Abstract
The Runt domain transcription factor AML1/RUNX1 is essential for the generation of hematopoietic stem cells and is the most frequent target of chromosomal translocations associated with leukemia. Here, we present a new AML1 translocation found in a patient with acute myeloid leukemia M4 with t(8;21)(q24;q22) at the time of relapse. This translocation generated an in-frame chimeric gene consisting of the N-terminal portion of AML1, retaining the Runt domain, fused to the entire length of TRPS1 on the C-terminus. TRPS1 encodes a putative multitype zinc finger (ZF) protein containing 9 C2H2 type ZFs and 1 GATA type ZF. AML1-TRPS1 stimulated proliferation of hematopoietic colony-forming cells and repressed the transcriptional activity of AML1 and GATA-1 by 2 different mechanisms: competition at their cognate DNA-binding sites and physical sequestrations of AML1 and GATA-1, suggesting that simultaneous deregulation of AML1 and GATA factors constitutes a basis for leukemogenesis.
Introduction
AML1/RUNX1 encodes the DNA-binding α subunit of the heterodimeric transcription factor PEBP2/CBF, which interacts with the partner β subunit (PEBP2β/CBFβ) through its evolutionarily conserved Runt domain.1 AML1 is one of the most frequently mutated genes in human leukemia,2–4 and was originally identified as a gene on chromosome 21 involved in t(8;21)(q22:q22).5 To date, 11 AML1-related translocations are known that produce chimeric proteins such as AML1-MTG8/ETO in t(8;21), AML1-EVI1 in t(3;21), and TEL-AML1 in t(12;21).2,6,7 All these AML1 chimeric proteins retain the Runt domain and inhibit transcriptional activity of wild-type AML1 in a dominant-negative manner.2–4 However, the functional contributions of partner moieties in leukemogenesis remain largely undetermined. Here, we report a new AML1 translocation, t(8;21)(q24;q22), in a patient with acute myeloid leukemia (AML), and present results from functional analyses of the chimeric protein AML1-TRPS1.
Patient, materials, and methods
Patient profile
A 56-year-old Japanese man was diagnosed with AML M4 with a normal karyotype in October 1997. He was treated with idarubicin and cytarabine, followed by postremission therapy according to the Japan Adult Leukemia Study Group (JALSG) AML97.8 In July 1999, his marrow showed 55.6% blasts with t(8;21)(q24;q22) at relapse. Bone marrow cells at diagnosis and relapse showed the similar morphology and immunophenotypes positive for CD13, CD33, CD4, and HLA-DR, but negative for CD34, which were consistent with AML M4.9 The study was approved by the Institutional Review Board of Kumamoto University School of Medicine, Japan, and informed consent was obtained from the patient, according to the Declaration of Helsinki.
Molecular cloning
The fusion partner gene was cloned by long-distance 3′ rapid amplification of cDNA ends (RACE) using the SMART RACE kit (Clontech Labs, Mountain View, CA).
Plasmid constructions
EMSA
Electrophoretic mobility shift assays (EMSAs) were performed using biotin-labeled probes containing the AML112 or GATA factor-binding sites.13 The specificity of the probes are shown in Figure S1, available on the Blood website (see the Supplemental Figure link at the top of the online article). Whole-cell extracts of COS7 cells (1 × 106) transfected with pEF-Bos expression vectors were subjected to the assay.
Transcription assay
The luciferase reporter constructs pBXH2-LTR-luc and pRBGP3-MαP were used to assay the transcriptional activities mediated by AML1 and GATA-1, respectively. pBXH2-LTR-luc contains the long terminal repeat (LTR) of the BXH2 retrovirus11 that includes a functional AML1 site, while the mouse α-1 globin gene promoter in pRBGP3-MαP14 contains a functional GATA site. The specificity of AML1 and GATA site-dependent activation of each reporter is shown in Figure S2.
Retroviral transduction and CFU-C assay
Retroviral transduction, sorting by FACS Vantage (Becton Dickinson, San Jose, CA), and colony-forming unit-culture (CFU-C) assay were performed as previously described.11
Immunoprecipitation
Immunoprecipitation of FLAG-tagged fusion proteins was performed according to the manufacturer's protocol (Sigma, St Louis, MO).
Results and discussion
Spectral karyotyping of the marrow cells at relapse confirmed a balanced translocation between chromosomes 8 and 21 (Figure 1A). Fluorescence in situ hybridization using cosmid clones containing the AML1 gene showed that this late-appearing translocation involved the AML1 gene (Figure 1B).15 We cloned the fusion partner gene with AML1 exon 6 by using 3′ RACE (Figure 1C). The t(8;21)(q24;q22) generated an in-frame chimeric gene that encoded a fusion protein consisting of the N-terminal portion of AML1, retaining the Runt domain but lacking its C-terminal transactivation domain, fused to the entire TRPS1/GC79 (Figure 1D). TRPS1 on chromosome 8q24.1 encodes a nuclear transcription factor with 10 zinc finger (ZF) domains, including a single GATA-type ZF.13,16,17 The gene is widely expressed in human tissues, including prostate, testis, ovary, kidney, lung, mammary gland, and hematopoietic cells.16,17 TRPS1 has been identified as a disease gene for tricho-rhino-phalangeal syndrome (TRPS) type I and type III (MIM190350 and MIM190351), which is a dominantly inherited disease characterized by craniofacial and skeletal abnormalities.16
Cloning of the AML1-TRPS1 that stimulates proliferation. (A) Spectral karyotyping (SKY) showed the balance translocation between chromosomes 8 and 21 (arrowheads). Representative SKY of chromosomes 8 and 21 is indicated as reverse DAPI (left) and SKY (right). (B) Fluorescence in situ hybridization using cosmid clones covering the whole AML1 gene showed that t(8;21)(q24;q22) involved the AML1 gene. Nearly half (55 [48.2%] of 114) of the cells had 3 signals of the AML1 gene. Images were acquired within an SD200 spectral imaging system (Applied Spectral Imaging, Migdal Haemek, Israel) attached to an Optiphot-2 epifluorescence microscope (Nikon, Tokyo, Japan) through a Splan Apo 100×/1.4 NA oil objective lens (Olympus, Tokyo, Japan) and analyzed with SkyView software (Applied Spectral Imaging). (C) The fusion partner gene with AML1 was cloned by long-distance 3′ RACE with 5′ primers on the AML1 exon 5 (AML1S5: 5′-cacagtggatgggccccgagaacctcg-3′) or exon 6 (AML1S6: 5′-tgcggcgcacagccatgagggtcagc-3′). The arrowhead indicates AML1a; the arrow shows the fusion gene. Lane 1, λ/HindIII DNA marker; lane 2, Human placental DNA; lane 3, AML cells (AML1S5); lane 4, AML cells (AML1S6). (D) N-terminal portion of AML1 retaining the Runt domain fused to the whole of TRPS1. An in-frame fusion gene consists of exon 6 of AML1 and TRPS1. In addition, sequencing analyses of subclones showed another transcript between exon 5 of AML1 and TRPS1, indicating skipping of exon 6 of AML1 and generating another in-frame fusion gene. The reciprocal TRPS1-AML1 transcript was not detected in the presenting case. (E) Structures of retrovirus constructs for control (MIG_mock) or AML1-TRPS1 (MIG_AT). AML1-TRPS1 was inserted into the indicated position of the plasmid MIG with internal ribosomal entry site (IRES) and the enhanced green fluorescent protein (EGFP) gene. (F) Schematic depiction of the CFU-C assay. Fetal liver cells from embryonic day–14.5 (E14.5) mouse embryo were infected with the MIG vector. EGFP-positive cells were sorted and subjected to the CFU-C assay, supplemented with interleukin-3, stem cell factor, erythropoietin, and granulocyte colony-stimulating factor, as described previously.11 (G) The number of all kinds of colony (total), granulocyte-macrophage (GM), and macrophage (M) are shown. Error bars indicate standard deviations for 3 independent experiments. Differences between AML1-TRPS1 (AT) and control (mock) transfectants were statistically significant (*P < .001, unpaired student t test) in all 3 categories.
Cloning of the AML1-TRPS1 that stimulates proliferation. (A) Spectral karyotyping (SKY) showed the balance translocation between chromosomes 8 and 21 (arrowheads). Representative SKY of chromosomes 8 and 21 is indicated as reverse DAPI (left) and SKY (right). (B) Fluorescence in situ hybridization using cosmid clones covering the whole AML1 gene showed that t(8;21)(q24;q22) involved the AML1 gene. Nearly half (55 [48.2%] of 114) of the cells had 3 signals of the AML1 gene. Images were acquired within an SD200 spectral imaging system (Applied Spectral Imaging, Migdal Haemek, Israel) attached to an Optiphot-2 epifluorescence microscope (Nikon, Tokyo, Japan) through a Splan Apo 100×/1.4 NA oil objective lens (Olympus, Tokyo, Japan) and analyzed with SkyView software (Applied Spectral Imaging). (C) The fusion partner gene with AML1 was cloned by long-distance 3′ RACE with 5′ primers on the AML1 exon 5 (AML1S5: 5′-cacagtggatgggccccgagaacctcg-3′) or exon 6 (AML1S6: 5′-tgcggcgcacagccatgagggtcagc-3′). The arrowhead indicates AML1a; the arrow shows the fusion gene. Lane 1, λ/HindIII DNA marker; lane 2, Human placental DNA; lane 3, AML cells (AML1S5); lane 4, AML cells (AML1S6). (D) N-terminal portion of AML1 retaining the Runt domain fused to the whole of TRPS1. An in-frame fusion gene consists of exon 6 of AML1 and TRPS1. In addition, sequencing analyses of subclones showed another transcript between exon 5 of AML1 and TRPS1, indicating skipping of exon 6 of AML1 and generating another in-frame fusion gene. The reciprocal TRPS1-AML1 transcript was not detected in the presenting case. (E) Structures of retrovirus constructs for control (MIG_mock) or AML1-TRPS1 (MIG_AT). AML1-TRPS1 was inserted into the indicated position of the plasmid MIG with internal ribosomal entry site (IRES) and the enhanced green fluorescent protein (EGFP) gene. (F) Schematic depiction of the CFU-C assay. Fetal liver cells from embryonic day–14.5 (E14.5) mouse embryo were infected with the MIG vector. EGFP-positive cells were sorted and subjected to the CFU-C assay, supplemented with interleukin-3, stem cell factor, erythropoietin, and granulocyte colony-stimulating factor, as described previously.11 (G) The number of all kinds of colony (total), granulocyte-macrophage (GM), and macrophage (M) are shown. Error bars indicate standard deviations for 3 independent experiments. Differences between AML1-TRPS1 (AT) and control (mock) transfectants were statistically significant (*P < .001, unpaired student t test) in all 3 categories.
To investigate whether the fusion gene AML1-TRPS1 affects proliferation and differentiation of hematopoietic cells, we introduced AML1-TRPS1 into mouse fetal liver cells and carried out a CFU-C assay (Figure 1E-F). As shown in Figure 1G, AML1-TRPS1 transfectants gave rise to a higher number of colonies than did the control transfectants, indicating that AML1-TRPS1 stimulated the proliferation of immature hematopoietic cells.
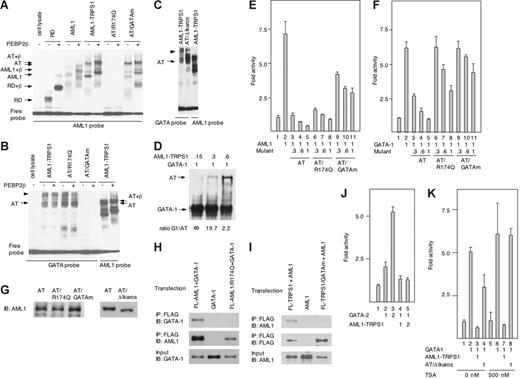
Since AML1-TRPS1 contains the Runt and GATA ZF domains, we evaluated the DNA-binding ability of AML1-TRPS1 to both AML1 and GATA sites. AML1-TRPS1 bound to the AML1 site and showed heterodimerization activity with the PEBP2β subunit (Figure 2A). AML1-TRPS1 also showed DNA-binding to the GATA site (Figure 2B). However, it did not form a heterodimer with the β subunit at this site. It is interesting to note that AML1-TRPS1 shifted as a doublet of bands with the GATA probe, and the upper band (arrowhead in Figure 2B) was much slower than that of AML1-TRPS1/PEBP2β complex bound to the AML1 probe (right end lane). Since the Ikaros-like ZF in the C-terminal TRPS1 moiety is known to serve as a domain to form homo- or heterodimers of factors containing this motif,18 the slower migrating band might indicate the formation of homodimer by AML1-TRPS1. In fact, the deletion of the Ikaros-like ZF resulted in a significant decrease in the intensity of the upper band, accompanied by an increase in that of the lower band (Figure 2C).
Functional analysis of AML1-TRPS1. (A-B) AML1-TRPS1 and their mutants were subjected to EMSA in the presence (+) or absence (−) of PEBP2β/CBFβ. RD and AT indicate the Runt domain and AML1-TRPS1, respectively. RD and AML1 served as positive controls. The position of the indicated factor or complex with DNA is shown. Mutant constructs of AML1-TRPS1 were generated by PCR-based mutagenesis, using the following primers: AT/R174Q, 5′-gtggatgggccccaagaacctcgaagaca-3′; and AT/GATAm, 5′-ggatatgtaggcaacgcgggtggcctctacc -3′. (C) AML1-TRPS1 seems to form a homodimer through the Ikaros-like ZF domain. The deletion of Ikaros-like ZF (AT/ΔIKAROS) resulted in a decrease in the upper band (arrowhead), which might indicate the homodimer of AML1-TRPS1, and an increase in the lower band (AT, AML1-TRPS1 monomer). The AT/ΔIKAROS mutant was made by PCR-based mutagenesis using 5′-gaagtactcaagatgaactttcaacataatgtgrgcactgtggc-3′ as a primer. (D) A fixed amount of GATA-1 (1 U) and increasing relative amounts (0.15, 0.3 and 0.6 U) of AML1-TRPS1 were subjected to EMSA. Relative amounts of expressed proteins were evaluated by Western blotting. (E-F) Wild-type AML1, GATA-1, AML1-TRPS1, or its mutants were cotransfected with pBXH2-LTR-luc or pRBGP3-MαP reporter at varying relative doses, as indicated, into NIH3T3 cells by the nonliposomal transfection reagent FuGENE6 (Roche Applied Science, Basel, Switzerland). Luciferase activities are expressed as fold changes relative to the control transfected with the backbone expression vector alone. The total input of plasmid DNAs was kept constant (0.6 μg) by supplementing appropriate amounts of the backbone pEF-Bos plasmid so as to avoid potential artifacts due to uneven overall DNA dosages. Error bars indicate standard deviations for 3 independent experiments. (G) Expression levels of AML1-TRPS1 and its mutants were comparable with each other. COS7 cells were transfected with expression plasmids for the indicated genes, and whole-cell extracts were prepared 48 hours after transfection and subjected to Western blotting using rabbit polyclonal anti-AML1 antibody (Active Motif, Carlsbad, CA). (H-I) The Runt domain of AML1 physically interacted with GATA-1 (H), while GATA ZF of TRPS1 interacted with AML1 (I) in immunoprecipitation (IP) assays. COS7 cells were cotransfected with pEF-Bos expression vectors for FLAG-tagged AML1 (FL-AML1), FLAG-tagged AML1 mutant R174Q (FL-AML1/R174Q), and GATA-1, or FLAG-tagged TRPS1 (FL-TRPS1), FLAG-tagged TRPS1 GATA ZF mutant (FL-TRPS1/GATAm), and AML1, as indicated. Cell lysates were immunoprecipitated using anti-FLAG antibody (M2 monoclonal antibody; Sigma). Immunoprecipitates were detected by immunoblotting (IB) using rat monoclonal anti-GATA1 antibody (N6; Santa Cruz Biotechnology, Santa Cruz, CA), rabbit polyclonal anti-AML1 antibody (Active Motif), or anti-FLAG antibody. (J) AML1-TRPS1 inhibits GATA-2–mediated transcription. GATA-2 and AML1-TRPS1 were cotransfected with MαP reporter at the indicated doses, into HL-60 cells. Luciferase activity is expressed as fold changes relative to the control. (K) HDAC inhibitor TSA did not greatly affect the repression mediated by intact AML1-TRPS1, but dramatically relieved that of AT/Δ Ikaros. GATA-1, AML1-TRPS1, or AT/ΔIKAROS were cotransfected into NIH3T3 cells with the MαP reporter at the indicated doses. Cells were treated with TSA (500 nM) for 36 hours from 12 hours after transfection. Luciferase activity is expressed as fold changes relative to the control. The results represent 3 independent experiments.
Functional analysis of AML1-TRPS1. (A-B) AML1-TRPS1 and their mutants were subjected to EMSA in the presence (+) or absence (−) of PEBP2β/CBFβ. RD and AT indicate the Runt domain and AML1-TRPS1, respectively. RD and AML1 served as positive controls. The position of the indicated factor or complex with DNA is shown. Mutant constructs of AML1-TRPS1 were generated by PCR-based mutagenesis, using the following primers: AT/R174Q, 5′-gtggatgggccccaagaacctcgaagaca-3′; and AT/GATAm, 5′-ggatatgtaggcaacgcgggtggcctctacc -3′. (C) AML1-TRPS1 seems to form a homodimer through the Ikaros-like ZF domain. The deletion of Ikaros-like ZF (AT/ΔIKAROS) resulted in a decrease in the upper band (arrowhead), which might indicate the homodimer of AML1-TRPS1, and an increase in the lower band (AT, AML1-TRPS1 monomer). The AT/ΔIKAROS mutant was made by PCR-based mutagenesis using 5′-gaagtactcaagatgaactttcaacataatgtgrgcactgtggc-3′ as a primer. (D) A fixed amount of GATA-1 (1 U) and increasing relative amounts (0.15, 0.3 and 0.6 U) of AML1-TRPS1 were subjected to EMSA. Relative amounts of expressed proteins were evaluated by Western blotting. (E-F) Wild-type AML1, GATA-1, AML1-TRPS1, or its mutants were cotransfected with pBXH2-LTR-luc or pRBGP3-MαP reporter at varying relative doses, as indicated, into NIH3T3 cells by the nonliposomal transfection reagent FuGENE6 (Roche Applied Science, Basel, Switzerland). Luciferase activities are expressed as fold changes relative to the control transfected with the backbone expression vector alone. The total input of plasmid DNAs was kept constant (0.6 μg) by supplementing appropriate amounts of the backbone pEF-Bos plasmid so as to avoid potential artifacts due to uneven overall DNA dosages. Error bars indicate standard deviations for 3 independent experiments. (G) Expression levels of AML1-TRPS1 and its mutants were comparable with each other. COS7 cells were transfected with expression plasmids for the indicated genes, and whole-cell extracts were prepared 48 hours after transfection and subjected to Western blotting using rabbit polyclonal anti-AML1 antibody (Active Motif, Carlsbad, CA). (H-I) The Runt domain of AML1 physically interacted with GATA-1 (H), while GATA ZF of TRPS1 interacted with AML1 (I) in immunoprecipitation (IP) assays. COS7 cells were cotransfected with pEF-Bos expression vectors for FLAG-tagged AML1 (FL-AML1), FLAG-tagged AML1 mutant R174Q (FL-AML1/R174Q), and GATA-1, or FLAG-tagged TRPS1 (FL-TRPS1), FLAG-tagged TRPS1 GATA ZF mutant (FL-TRPS1/GATAm), and AML1, as indicated. Cell lysates were immunoprecipitated using anti-FLAG antibody (M2 monoclonal antibody; Sigma). Immunoprecipitates were detected by immunoblotting (IB) using rat monoclonal anti-GATA1 antibody (N6; Santa Cruz Biotechnology, Santa Cruz, CA), rabbit polyclonal anti-AML1 antibody (Active Motif), or anti-FLAG antibody. (J) AML1-TRPS1 inhibits GATA-2–mediated transcription. GATA-2 and AML1-TRPS1 were cotransfected with MαP reporter at the indicated doses, into HL-60 cells. Luciferase activity is expressed as fold changes relative to the control. (K) HDAC inhibitor TSA did not greatly affect the repression mediated by intact AML1-TRPS1, but dramatically relieved that of AT/Δ Ikaros. GATA-1, AML1-TRPS1, or AT/ΔIKAROS were cotransfected into NIH3T3 cells with the MαP reporter at the indicated doses. Cells were treated with TSA (500 nM) for 36 hours from 12 hours after transfection. Luciferase activity is expressed as fold changes relative to the control. The results represent 3 independent experiments.
To assess the effect of AML1-TRPS1 on transcription, we next performed a reporter assay. While AML1 and GATA-1 activated BXH2-LTR and MαP reporters, respectively, AML1-TRPS1 inhibited transactivation activity of both AML1 and GATA-1 in a dose-dependent manner (Figure 2E-F; bars 3-5). AT/R174Q has an amino acid substitution in the Runt domain and resulted in the loss of AML1 site-binding activity (Figure 2A).19 AT/R174Q showed reduced ability to suppress wild-type AML1 in cotransfection assays (Figure 2E; bars 6-8 vs 3-5), suggesting that AML1-TRPS1 inhibits wild-type AML1 transactivation through DNA binding of its Runt domain. Likewise, AT/GATAm lost its DNA-binding function for the GATA site (Figure 2B) due to changes in 2 amino acids that contribute to its structural integrity,13 and demonstrated weaker inhibition of wild-type GATA-1 transactivation than did AML1-TRPS1 (Figure 2F; bars 9-11 vs 3-5). As the DNA-binding affinity of AML1-TRPS1 was comparable with that of GATA-1 (Figure 2D), this competition for GATA-binding sites is considered to play a significant role in transcriptional suppression mediated by AML1-TRPS1. However, these DNA-binding mutants still retained significant repressive activity (compare bars 2, 5, and 8 in Figure 2E and bars 2, 5, and 11 in Figure 2F), and AT/R174Q and AT/GATAm even showed decreased suppression in the MαP and BXH2-LTR reporter assays, respectively, in a dose-dependent manner (bars 6-8 in Figure 2F and bars 9-11 in Figure 2E). Since AML1 is known to physically interact with GATA-1 through its Runt domain,20–22 the chimera may also bind to GATA-1. As expected, in an immunoprecipitation experiment, wild-type AML1 interacted with GATA-1, while the AML1 R174Q mutant did not (Figure 2H). On the other hand, GATA ZF in GATA-1 is one of the domains responsible for the interaction with the Runt domain.21 It is therefore plausible that GATA ZF in TRPS1 is also capable of interacting with AML1. Indeed, we demonstrated that wild-type TRPS1 bound to AML1, whereas the GATA ZF mutant of TRPS1 did not (Figure 2I). Taken together, we postulate that AML1-TRPS1 compromises AML1 and GATA-1 functions through 2 distinct mechanisms: (1) as a dominant-negative protein competing for their cognate binding sites; and (2) via the physical sequestration of both AML1 and GATA-1 proteins. Collectively, these 2 functions may enable AML1-TRPS1 to simultaneously disrupt AML1 and GATA factor-driven genetic programs. Since AML1-TRPS1 showed the inhibition of GATA-2 mediated transactivation (Figure 2J), the inhibitory mechanism may be extended further to other GATA factors.
In an attempt to further investigate the AML1-TRPS1–mediated transcriptional inhibitory mechanism, we carried out a reporter assay in the presence of histone deacetylase (HDAC) inhibitor trichostatin A (TSA). TSA did not affect greatly the repression mediated by intact AML1-TRPS1 (Figure 2K; bar 3 vs bar 7), but dramatically relieved that of AT/Δ Ikaros (Figure 2K; bar 4 vs bar 8), suggesting that the dimerization of AML1-TRPS1 through Ikaros-like ZF confers potent recruitment of corepressors such as HDAC. AML1-TRPS1 seems to serve as another example confirming the emerging hypothesis that most of the chimeric proteins possess the property of forming a dimer (or a multimer), thereby leading to aberrant transcriptional regulation.2,23
Although there is only 1 additional reported patient with AML with t(8;21)(q24;q22) probably carrying AML1-TRPS1,6,7 a similar fusion, AML1-FOG2, that represses both AML1- and GATA-1–mediated transactivation, was found in a patient with myelodysplastic syndrome.24 Moreover, the most pervasive fusion gene in AML, AML1-MTG8, has also been shown to repress GATA-1 function in addition to its dominant-negative effect on AML1.20,25 Collectively, the concurrent transcriptional deregulation of AML1 and GATA factors in leukemia seems relatively common. This novel chimeric gene AML1-TRPS1 could serve as a tool for elucidating the details of the interplay between AML1 and GATA factors and its disruption in leukemogenesis.
Authorship
Author contributions: N.A. and M. Yanagida contributed equally to this study. N.A. participated in the study design, molecular cloning, and writing of the manuscript. M. Yanagida performed the molecular analyses and wrote the manuscript. L.H. and M. Yamamoto provided vital analysis tools. K.S., H.M., and Y.I. contributed to the writing of the manuscript and discussions on the experimental design. M.O. participated in the study design, analysis of the experiments, and writing of the manuscript. All authors have reviewed the manuscript.
Conflict-of-interest statement: All authors declare no competing financial interests.
Correspondence: Norio Asou, Department of Hematology, Kumamoto University School of Medicine, 1-1-1 Honjo, Kumamoto 860-8556, Japan; e-mail: ktcnasou@gpo.kumamoto-u.ac.jp.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
The authors are very grateful to Hiromi Ogata-Aoki for technical assistances and Dominic C. Voon for critical reading of the manuscript.
Supported in part by Grants-in-Aid for Scientific Research from the Japanese Ministry of Education, Culture, Sport, Science and Technology; Grants-in-Aid for Cancer Research from the Japanese Ministry of Health, Labor and Welfare; and Agency of Science, Technology and Research (A*STAR), Singapore.

![Figure 1. Cloning of the AML1-TRPS1 that stimulates proliferation. (A) Spectral karyotyping (SKY) showed the balance translocation between chromosomes 8 and 21 (arrowheads). Representative SKY of chromosomes 8 and 21 is indicated as reverse DAPI (left) and SKY (right). (B) Fluorescence in situ hybridization using cosmid clones covering the whole AML1 gene showed that t(8;21)(q24;q22) involved the AML1 gene. Nearly half (55 [48.2%] of 114) of the cells had 3 signals of the AML1 gene. Images were acquired within an SD200 spectral imaging system (Applied Spectral Imaging, Migdal Haemek, Israel) attached to an Optiphot-2 epifluorescence microscope (Nikon, Tokyo, Japan) through a Splan Apo 100×/1.4 NA oil objective lens (Olympus, Tokyo, Japan) and analyzed with SkyView software (Applied Spectral Imaging). (C) The fusion partner gene with AML1 was cloned by long-distance 3′ RACE with 5′ primers on the AML1 exon 5 (AML1S5: 5′-cacagtggatgggccccgagaacctcg-3′) or exon 6 (AML1S6: 5′-tgcggcgcacagccatgagggtcagc-3′). The arrowhead indicates AML1a; the arrow shows the fusion gene. Lane 1, λ/HindIII DNA marker; lane 2, Human placental DNA; lane 3, AML cells (AML1S5); lane 4, AML cells (AML1S6). (D) N-terminal portion of AML1 retaining the Runt domain fused to the whole of TRPS1. An in-frame fusion gene consists of exon 6 of AML1 and TRPS1. In addition, sequencing analyses of subclones showed another transcript between exon 5 of AML1 and TRPS1, indicating skipping of exon 6 of AML1 and generating another in-frame fusion gene. The reciprocal TRPS1-AML1 transcript was not detected in the presenting case. (E) Structures of retrovirus constructs for control (MIG_mock) or AML1-TRPS1 (MIG_AT). AML1-TRPS1 was inserted into the indicated position of the plasmid MIG with internal ribosomal entry site (IRES) and the enhanced green fluorescent protein (EGFP) gene. (F) Schematic depiction of the CFU-C assay. Fetal liver cells from embryonic day–14.5 (E14.5) mouse embryo were infected with the MIG vector. EGFP-positive cells were sorted and subjected to the CFU-C assay, supplemented with interleukin-3, stem cell factor, erythropoietin, and granulocyte colony-stimulating factor, as described previously.11 (G) The number of all kinds of colony (total), granulocyte-macrophage (GM), and macrophage (M) are shown. Error bars indicate standard deviations for 3 independent experiments. Differences between AML1-TRPS1 (AT) and control (mock) transfectants were statistically significant (*P < .001, unpaired student t test) in all 3 categories.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/9/10.1182_blood-2006-01-031781/4/m_zh80090700300001.jpeg?Expires=1769113890&Signature=Qw2BpM2D-JEhnDALyInud1~5soMF8RVnN4l25QCrtGMYOnAN1H8K3A81yInt12C8vS6leJ7lpFJ5dLGCGly8a7xZGMol2mDa777T5mpUqTe70LsFi8wAXvQqiEhI6j1A4o4RSYaUZfz-s3dGL0OPi16yHBUXdSufm5FTI35gRfzbNIvb4xDBN4NFKoCFFsddovtCLMU9lVe2DS~aBohOvpsivkA1kT3JqmM~1TVYUmJ0-SQNbjFMIhD~LQwtrj1neb7S5HvXKknKqFIxb2nCoz6a~3UOxFxiFK0CaBuCwOXiQlPI-tNwQO3aWnWzjnGonUtr4xE1FnsRW2H0XR~P0A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal