 3E,H). Of importance, overexpression of Cdk4 alone had little effect on either cell size or polyploidization (Figure 3C,G), indicating that overexpression of cyclin D1 requires Cdk4 to increase polyploidization and cell growth in differentiating megakaryocytes. Further, we noted a minimal effect on polyploidization when cyclin D1 and Cdk4 were overexpressed at comparable levels in wild-type cells cultured under similar conditions (Figure S3). This latter observation suggests that cyclin D–Cdk4 kinase activity is specifically limiting in GATA-1–deficient cells.
Ectopic expression of cyclin D1 and Cdk4 results in an increase in polyploidization and cell size of GATA-1 KD megakaryocytes. (A-D) Ploidy analysis of BSA gradient–purified GATA-1 KD megakaryocytes was performed using flow cytometry of DAPI-stained cells. Cells were doubly infected with the MigR1 containing EGFP and pCS containing hCD25. Cyclin D1 was cloned into MigR1 and Cdk4 was cloned into pCS allowing for double infection and expression of cyclin D1 alone, Cdk4 alone, or cyclin D1 and Cdk4. Double-infected cells were selected through gating of EGFP-positive and CD25 double-positive cells. Bars indicate the percentage of cells that have DNA content of 8N or greater. One representative of 3 experiments is shown. (E-H) Cell size analysis performed using flow cytometry of the infected cells described in panels A-D. Double-positive cells were analyzed using histograms of forward scatter–area to compare cell size. Values for the mean FSC-A are indicated for each population of doubly infected cells. One representative of 3 experiments is shown.
Ectopic expression of cyclin D1 and Cdk4 results in an increase in polyploidization and cell size of GATA-1 KD megakaryocytes. (A-D) Ploidy analysis of BSA gradient–purified GATA-1 KD megakaryocytes was performed using flow cytometry of DAPI-stained cells. Cells were doubly infected with the MigR1 containing EGFP and pCS containing hCD25. Cyclin D1 was cloned into MigR1 and Cdk4 was cloned into pCS allowing for double infection and expression of cyclin D1 alone, Cdk4 alone, or cyclin D1 and Cdk4. Double-infected cells were selected through gating of EGFP-positive and CD25 double-positive cells. Bars indicate the percentage of cells that have DNA content of 8N or greater. One representative of 3 experiments is shown. (E-H) Cell size analysis performed using flow cytometry of the infected cells described in panels A-D. Double-positive cells were analyzed using histograms of forward scatter–area to compare cell size. Values for the mean FSC-A are indicated for each population of doubly infected cells. One representative of 3 experiments is shown.
Increased polyploidization occurs independently of megakaryocyte terminal maturation
It remains controversial whether polyploidization and platelet production are inextricably linked during megakaryocyte differentiation. We noted that although expression of cyclin D1 and Cdk4 resulted in increased size and DNA content, terminal differentiation was not restored in the G1KD cells, as evidenced by a lack of proplatelet forms (data not shown). To address whether polyploidization impacts expression of genes associated with terminal maturation, we isolated mRNA from gradient-purified G1KD megakaryocytes engineered to express cyclin D1 and/or Cdk4 and performed qRT-PCR on a set of megakaryocyte-specific genes, including β1-tubulin, NF-E2, and platelet factor 4 (PF4). Each of these genes is dependent upon GATA-1 for expression and all show reduced expression in G1KD cells.20 Of note, the expression of β1-tubulin, NF-E2, and PF4 were unchanged in cells that overexpress cyclin D1 and Cdk4 (Figure 4). These data indicate that polyploidization can be augmented through the expression of cyclin D1–Cdk4 without affecting terminal differentiation.
Ectopic cyclin D1–Cdk4 does not rescue megakaryocyte terminal differentiation. mRNA was extracted from BSA gradient–purified megakaryocytes derived from wild-type fetal livers or G1KD fetal livers infected with cyclin D1 and Cdk4 encoding retroviruses (described in Figures 3 and 5). Expression levels of β1-tubulin, NF-E2, and PF4 are shown relative to wild-type expression levels. The mean ± 1 SD is shown for each study. As shown, overexpression of cyclin D1, Cdk4, or both does affect the expression of these genes.
Ectopic cyclin D1–Cdk4 does not rescue megakaryocyte terminal differentiation. mRNA was extracted from BSA gradient–purified megakaryocytes derived from wild-type fetal livers or G1KD fetal livers infected with cyclin D1 and Cdk4 encoding retroviruses (described in Figures 3 and 5). Expression levels of β1-tubulin, NF-E2, and PF4 are shown relative to wild-type expression levels. The mean ± 1 SD is shown for each study. As shown, overexpression of cyclin D1, Cdk4, or both does affect the expression of these genes.
Polyploidization is moderately reduced in megakaryocytes derived from cyclin D1 knockout mice
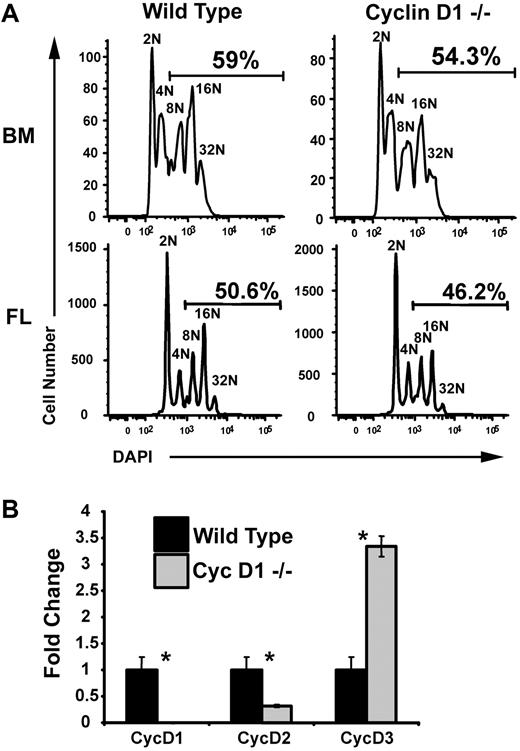
Given the dependence of polyploidization on cyclin D–Cdk4 activity, one might predict defects in megakaryopoiesis in cyclin D1 knockout mice. To test this, lineage-depleted hematopoietic progenitor cells were isolated from bone marrow of adult cyclin D1 knockout mice and fetal livers of cyclin D1–deficient embryos and grown in vitro. After 3 days of differentiation, the percentage of polyploid megakaryocytes from both the bone marrow and fetal liver were consistently, but not significantly, reduced compared to wild-type cells (Figure 5A). The lack of a more striking phenotype in the cyclin D1–deficient megakaryocytes may be due to compensation by other cyclin D proteins. To assay this possibility, we extracted mRNA from wild-type and cyclin D1−/− megakaryocytes, generated cDNA, and performed qRT-PCR. This analysis revealed that there was 3.5-fold increase in expression of cyclin D3 in cyclin D1–deficient megakaryocytes, in comparison to wild-type counterparts (Figure 5B). Therefore, the lack of a striking phenotype in the cyclin D1–deficient megakaryocytes is likely due to compensation by cyclin D3.
Loss of cyclin D1 alone does not significantly inhibit polyploidization. (A) Megakaryocytes were derived from wild-type and cyclin D1−/− bone marrow (BM) and fetal liver (FL). After enrichment on a BSA gradient, cells were stained with DAPI for DNA content and analyzed by flow cytometry. The percentages of cells with DNA content of 8N or more are shown. A reproducible reduction in polyploid cells was observed in the cyclin D1–deficient megakaryocytes. One representative of 2 experiments is shown. (B) Real-time quantitative PCR for cyclins D1, D2, and D3. mRNA was isolated from BSA gradient–purified megakaryocytes generated from wild-type and cyclin D1−/− bone marrow–derived megakaryocytes. Expression of cyclins D1, D2, and D3 were determined using primer sets specific for each transcript. Expression levels are shown relative to wild-type mRNA levels. For each study, the mean ± 1 SD is shown. Asterisks indicate P values determined by Student t test to be of .05 or less. One representative of 2 experiments, each in triplicate, is shown.
Loss of cyclin D1 alone does not significantly inhibit polyploidization. (A) Megakaryocytes were derived from wild-type and cyclin D1−/− bone marrow (BM) and fetal liver (FL). After enrichment on a BSA gradient, cells were stained with DAPI for DNA content and analyzed by flow cytometry. The percentages of cells with DNA content of 8N or more are shown. A reproducible reduction in polyploid cells was observed in the cyclin D1–deficient megakaryocytes. One representative of 2 experiments is shown. (B) Real-time quantitative PCR for cyclins D1, D2, and D3. mRNA was isolated from BSA gradient–purified megakaryocytes generated from wild-type and cyclin D1−/− bone marrow–derived megakaryocytes. Expression of cyclins D1, D2, and D3 were determined using primer sets specific for each transcript. Expression levels are shown relative to wild-type mRNA levels. For each study, the mean ± 1 SD is shown. Asterisks indicate P values determined by Student t test to be of .05 or less. One representative of 2 experiments, each in triplicate, is shown.
p16 inhibits polyploidization
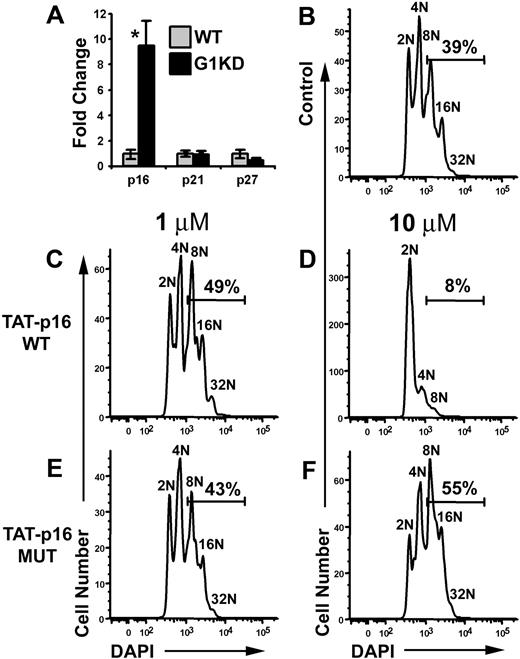
Since cyclin D1, but not Cdk4, was down-regulated in GATA-1 KD megakaryocytes, we investigated why overexpression of both cyclin D1 and Cdk4 was required to increase polyploidization and cell size. One possibility was that levels of cell cycle inhibitors might be elevated. Thus, we used qRT-PCR to gauge the expression levels of G1 inhibitors p16, p21, and p27. This analysis revealed that p16 was up-regulated nearly 10-fold in G1KD megakaryocytes compared to WT (Figure 6A). In contrast, p21 and p27 transcript levels were not significantly affected by the absence of GATA-1. This finding suggested to us that perhaps p16 restricts the endomitotic cell cycle.
p16 inhibits polyploidization. (A) qRT-PCR of mRNA isolated from wild-type and GATA-1 KD megakaryocytes for p16, p21, and p27. GATA-1 KD mRNA levels are shown relative to wild-type expression. P values determined using Student t test. Asterisk indicates P ≤ .01. One representative of 2 experiments, each in triplicate, is shown. For each study, the mean ± 1 SD is shown. (B) Purified, untreated wild-type megakaryocytes were differentiated for 3 days and stained with DAPI. (C,E) Wild-type megakaryocytes were treated with either wild-type TAT-p16 fusion peptide or a charge-matched mutant TAT-p16 peptide at a concentration of 1 μM. Peptide was added 3 times during the second day of differentiation due to instability of the TAT peptides. Cells were fixed and stained with DAPI on day 3 of differentiation. (D,F) Wild-type megakaryocytes were treated with the peptides used in panels C and E at a concentration of 10 μM. Cells were fixed and stained with DAPI on day 3 of differentiation. Gates indicate the percentage of cells at a ploidy class of 8N or greater. One representative of 2 experiments is shown.
p16 inhibits polyploidization. (A) qRT-PCR of mRNA isolated from wild-type and GATA-1 KD megakaryocytes for p16, p21, and p27. GATA-1 KD mRNA levels are shown relative to wild-type expression. P values determined using Student t test. Asterisk indicates P ≤ .01. One representative of 2 experiments, each in triplicate, is shown. For each study, the mean ± 1 SD is shown. (B) Purified, untreated wild-type megakaryocytes were differentiated for 3 days and stained with DAPI. (C,E) Wild-type megakaryocytes were treated with either wild-type TAT-p16 fusion peptide or a charge-matched mutant TAT-p16 peptide at a concentration of 1 μM. Peptide was added 3 times during the second day of differentiation due to instability of the TAT peptides. Cells were fixed and stained with DAPI on day 3 of differentiation. (D,F) Wild-type megakaryocytes were treated with the peptides used in panels C and E at a concentration of 10 μM. Cells were fixed and stained with DAPI on day 3 of differentiation. Gates indicate the percentage of cells at a ploidy class of 8N or greater. One representative of 2 experiments is shown.
To determine the role of p16 in the regulation of polyploidization, we increased the level of p16 in wild-type megakaryocytes by treating cells with a TAT-p16 fusion polypeptide.29 Wild-type fetal liver progenitors were cultured ex vivo in the presence of TPO to generate megakaryocytes. After one day of culture, TAT-p16 or TAT-p16 MUT, a charge-matched p16 mutant that fails to interact with Cdk4,29 was added to the cells at final concentrations of 1 μM or 10 μM, and then incubated for an additional day. Next, cells were stained with DAPI and CD41, and the DNA content of the megakaryocytes was determined by flow cytometry. Treatment with 10 μM TAT-p16 dramatically inhibited polyploidization, with only 8% of the cells reaching a ploidy class of 8N or greater compared to 39% in untreated control cultures (Figure 6B,D). In contrast, addition of TAT-p16 MUT at either 1- or 10-μM concentrations had no effect on the ability of wild-type progenitors to generate polyploid megakaryocytes (Figure 6E-F). Similarly, treatment of wild-type cells with TAT-p16 at 1 μM did not affect the percentage of cells entering endomitosis (Figure 6C). These data indicate that inhibition of cyclin D–Cdk4/6 activity leads to a striking block in the ability of megakaryocytes to undergo endomitosis and suggest that the reduction in polyploidization in G1KD cells may be due to the combination of decreased cyclin D1 and increased p16 expression. Further, these data suggest that there is a strict requirement for cyclin D-Cdk4 for polyploidization that cannot be compensated by cyclin E-Cdk2.
Discussion
Cyclin D and Cdk4 regulate size and polyploidization in differentiating megakaryocytes
Of the G1 regulators, several studies have established that both cyclin D1 and cyclin D3 are implicated in megakaryocyte polyploidization.30–32 For example, when the F36 megakaryocytic cell line was differentiated upon exposure to TPO, a robust up-regulation of both cyclins D1 and D3 was observed.33 Furthermore, human megakaryocytes, derived from CD34+ hematopoietic progenitors cultured in the presence of TPO, displayed increased expression of cyclin D1 and Cdk4 during their maturation.34 Primary murine megakaryocytes also express both cyclins D1 and D3, but have been reported not to express cyclin D2.30,31 Of interest, human F36 cells were shown to express cyclin D2 both at the mRNA and protein levels.33 Consistent with a role for D-type cyclins in endomitosis, transgenic mice engineered to overexpress cyclin D3 harbor megakaryocytes that undergo excessive rounds of polyploidization.32 Megakaryocytes in transgenic mice engineered to overexpress cyclin D1 also showed an increased ploidy, although this change was not as striking as that in the cyclin D3 transgenics, possibly due to modest expression of the cyclin D1 transgene.35 Several other lines of evidence suggest that cyclins D1 and D3 contribute to endomitosis. Overexpression of cyclin D1 in the DAMI megakaryocytic cell line led to proliferation arrest and, in the presence of PMA, enhanced polyploidization.36 Recently, two reports have shown that the mTOR pathway plays a critical role in both growth and polyploidization of megakaryocytes, likely in part by regulating expression and localization of cyclin D3.37,38 Thus, it is likely that one function of the D-type cyclins in megakaryocytes is to promote cell cycle progression in a manner analogous to its role in dividing cells. However, since megakaryocytes bypass the late stages of mitosis, the end product is a cell with increased DNA content as opposed to an increase in the numbers of descendants.
Apart from the increase in DNA content, megakaryocytes are noted for becoming very large. A role for cyclin D–Cdk4 in accumulation of cellular mass has been previously suggested. In Drosophila, progression through the cell cycle can occur in the absence of Cdk4, but growth of cells is reduced, suggesting that Cdk4 kinase activity has an important function outside of the cell cycle.39 In addition, elegant genetic studies have shown that cyclin D–Cdk4 primarily functions to promote cellular growth and leads to differential effects in distinct cell types.40 For example, ectopic expression of cyclin D and Cdk4, but not cyclin D alone, in endoreplicating salivary glands caused excessive DNA replication and hypertrophy. In differentiating eye cells, cyclin D–Cdk4 promoted cell enlargement, while in proliferating wing imaginal cells, the kinase complex led to accelerated cell division. Taken together, these results demonstrate that cyclin D–Cdk4 has growth-promoting activity in addition to its role in cell cycle progression. The mechanism of cellular growth in flies appears to include the induction of Hif-1 prolyl hydroxylase activity and to require the mitochondrial ribosomal protein, mRpL12.41,42 Although we observed a dramatic increase in cellular size and DNA content following overexpression of cyclin D1–Cdk4 in G1KD cells, we did not observe a difference in expression of any Hif-1 downstream targets, including VEGF and Flt1 (data not shown). This suggests that cyclin D1–Cdk4 may be acting through a unique set of target genes in megakaryocytes.
The role and regulation of cell-cycle inhibitors in endomitosis
Megakaryocytes express p21 and p27 during differentiation,43 with the highest levels detected within megakaryocytes with high ploidy.44 While overexpression of p21 was found to inhibit polyploidization, loss of p21 either had no effect44 or was associated with an increased state of megakaryocyte polyploidization.45 In contrast, less is known about the role of INK4 family members in endomitosis. One study revealed that p15ink4b expression was up-regulated in CD41+ cells generated ex vivo from human CD34+ cells,46 while another reported that p16 decreased upon megakaryocytic differentiation.47 Our observation that GATA-1–deficient megakaryocytes express nearly 10-fold less p16 suggests that GATA-1 may directly repress p16 transcription in the course of megakaryocyte maturation. In support of this hypothesis, we have found that ectopic expression of GATA-1 in GATA-1–deficient G1ME cells induces terminal differentiation and polyploidization that is accompanied by a decrease in p16 expression (Z. Huang and J.D.C., unpublished observations, December, 2006). With respect to the function, we show that p16 potently inhibits endomitosis. This is a consequence of inhibition of the cyclin D–Cdk4/6 complex, as a mutant in p16 that fails to interact with Cdk4 did not block polyploidization. Thus, although cyclin D–Cdk4/6 is largely dispensable for progression of proliferating cells, its activity is specifically required for endomitosis and cannot be compensated by cyclin E–Cdk2.
GATA-1 regulates polyploidization and terminal maturation through different target genes
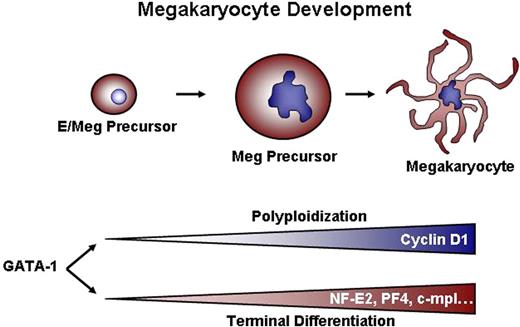
Once a megakaryocyte precursor commits to terminal differentiation and ceases dividing, multiple changes occur, including alterations in the expression of cell cycle regulators, such that the cell can progress through mitosis without cell division. In addition to cell cycle changes, megakaryocyte-specific genes are expressed, such as PF4, β1-tubulin, and NF-E2, that are necessary for platelet synthesis and function. Although DNA content and cellular mass were increased in GATA-1–deficient cells in response to overexpression of cyclin D1 and Cdk4, other deficiencies in megakaryocyte terminal maturation were not rescued, as evidenced by a lack of proplatelet production and a failure to up-regulate megakaryocyte-specific genes (Figure 4 and data not shown). These results suggest that GATA-1 regulates the process of polyploidization independently of terminal differentiation, likely by controlling a distinct set of target genes (Figure 7).
Dual requirements for GATA-1–dependent megakaryocyte development. We conclude that GATA-1 controls 2 sets of genes: One group, including cyclin D1, regulates growth and polyploidization. A second class of targets, which includes genes such as NF-E2, PF4, and c-mpl, drives terminal maturation and platelet biosynthesis. The observation that ectopic cyclin D1–cdk4 can cause increased size and DNA content without concomitant expression of NF-E2, PF-4, and c-mpl indicates that the 2 processes (ie, polyploidization and terminal maturation) can be uncoupled from one another.
Dual requirements for GATA-1–dependent megakaryocyte development. We conclude that GATA-1 controls 2 sets of genes: One group, including cyclin D1, regulates growth and polyploidization. A second class of targets, which includes genes such as NF-E2, PF4, and c-mpl, drives terminal maturation and platelet biosynthesis. The observation that ectopic cyclin D1–cdk4 can cause increased size and DNA content without concomitant expression of NF-E2, PF-4, and c-mpl indicates that the 2 processes (ie, polyploidization and terminal maturation) can be uncoupled from one another.
GATA1 regulates cyclin D1 expression in megakaryocytes, but not erythroid cells
In this study, we show that GATA-1 binds and regulates the cyclin D1 promoter in megakaryocytes, but not erythroid cells. Since GATA-1 is expressed in both lineages, this begs the question of how it can bind differentially to the same promoter. It is likely that the binding of GATA-1 to DNA is modulated by its association with different protein partners. For example, GATA-1 has been shown to synergize with Ets transcription factors bound to adjacent sites on megakaryocyte promoters to regulate gene expression.48 Of note, we identified an Ets-binding site 25 base pairs from the GATA-binding site at +118. Another attractive candidate for promoting the binding of GATA-1 to the cyclin D1 promoter in the megakaryocyte lineage is RUNX1, which interacts with GATA-1 and is up-regulated during megakaryocyte, but not erythroid, differentiation.49 RUNX1 has been reported to induce cyclin D3 expression in myeloid and lymphoid cell lines and also bind the cyclin D3 promoter.50 Of interest, RUNX1-deficient megakaryocytes are also deficient in polyploidization.51 Thus, an attractive model is that GATA-1 and RUNX1 cooperate with one another to execute the megakaryocyte program of endomitosis and growth.
Human GATA1 mutations have differential effects on megakaryocyte size and ploidy
Mutations in GATA1 have been found associated with multiple hematopoietic diseases. For example, humans who inherit mutations in GATA1 that disrupt the interaction between GATA-1 and its cofactor FOG-1 suffer from thrombocytopenia and anemia.52 In particular, patients with the G208S alteration in GATA-1 accumulate large, immature megakaryocytes within their bone marrow and suffer from macrothrombocytopenia.53 Similarly, expression of the V205G mutant of GATA-1 in place of the wild-type protein also leads to the generation of large, abnormal megakaryocytes both in vivo and in vitro.20,54 As shown in this report, these latter mutant megakaryocytes express cyclin D1 at more than 20 times the amount found in GATA-1–deficient counterparts. These data point to a striking correlation between cyclin D1 expression levels and cellular size, where high levels of cyclin D1 are associated with abnormally large cells. Based on the transient reporter assays and the in vivo expression data, we predict that FOG-1 acts as a rheostat to maintain proper expression of cyclin D1 in maturing megakaryocytes.
A recent study has described a family with a germ-line mutation in GATA1 that results in the exclusive synthesis of GATA-1s.55 These patients display macrocytic anemia, but have normal platelet counts; however, the patients harbor a preponderance of micromegakaryocytes in their bone marrow. This finding is consistent with our observations that GATA-1s cannot drive expression of the cyclin D1 promoter and that G1KD megakaryocytes reconstituted with the GATA-1s cDNA are smaller than normal and also fail to express cyclin D1 mRNA above that detected in GATA-1–deficient cells (Figure 2E). Taken together, these observations made in human patients and in mouse models argue that one of the main functions of GATA-1 is to promote expression of cyclin D1 in committed megakaryocytes to promote both increased cell growth and DNA content.
The online version of this manuscript contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported, in part, by a grant from the Elsa U. Pardee Foundation (J.D.C.), and NIH grants R01 CA101774 and DK61464. J.D.C. is a Scholar of the Leukemia and Lymphoma Society.
The authors thank Dr Barbara Kee for the pCS retrovirus and the cyclin D1 cDNA, Dr Hiroaki Kiyokawa for the Cdk4 cDNA, Dr Wei Du for the cyclin D1 knockout mice, and Dr Stuart Orkin and Dr Paresh Vyas for the G1KD mice. Additional thanks to Dr Harinder Singh and Dr Sandeep Gurbuxani for helpful suggestions.
National Institutes of Health
Authorship
Contribution: A.G.M. designed and performed the experiments, interpreted the data, and wrote the paper; L.P. designed and performed the experiments, and interpreted the data; M.P. designed experiments, interpreted the data, and assisted with the paper; S.F.D. contributed valuable reagents and expertise; G.A.B. designed experiments, interpreted the data, and assisted with the paper; J.D.C. designed experiments, interpreted the data, and assisted with the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: John Crispino, Northwestern University, Division of Hematology/Oncology, 303 East Superior St, Lurie 5-113, Chicago, IL 60611; e-mail: j-crispino@northwestern.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal