Abstract
ADAMTS13 cleaves von Willebrand factor (VWF) between Tyr1605 and Met1606 residues at the central A2 subunit. The amino-terminus of ADAMTS13 protease appears to be sufficient to bind and cleave VWF under static and denatured condition. However, the role of the carboxyl-terminus of ADAMTS13 in substrate recognition remains controversial. Present study demonstrates that ADAMTS13 cleaves VWF in a rotation speed– and protease concentration–dependent manner on a mini vortexer. Removal of the CUB domains (delCUB) or truncation after the spacer domain (MDTCS) significantly impairs its ability to cleave VWF under the same condition. ADAMTS13 and delCUB (but not MDTCS) bind VWF under flow with dissociation constants (KD) of about 50 nM and about 274 nM, respectively. The isolated CUB domains are neither sufficient to bind VWF detectably nor capable of inhibiting proteolytic cleavage of VWF by ADAMTS13 under flow. Addition of the TSP1 5-8 (T5-8CUB) or TSP1 2-8 repeats (T2-8CUB) to the CUB domains restores the binding affinity toward VWF and the inhibitory effect on cleavage of VWF by ADAMTS13 under flow. These data demonstrate directly and quantitatively that the cooperative activity between the middle carboxyl-terminal TSP1 repeats and the distal carboxyl-terminal CUB domains may be crucial for recognition and cleavage of VWF under flow.
Introduction
ADAMTS13 controls the sizes of von Willebrand factor (VWF) multimers by cleaving VWF at the Tyr1605-Met1606 bond at the central A2 domain.1 Deficiency of plasma ADAMTS13 activity, due to either inherited mutations of the ADAMTS13 gene2–9 or acquired autoantibodies against the ADAMTS13 protein,10,11 results in thrombotic thrombocytopenic purpura (TTP).
ADAMTS13 is primarily synthesized in hepatic stellate cells,12–14 endothelial cells,15,16 and megakaryocytes or platelets.17,18 The plasma ADAMTS13 in healthy individuals ranges from 0.5 to 1 mg/L.19,20 ADAMTS13 consists of metalloprotease, disintegrin, first thrombospondin type 1 (TSP1) repeat, and Cys-rich and spacer domains.2,21 The C-terminus of ADAMTS13 has additional TSP1 repeats and 2 CUB (Cis/Cis/Urinary epidermal growth factor, Bone morphogenetic protein) domains.2,21 Previous studies have shown that the N-terminus of ADAMTS13 is required and sufficient for recognition and cleavage of denatured multimeric VWF22–24 or peptide substrate (GST-VWF73 or FRETS-VWF73).22 More recent studies have demonstrated that the spacer domain of ADAMTS13 binds the exosite (E1660APDLVLQR1668) near the C-terminus of the VWF-A2 domain.25,26
However, the role of the middle and distal C-terminal domains of ADAMTS13 in substrate recognition remains controversial. On the one hand, ADAMTS13 mutant lacking the CUB domains or truncated after the spacer domain cleaved multimeric VWF with similar efficiency as the full-length ADAMTS13 under static and denatured conditions23,24 ; the mutant truncated after the spacer domain, when mixed with ADAMTS13-depleted plasma, was found to be “hyperactive” in cleaving a “stringlike” structure, which represents platelets attached to the newly released VWF on the endothelial cell surface in a parallel-flow chamber–based assay.27 These data suggest that the distal portion of ADAMTS13 molecule may be dispensable under static and denatured conditions but may play a role in modulating ADAMTS13-VWF interaction under flow. On the other hand, synthetic peptides or recombinant fragments derived from the CUB domains28 appeared to block the cleavage of the stringlike structure on endothelial cells, suggesting that the CUB domains may directly participate in binding or recognition of VWF under flow. Although the parallel-flow chamber assay may mimic physiological condition, its complexity involving live endothelial cells, histamine stimulation, and platelets makes the quantitation less accurate and kinetic determination of ADAMTS13 and VWF interaction impossible.
In the present study, we have developed a simple flow assay based on mechanical-induced shear stress on a mini vortexer or a laminar flow in a BIAcore (Uppsala, Sweden) system to determine the role of the C-terminal ADAMTS13 in recognition and cleavage of multimeric VWF. Our data demonstrate directly and quantitatively that the cooperative activity between the middle C-terminal TSP1 repeats and the distal C-terminal CUB domains of ADAMTS13 may be crucial for productive binding and cleavage of VWF under flow.
Materials and methods
The human study has been approved by the Institutional Review Board of The Children's Hospital of Philadelphia and the University of Pennsylvania.
Constructs
The plasmids containing full-length ADAMTS13 (FL-A13) and variants truncated after the eighth TSP1 repeat (delCUB) or after the spacer domain (MDTCS) or the metalloprotease domain (M) were described previously.22,24,29 The cDNA fragments encoding the CUB domains (CUB), TSP1 2-8 (T2-8), TSP1 5-8 (T5-8), TSP1 5-8 repeats plus CUB domains (T5-8CUB), and TSP1 2-8 plus CUB domains (T2-8CUB) were amplified by polymerase chain reaction (PCR) using pcDNA3.1-FL-A13 as a template and cloned into pSecTag/FRT/V5-HisTOPO (Invitrogen, Carlsbad, CA) according to manufacturer's recommendation. The constructs CUB, T2-8, T5-8, T5-8CUB, and T2-8CUB were tagged at their N-termini with a linker sequence and a flag epitope (AAQPARRARRTKLALDTKDDDDKHVWTPVA) (underlined) and at their C-termini with V5-His epitope. The plasmids were sequenced to confirm the accuracy.
Cell culture and transfection
The human embryonic kidney cells (HEK-293) grown in Dulbecco modified Eagle medium (DMEM) (Invitrogen) containing 10% of FetalPlex (Gemini BioProducts, West Sacramento, CA) were transfected with a mixture of LipofectAMINE2000 and plasmids (3:1 vol/wt) in serum-free Opti-MEM. Constructs in pSecTag/FRT/V5-His vector were cotransfected with pcDNA3.1 vector (Invitrogen) to obtain the neo gene for stable selection. After 72 hours of transfection, the stable clones were selected by treating the cells with 0.5 mg/mL geneticin (G418) (Invitrogen) and identified by Western blotting with anti-V5 IgG (Invitrogen) as described previously.22,24
Preparation of recombinant proteins
Stably transfected HEK-293 cells expressing ADAMTS13 and variants were cultured on 10-layer cell factories (Fisher Scientific, Hampton, NH) in Opti-MEM (Invitrogen) or serum-free DMEM supplemented with 5 mg/mL of insulin transferring selenium (ITS) (Roche Applied Science, Indianapolis, IN) supplement. The conditioned medium (about 2 L) was collected every 24 to 48 hours, and the cell debris was removed by centrifugation at 1500g for 10 minutes and filtration through coarse filter paper (Fisher Scientific). After addition of 5 mM benzamidine and 1 mM phenylmethylsulfonyl fluoride (PMSF) (Sigma, St Louis, MO), the conditioned medium was frozen and stored at −80°C until use.
The conditioned medium was thawed at room temperature and diluted (1:3) with distilled water. The pH was adjusted to 8.0 by adding 2 M Tris-HCl, pH 8.0. The diluted conditioned medium was loaded onto Q-fast flow ion exchange column (125 mL) at 4°C overnight. After being washed with 20 mM Tris-HCl, pH 8.0, the protein was eluted with 5 to 10 column volumes of 1 M NaCl in 20 mM Tris-HCl, pH 8.0. The fractions containing proteins were pooled and then loaded onto 10 to 80 mL Ni-NTA affinity column (Invitrogen). After being washed with 20 mM Tris-HCl, pH 8.0, 400 mM NaCl in presence of 10 mM imidazole, the bound proteins were eluted with 60 mM and 250 mM imidazole in 20 mM Tris-HCl, pH 8.0, and 400 mM NaCl. The fractions (4 mL each) were collected, and the peak fractions containing recombinant proteins of interest were pooled and concentrated with Centri-Prep30 (Millipore, Billerica, MA). The proteins were further separated by Superose 6 10/300GL gel filtration chromatography (GE Biosciences, Piscataway, NJ) at 0.5 mL/min with 20 mM Tris-HCl, 150 mM NaCl, pH 7.5, as described previously.22 The SDS–polyacrylamide gel electrophoresis (PAGE) and Coomassie blue staining determined the molecular weight and purity of the purified proteins. The amount of the purified proteins was determined by absorbance at 280 nm (corrected with light scattering at 340 nm) with absorbance coefficients of 0.68 (FL-A13), 0.71 (delCUB), 0.91 (MDTCS), 0.63 (CUB), 0.62 (T2-8), 0.81 (T5-8), 0.68 (T5-8CUB), and 0.60 (T2-8CUB) mg mL−1 cm−1.30,31 The amount of specific ADAMTS13 antigen was also verified by Western blot with anti-V5 using Positope (Invitrogen) as a standard.
Cleavage of VWF under flow and static condition
Purified plasma-derived VWF (37.5 μg/mL or 150 nM, final concentration)22,24 was incubated with ADAMTS13 and variants at concentrations indicated in each figure and figure legend in 50 mM HEPES buffer containing 0.25% BSA, 5 mM CaCl2, and 0.25 mM ZnCl2 (total volume, 20 μL) in a 0.2 mL thin-walled PCR tube with dome caps (Fisher Scientific) for 1 minute. Here the molar concentration of VWF was calculated using a molecular weight of 250 kDa for each VWF polypeptide as described previously30 . The reaction mixture was subjected to vortexing at a fixed rotation rate of about 2500 rpm (set “8”) or various rotation speeds between 0 and about 3200 rpm for 3 minutes on a mini vortexer (Fisher Scientific).32
Alternatively, purified plasma-derived VWF was incubated with 1.5 M guanidine-HCl at 37°C for 2 hours.1,10 The denatured VWF was diluted 1:10 with 50 mM HEPES buffer containing 0.25% BSA, 5 mM CaCl2, and 0.25 mM ZnCl2.30 Denatured VWF (37.5 μg/mL or 150 nM) was incubated with about 60 nM of ADAMTS13 (or variants) at 37°C for 1 hour. The reaction was quenched by heating the samples at 100°C for 10 minutes after addition of sample buffer (0.625 mM Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, and 0.01% bromphenol blue). The cleavage products were detected by Western blot with peroxidase-conjugated anti-VWF IgG (p0226; DAKO, Carpinteria, CA) (1:3000) in 1% casein (Sigma) or anti-VWF IgG (p082; DAKO) followed by peroxidase-conjugated antirabbit IgG (1:5000), and SuperSignal Chemiluminescent reagents (Pierce, Rockford, IL).
Cleavage of GST-VWF73-H by ADAMTS13 and variants
The proteolytic cleavage of GST-VWF73 was determined by Western blotting with rabbit anti-GST IgG (Molecular Probes) as described,22 followed by Alexa Fluor680–conjugated antirabbit IgG (Molecular Probes, Carlsbad, CA) (1:12 500). The bound fluorescent antibody was quantified by Odyssey infrared fluorescent image system (LI-COR Bioscience, Lincoln, NE).
Binding of VWF to ADAMTS13 and variants under flow
In contrast to a mini vortexer that generates turbulent flow, a BIAcore system produces laminar flow. The shear rate at the inner surface of the injection tube (with diameter of 0.2 mm) can be calculated with a simple equation: shear rate ≈ 1.27f/πR3, where f is injection flow rate (μL/min) and R is the diameter of the tube (mm). In the microfluidic cells, the shear rate can also be calculated: shear rate ≈ f/10wh2, where f is also the injection flow rate (μL/mi), w is the side length (mm), and h is the height (mm) of the microfluidic cell. In BIAcore2000 (BIAcore), the dimension of the fluidic cell is 2.4 mm in length, 0.5 mm in width, and 0.05 mm in height, with a total volume of 60 nL. Accordingly, at the injection rate of 1 μL/min, about 50 s−1 shear rate in the inner surface of tube and 80 s−1 shear rate in the microfluidic cells can be generated. Therefore, the BIAcore system provides us with a unique opportunity to accurately and quantitatively determine the interaction between VWF and ADAMTS13 (or variants) at the single-molecule level in real time under flow shear stress.
Briefly, the surface of a carboxymethylated dextran (CM5) chip was activated by injection of 35 μL mixture (1:1 vol/vol) of 0.4 M 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide and 0.1 M N-hydroxysuccinimide according to manufacturer's instruction (BIAcore). Approximately 2000 to 8000 response units (RU) (2 to 10 ng/mm2) of purified recombinant proteins were covalently attached onto the activated CM5 chip surface. The control surface was activated similarly but not immobilized by protein or immobilized by same amount of bovine serum albumin (BSA) (Sigma, St. Louis, MO). The reactive groups on the dextran surface were blocked by injection of 35 μL of 1 M ethanolamine (pH 8.5) at flow rate of 5 μL/min for 7 minutes. Then, purified plasma VWF at various concentrations (0 to 250 μg/mL or 0 to 1000 nM as in Figure 3; 0 to 125 μg/mL or 0 to 500 nM as in Figure 4) in 10 mM HEPES, 150 mM NaCl, pH 7.5, containing 0.005% Tween 20 and 2 mg/mL BSA (HBS-T) was injected and passed over the surface at injection rates of 10 to 100 μL/min or 20 μL/min for 3 to 5 minutes. The HBS-T replaced the protein solution and continued to flow for approximately 4 minutes; further washing with HBS-T for 20 to 30 minutes regenerated the surfaces prior to the next injection. The dissociation constants, KD (S) at the equilibrium were determined by fitting the data from the binding isotherm using a nonlinear regression curve on the PRISM4 software (GraphPad Software, San Diego, CA).
Binding of ADAMTS13 or variants to VWF immobilized on solid surfaces
The binding of ADAMTS13 and variants to immobilized VWF on a microtiter plate was performed as described previously.29 The specific binding was obtained after subtraction of absorbance in the control wells without VWF ligand. The kinetic parameters were determined by fitting the data into nonlinear regression equation.
The binding of ADAMTS13 to immobilized Affi-gel 10 was described previously.22 Briefly, purified VWF (5 mg) was covalently coupled onto 2 mL activated Affi-gel-10 (Bio-Rad, Hercules, CA) in HEPES buffer, pH 7.5, at 4°C for 5 hours. The residual reactive groups on the Affi-gel-10 beads were blocked with 0.1 M glycine ethyl ester (Sigma), pH 6.5, and 2.5% BSA fraction V (Sigma) for 2 hours. The VWF-coupled Affi-gel was stored at 4°C in 5 mM Tris-HCl, pH 8.0, containing 0.02% sodium azide until use. Ten microliters of VWF–Affi-gel (2.5 μg VWF per microliter gel) or control Affi-gel that was not coupled with VWF were incubated with approximately 200 nM FL-A13 (or variants) in 20 mM HEPES, pH 7.5, 150 mM NaCl in presence of 0.25% BSA at 25°C for 30 minutes. The beads were washed 3 times with 10 volumes of 20 mM HEPES, pH 7.5, 150 mM NaCl and once with 500 mM NaCl. The bound FL-A13 and variants were eluted from the beads by boiling them at 100°C for 10 minutes and detected by Western blotting with anti-V5 IgG as described previously.22,24,29
The C-terminal fragments of ADAMTS13 block cleavage of VWF by ADAMTS13 under flow
Purified plasma VWF (37.5 μg/mL or 150 nM) was incubated in absence or presence of 0 to 150 nM recombinant CUB, T2-8, T5-8, T5-8CUB, and T2-8CUB in 50 mM HEPES buffer containing 5 mM CaCl2, 0.25 mM ZnCl2, and 2 mg/mL BSA for 60 minutes. Then ADAMTS13 (about 50 nM) was added, and the mixture was subjected to vortexing at 2500 rpm (set “8”) for 3 minutes at 22°C. The reaction was quenched by heating the sample in 1× SDS sample buffer at 100°C for 5 minutes. Western blotting as described in “Cleavage of VWF under flow and static condition” determined the cleavage of VWF.
Results
Purification of recombinant ADAMTS13 and variants
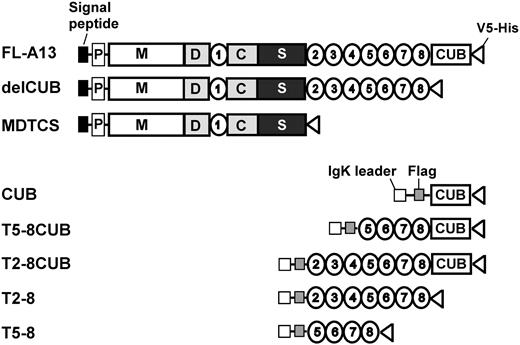
To determine the kinetic interactions between VWF and ADAMTS13 or variants in a purified system, we expressed and purified full-length ADAMTS13 and variants or C-terminal fragments. The domain composition of each construct is listed in Figure 1. The proteins were purified to homogeneity by 3 sequential column chromatographies: Q-fast flow ion exchange, Ni-NTA affinity column, and Superose 6 gel filtration as described previously.22 Typically, approximately 0.2 to 1.0 mg with about 90% to 95% purity of recombinant proteins were obtained from 2 to 10 L of conditioned medium. The molecular weights of FL-A13, delCUB, and MDTCS are estimated to be about 195 kDa, about 150 kDa, and about 95 kDa, respectively, on SDS-PAGE under denatured and reduced condition (data not shown). The molecular weights of the constructs CUB, T2-8, T5-8, T5-8CUB, and T2-8CUB, however, are about 50 kDa, about 100 kDa, about 52 kDa, about 95 kDa, and about 116 kDa, respectively (data not shown).
Constructs of ADAMTS13 and truncated variants. The full-length ADAMTS13 (FL-A13) and the variants truncated after the eighth TSP1 repeat (delCUB) and after the spacer domain (MDTCS) were cloned into pcDNA3.1 V5-His TOPO vector. The original signal peptide and propeptide of ADAMTS13 were included. The CUB domains (CUB, C1192-T1427), T2-8 repeats (T2-8, W686-W1076), T5-8 repeats (H884-W1076), the CUB domains plus the TSP1 5-8 repeats (T5-8CUB, H884-A1191), and the CUB domains plus the TSP1 2-8 repeats (T2-8CUB, W686-A1191) were cloned into pSecTag/FRT/V5-His TOPO, in which an IgK secretion peptide and a Flag epitope (-DDDDK-) were engineered at the N terminus of the CUB, T2-8, T5-8, T5-8CUB, and T2-8CUB. All constructs contain V5-His epitopes at their C termini to facilitate purification and detection.
Constructs of ADAMTS13 and truncated variants. The full-length ADAMTS13 (FL-A13) and the variants truncated after the eighth TSP1 repeat (delCUB) and after the spacer domain (MDTCS) were cloned into pcDNA3.1 V5-His TOPO vector. The original signal peptide and propeptide of ADAMTS13 were included. The CUB domains (CUB, C1192-T1427), T2-8 repeats (T2-8, W686-W1076), T5-8 repeats (H884-W1076), the CUB domains plus the TSP1 5-8 repeats (T5-8CUB, H884-A1191), and the CUB domains plus the TSP1 2-8 repeats (T2-8CUB, W686-A1191) were cloned into pSecTag/FRT/V5-His TOPO, in which an IgK secretion peptide and a Flag epitope (-DDDDK-) were engineered at the N terminus of the CUB, T2-8, T5-8, T5-8CUB, and T2-8CUB. All constructs contain V5-His epitopes at their C termini to facilitate purification and detection.
Cleavage of VWF by ADAMTS13 and variants under flow
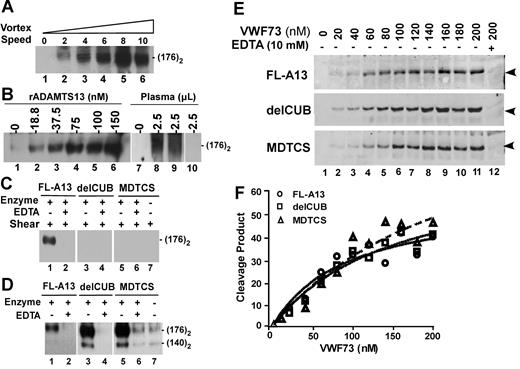
To determine whether the C-terminal domains of ADAMTS13 are required for cleavage of VWF under flow, we developed a simple flow-based assay using a mini vortexer as described elsewhere.32 Unique to vortex rotation, turbulent flow that mimics the flow condition in the branching of the vessels or downstream of partially occluded vessels is generated.33,34 When vortexing at rotation rates between 640 to 3200 rpm (set “2-8”), VWF was readily cleaved within 3 minutes by full-length ADAMTS13 in a rotation rate–dependent manner; the cleavage product (a dimer of 176 kDa) reached the plateau at rotation rate of about 2500 rpm (with estimated shear rate more than 12 000 s−1)33,34 (Figure 2A); the cleavage of VWF was also ADAMTS13 concentration dependent at a fixed rotation rate of about 2500 (Figure 2B); even 2.5 μL of normal human plasma was sufficient to cleave VWF in presence of 30 to 60 μg/mL heparin under this condition (Figure 2B). Addition of more heparin (500 μg/mL) and barium chloride (10 mM) increased VWF cleavage product by plasma ADAMTS13 (Sara Meyer and X.L.Z., unpublished data, May 2007). The specificity was confirmed by lack of VWF cleavage product after addition of 10 mM EDTA into the reaction or omitting of ADAMTS13 enzyme or using of TTP-patient plasma (Figure 2).
Proteolytic cleavage of VWF and VWF73 under flow or static condition by ADAMTS13 and C-terminal truncated variants. (A) Rotation speed–dependent cleavage of VWF by ADAMTS13. Native plasma VWF (37.5 μg/mL or 150 nM) was incubated with ADAMTS13 (about 60 nM) for 1 minute and then vortexed for 3 minutes at 22°C at rotation speeds from 0 to about 3200 rpm (set at “0-10”). (B) Dose-dependent cleavage of VWF by ADAMTS13. VWF (18.75 μg/mL or 75 nM) was vortexed for 3 minutes without (lanes 1 and 7) or with various concentrations of rADAMTS13 (lane 2-6) or 2.5 μL of normal human plasma with 30 μg/mL (lane 8) or 60 μg/mL (lane 9) heparin or TTP patient plasma (lane 10). (C) Cleavage of VWF by ADAMTS13 and variants. VWF (18.75 μg/mL or 75 nM) was incubated and vortexed for 3 minutes without (−) or with (+) about 60 nM of FL-A13, delCUB, and MDTCS in absence (−) or presence (+) of 10 mM EDTA. (D) Cleavage of guanidine-HCl–denatured VWF by ADAMTS13 and variants. Denatured VWF (37.5 μg/mL or about 150 nM) was incubated without (−) or with (+) about 60 nM of purified FL-A13, delCUB, and MDTCS in absence (−) or presence (+) of 10 mM EDTA for 1 hour. All the reactions above were quenched by addition of SDS sample buffer and heated at 100°C for 5 minutes. The cleavage product (dimer of 176 kDa) was determined by Western blot with peroxidase-conjugated rabbit anti-VWF IgG, followed by chemiluminescent ECL reagents. The signal was obtained by exposure to X-ray film within 5 to 30 seconds. (E) Cleavage of GST-VWF73-H by ADAMTS13 and variants. GST-VWF73-H at various concentrations (0 to 200 nM) was incubated with about 60 nM of FL-A13, delCUB, and MDTCS for 10 minutes at 37°C. The cleavage product (34.4 kDa, arrowheads indicated) was determined by Western blot with rabbit anti-GST IgG and Alexa Fluor680–conjugated antirabbit IgG. (F) The plot of the fluorescent signal; obtained by Odyssey infrared fluorescent image system against concentrations of GST-VWF73 substrate.
Proteolytic cleavage of VWF and VWF73 under flow or static condition by ADAMTS13 and C-terminal truncated variants. (A) Rotation speed–dependent cleavage of VWF by ADAMTS13. Native plasma VWF (37.5 μg/mL or 150 nM) was incubated with ADAMTS13 (about 60 nM) for 1 minute and then vortexed for 3 minutes at 22°C at rotation speeds from 0 to about 3200 rpm (set at “0-10”). (B) Dose-dependent cleavage of VWF by ADAMTS13. VWF (18.75 μg/mL or 75 nM) was vortexed for 3 minutes without (lanes 1 and 7) or with various concentrations of rADAMTS13 (lane 2-6) or 2.5 μL of normal human plasma with 30 μg/mL (lane 8) or 60 μg/mL (lane 9) heparin or TTP patient plasma (lane 10). (C) Cleavage of VWF by ADAMTS13 and variants. VWF (18.75 μg/mL or 75 nM) was incubated and vortexed for 3 minutes without (−) or with (+) about 60 nM of FL-A13, delCUB, and MDTCS in absence (−) or presence (+) of 10 mM EDTA. (D) Cleavage of guanidine-HCl–denatured VWF by ADAMTS13 and variants. Denatured VWF (37.5 μg/mL or about 150 nM) was incubated without (−) or with (+) about 60 nM of purified FL-A13, delCUB, and MDTCS in absence (−) or presence (+) of 10 mM EDTA for 1 hour. All the reactions above were quenched by addition of SDS sample buffer and heated at 100°C for 5 minutes. The cleavage product (dimer of 176 kDa) was determined by Western blot with peroxidase-conjugated rabbit anti-VWF IgG, followed by chemiluminescent ECL reagents. The signal was obtained by exposure to X-ray film within 5 to 30 seconds. (E) Cleavage of GST-VWF73-H by ADAMTS13 and variants. GST-VWF73-H at various concentrations (0 to 200 nM) was incubated with about 60 nM of FL-A13, delCUB, and MDTCS for 10 minutes at 37°C. The cleavage product (34.4 kDa, arrowheads indicated) was determined by Western blot with rabbit anti-GST IgG and Alexa Fluor680–conjugated antirabbit IgG. (F) The plot of the fluorescent signal; obtained by Odyssey infrared fluorescent image system against concentrations of GST-VWF73 substrate.
Strikingly, a removal of the CUB domains (delCUB) or truncation after the spacer domain (MDTCS) significantly impaired the ability of ADAMTS13 to cleave VWF under the flow condition at the rotation rate of about 2500 rpm (Figure 2C). The same amount of the C-terminal truncated mutants was able to cleave guanidine-HCl–denatured VWF even more efficiently than full-length ADAMTS13 (Figure 2D). The constructs FL-A13, delCUB, and MDTCS all cleaved GST-VWF73-H (Figure 2E-F) or FRETS-VWF73 substrate (data not shown) with similar efficacy. The data suggest that the CUB domains of ADAMTS13 are required for cleavage of VWF under turbulent flow.
Binding of VWF to ADAMTS13 (or variants) under flow
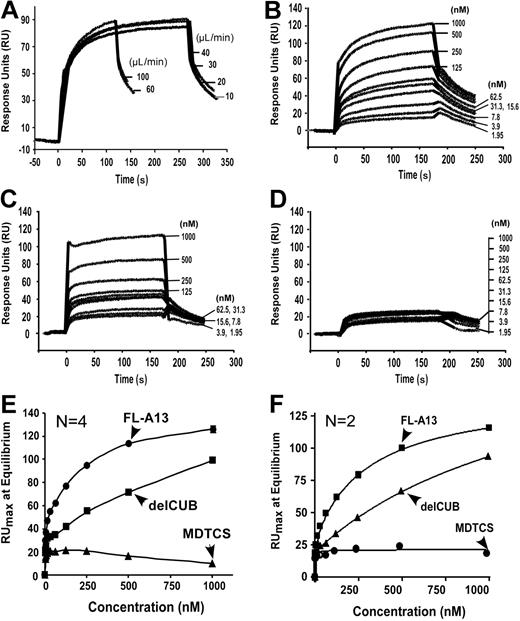
To determine the binding interaction between VWF and ADAMTS13 (or variants) under laminar flow, we employed the BIAcore technology based on measurement of surface plasmon resonance. We chose to attach full-length ADAMTS13 or C-terminal truncated variants covalently onto the CM5 surface to avoid VWF activation induced by amine coupling. We then passed purified plasma VWF in the binding buffer at various concentrations (0 to 1000 nM) over the ADAMTS13 immobilized surfaces. Because plasma VWF multimers vary in sizes and are sensitive to shear stress, injection flow rate may affect the molecule diffusion rate and conformation. To determine diffusion effect or effect of flow rate on VWF-ADAMTS13 binding, a fixed concentration of plasma VWF (12.5 μg/mL or 50 nM) was injected over the surface immobilized by full-length ADAMTS13 at various flow rates (10 to 100 μL/min) (estimated shear rates between about 250 s−1 and about 5000 s−1). We found that VWF at various flow rates was able to bind ADAMTS13 with similar association and dissociation kinetics (Figure 3A). These data suggest that VWF binds ADAMTS13 in high affinity at various flow shear rates. The data also indicate that the VWF-ADAMTS13 binding is not diffusion limited.
Kinetic binding interaction between VWF and ADAMTS13 (or variants) under flow. (A) Effect of flow rates on binding of VWF to ADAMTS13. Purified VWF (18.75 μg/mL or 50 nM) was injected at various flow rates for 3 to 5 minutes over the CM5 surface immobilized with FL-A13 in absence of EDTA. (B-D) Binding of VWF to ADAMTS13 and C-terminal truncated variants. Purified VWF at various concentrations (0 to 250 μg/mL or 0 to 1000 nM) was injected over the surfaces immobilized by FL-A13 (B), delCUB (C), and MDTCS (D). After equilibrium was established, the HBS-T buffer was then injected over the surface to allow the dissociation to occur. The representative sensograms in absence of EDTA are shown in panel A-D. The maximal response units (RUmax) at equilibrium (y-axis) were obtained from the sensograms and plotted against various concentrations of VWF injected (x-axis). The entries in panels E and F are the mean of 2 to 4 repeats in absence (E) or presence (F) of 10 mM EDTA. The equilibrium dissociation constant, KD, was calculated by fitting the data to the binding isotherm using nonlinear regression.
Kinetic binding interaction between VWF and ADAMTS13 (or variants) under flow. (A) Effect of flow rates on binding of VWF to ADAMTS13. Purified VWF (18.75 μg/mL or 50 nM) was injected at various flow rates for 3 to 5 minutes over the CM5 surface immobilized with FL-A13 in absence of EDTA. (B-D) Binding of VWF to ADAMTS13 and C-terminal truncated variants. Purified VWF at various concentrations (0 to 250 μg/mL or 0 to 1000 nM) was injected over the surfaces immobilized by FL-A13 (B), delCUB (C), and MDTCS (D). After equilibrium was established, the HBS-T buffer was then injected over the surface to allow the dissociation to occur. The representative sensograms in absence of EDTA are shown in panel A-D. The maximal response units (RUmax) at equilibrium (y-axis) were obtained from the sensograms and plotted against various concentrations of VWF injected (x-axis). The entries in panels E and F are the mean of 2 to 4 repeats in absence (E) or presence (F) of 10 mM EDTA. The equilibrium dissociation constant, KD, was calculated by fitting the data to the binding isotherm using nonlinear regression.
Because plasma-derived VWF varies in length and exhibited very fast-association (on) and fast-dissociation (off) rates, the kon and koff could not be accurately determined. Fitting the data directly using BIAcore evaluation software may overestimate the binding affinity between VWF and ADAMTS13 due to the heterogeneity of VWF molecules. We therefore only report the dissociation constants, KD (S) at equilibrium here. Under the laminar flow, VWF bound full-length ADAMTS13 in a dose- and time-dependent manner (Figure 3B), with a KD of 50 plus or minus 9.0 nM. A removal of the CUB domains (delCUB) reduced its affinity by 5-fold (KD = 274 ± 92 nM; mean ± SEM) (Table 1; Figure 3C). Further removal of the TSP1 2-8 repeats (MDTCS) abolished its affinity toward flowing VWF (Figure 3D). The binding affinity was independent of divalent cations, because addition of 10 mM EDTA into the binding buffer did not affect the binding kinetics or KD values (Figure 3F). These data demonstrate quantitatively that the distal C-terminal TSP1 repeats and CUB domains may be required for recognition of VWF under laminar flow.
Kinetic determination of VWF binding to ADAMTS13 (or variants) by SPR
| VWF; KD, ×10−9 M (n) . | VWF;*KD ×10−9 M (n) . |
|---|---|
| FL-A13 | |
| 50±9 (6) | 83 ± 17 (6) |
| 77±26 (4)† | — |
| delCUB | |
| 274±92 (6) | 242 ± 73 (6) |
| 468±131 (4)† | — |
| MDTC | |
| No binding | 337 ± 186 (6) |
| No binding† | — |
| VWF; KD, ×10−9 M (n) . | VWF;*KD ×10−9 M (n) . |
|---|---|
| FL-A13 | |
| 50±9 (6) | 83 ± 17 (6) |
| 77±26 (4)† | — |
| delCUB | |
| 274±92 (6) | 242 ± 73 (6) |
| 468±131 (4)† | — |
| MDTC | |
| No binding | 337 ± 186 (6) |
| No binding† | — |
The entries are means plus and minus a standard error.
FL-A13 is the full-length ADAMTS13; delCUB, the ADAMTS13 variant truncated after the eighth TSP1 repeat; MDTCS, the variant truncated after the spacer domain; n, the number of repeats performed; —, not applicable.
The VWF substrate was denatured at 37°C for 2 hours with 1.5 M quanidine HCl prior to binding experiments.
Data obtained from experiments performed in presence of 10 mM EDTA.
Binding of denatured VWF to ADAMTS13 (or variants) under flow
It has been shown that addition of 1.5 M guanidine-HCl1,10 or 1.5 M urea1 significantly accelerates VWF proteolysis by ADAMTS13. To determine whether predenatured VWF increases its interaction with ADAMTS13 (or variants) under flow, the denatured plasma VWF at various concentrations (0 to 500 nM) was passed over full-length ADAMTS13 and variants surface. We showed that predenatured VWF was able to bind the short construct MDTCS with an increased affinity (KD of 337 ± 186 nM; mean ± SEM) (n = 6) (Figure 4C,D; Table 1), but the affinity between the denatured VWF and FL-A13 (or delCUB) was not significantly altered with the KD (S) of 83 (± 17) nM and 242 (± 73) nM (mean ± SEM), respectively (Table 1). These data suggest that additional cryptic binding sites that are potentially recognized by the N-terminal domains of ADAMTS13 may be exposed upon predenaturization of VWF plus flow shear stress.
Binding of denatured VWF to ADAMTS13 and C-terminal truncated variants. Purified VWF pretreated for 2 hours with 1.5 M guanidine-HCl at 37°C was diluted (1:10) with HBS-T buffer with (not shown) or without EDTA into various concentrations (0 to 125 μg/mL or 0 to 500 nM). The diluted VWF was then injected at 20 μL/min for 3 minutes over the CM5 chips covalently coupled by FL-A13 (A), delCUB (B), and MDTCS (C). After the equilibrium was established, the HBS-T buffer without VWF was flowed over the surface to allow the dissociation phase to be recorded. The equilibrium constant, KD, was determined similarly as described for Figure 3. The entries in panel D represent the means (± SD) of 6 repeats.
Binding of denatured VWF to ADAMTS13 and C-terminal truncated variants. Purified VWF pretreated for 2 hours with 1.5 M guanidine-HCl at 37°C was diluted (1:10) with HBS-T buffer with (not shown) or without EDTA into various concentrations (0 to 125 μg/mL or 0 to 500 nM). The diluted VWF was then injected at 20 μL/min for 3 minutes over the CM5 chips covalently coupled by FL-A13 (A), delCUB (B), and MDTCS (C). After the equilibrium was established, the HBS-T buffer without VWF was flowed over the surface to allow the dissociation phase to be recorded. The equilibrium constant, KD, was determined similarly as described for Figure 3. The entries in panel D represent the means (± SD) of 6 repeats.
Binding of ADAMTS13 (or variants) to immobilized VWF on solid surfaces
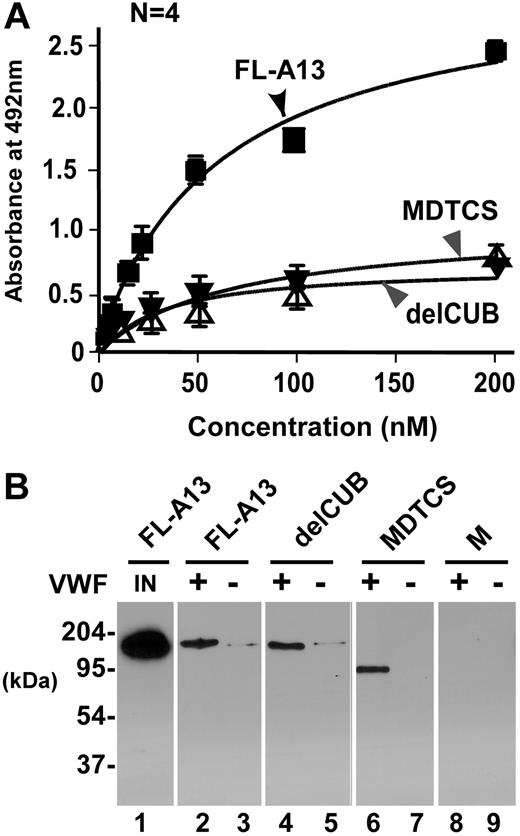
VWF can be activated by adsorption onto the solid surfaces.29 To validate the specificity of VWF-ADAMTS13 interaction seen in the BIAcore system and to be sure that the purified ADAMTS13 and variants behave as expected in recognition of VWF under a static condition, we determined the binding on a microtiter plate. Consistent with the data reported by Majerus et al,29 our recombinant FL-A13, delCUB, and MDTCS bound immobilized VWF with a KD (S) of 50 (± 6) nM, 70 (± 23) nM, and 56 (± 30) nM, respectively (Figure 5A). The binding interaction was not disrupted by 0.5 M sodium chloride (Figure 5B), confirming the high-affinity binding. The metalloprotease domain alone did not bind immobilized VWF on microtiter plate (data not shown)29 or on VWF–Affi-gel 10 detectably (Figure 5B), confirming that the exosites beyond the catalytic domain mediate the ADAMTS13 interaction with immobilized/activated VWF on solid surfaces.
Binding of ADAMTS13 (or variants) to VWF immobilized on solid surfaces. (A) Binding of ADAMTS13 and variants to VWF immobilized on a microtiter plate. FL-A13, delCUB, and MDTCS (0 to 200 nM) were incubated without (control) or with VWF (10 μg/mL, 100 μL per well) immobilized on a microtiter plate for 1 hour. The bound ADAMTS13 and variants were determined by mouse anti-V5 IgG, followed by rabbit antimouse IgG, peroxidase-conjugated and OPD-H2O2. The KD (S) was determined by fitting the data into nonlinear regression. (B) Binding of ADAMTS13 and variants to immobilized VWF on Affi-gel 10. FL-A13, delCUB, MDTCS, and metalloprotease domain (M) (about 50 nM) were incubated at 37°C for 1 hour without (−) or with (+) VWF covalently immobilized onto the Affi-gel 10. After extensive washing with TBS and 20 mM Tris-HCl, pH 7.5, 500 nM NaCl, the bound ADAMTS13 and variants were eluted from the beads with SDS-gel sample buffer and determined by Western blot with anti-V5. The amount of input FL-A13, delCUB, and MDTCS is the same with the signal of only FL-A13 shown in lane 1 (IN).
Binding of ADAMTS13 (or variants) to VWF immobilized on solid surfaces. (A) Binding of ADAMTS13 and variants to VWF immobilized on a microtiter plate. FL-A13, delCUB, and MDTCS (0 to 200 nM) were incubated without (control) or with VWF (10 μg/mL, 100 μL per well) immobilized on a microtiter plate for 1 hour. The bound ADAMTS13 and variants were determined by mouse anti-V5 IgG, followed by rabbit antimouse IgG, peroxidase-conjugated and OPD-H2O2. The KD (S) was determined by fitting the data into nonlinear regression. (B) Binding of ADAMTS13 and variants to immobilized VWF on Affi-gel 10. FL-A13, delCUB, MDTCS, and metalloprotease domain (M) (about 50 nM) were incubated at 37°C for 1 hour without (−) or with (+) VWF covalently immobilized onto the Affi-gel 10. After extensive washing with TBS and 20 mM Tris-HCl, pH 7.5, 500 nM NaCl, the bound ADAMTS13 and variants were eluted from the beads with SDS-gel sample buffer and determined by Western blot with anti-V5. The amount of input FL-A13, delCUB, and MDTCS is the same with the signal of only FL-A13 shown in lane 1 (IN).
Direct binding interaction between VWF and the C-terminal fragments of ADAMTS13 under flow
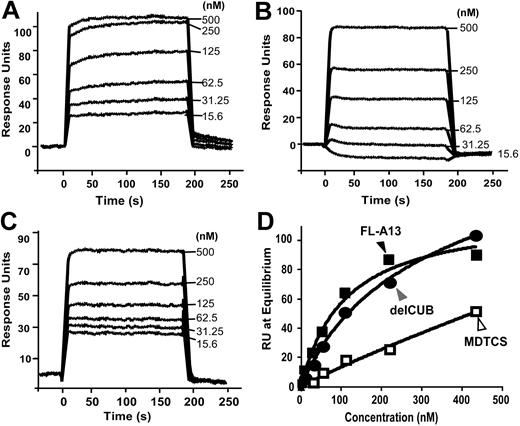
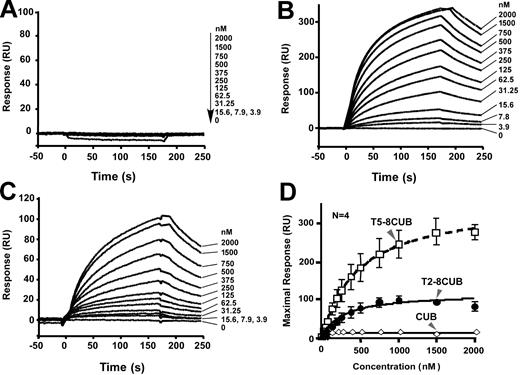
To further determine whether the isolated C-terminal fragments of ADAMTS13 are sufficient to interact with VWF under flow, we injected plasma VWF at various concentrations (0 to 1000 nM) and passed it over the surfaces that were covalently attached by CUB, T5-8CUB, and T2-8CUB. Surprisingly, VWF did not bind the isolated CUB domains detectably but bound the constructs T5-8CUB and T2-8CUB with the KD values of 212 (± 50) nM and 140 (± 36) (means ± SEM), respectively (Figure 6), suggesting that the cooperative activity between the distal TSP1 repeats and the CUB domains may be required for productive binding VWF under flow.
Binding of VWF to the C-terminal fragments of ADAMTS13 under flow. Purified VWF in HBS-T (0 to 500 μg/mL or 0 to 2000 nM) was injected at 20 μL/min for 3 minutes over the CM5 surface covalently coupled to CUB (A), T5-8CUB (B), or T2-8CUB (C). After equilibrium was established, the HBS-T was injected to allow the dissociation phase to be recorded. The KD was determined similarly as described in “Materials and methods.” The entries in panel D represent the means ± SD of 4 repeats (n = 4).
Binding of VWF to the C-terminal fragments of ADAMTS13 under flow. Purified VWF in HBS-T (0 to 500 μg/mL or 0 to 2000 nM) was injected at 20 μL/min for 3 minutes over the CM5 surface covalently coupled to CUB (A), T5-8CUB (B), or T2-8CUB (C). After equilibrium was established, the HBS-T was injected to allow the dissociation phase to be recorded. The KD was determined similarly as described in “Materials and methods.” The entries in panel D represent the means ± SD of 4 repeats (n = 4).
The C-terminal fragments of ADAMTS13 inhibit cleavage of VWF by ADAMTS13 under flow
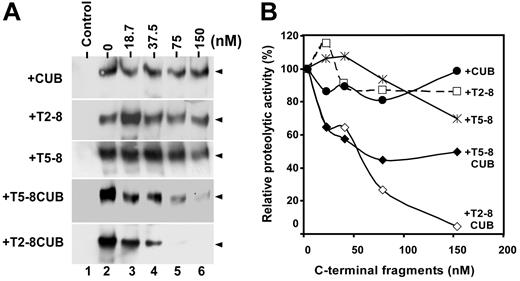
A 5-fold reduction in affinity after removal of the CUB domains suggests these domains play a role in recognition of VWF under flow (Figure 3; Table 1). However, the immobilized CUB domains alone failed to bind the flowing VWF detectably (Figure 6A). The discrepancy may be caused by partial deletion of the binding site within the distal TSP1 repeats or at the junction between the eighth TSP1 repeat and the first CUB domain, which cooperates with the CUB domains for binding VWF; it may be also caused by the unfavorable orientation of the isolated CUB fragment immobilized on the sensor surface. To resolve this discrepancy, we performed a functional inhibition assay on a mini vortexer. Clearly, when added to the reaction, the CUB domains, TSP1 2-8, or TSP1 5-8 fragment did not significantly inhibit cleavage of VWF by full-length ADAMTS13 (Figure 7). However, the T5-8CUB and T2-8CUB blocked cleavage of VWF by ADAMTS13 in a dose-dependent manner (Figure 7). At 150 nM, however, the constructs T5-8CUB and T2-8CUB inhibited proteolytic cleavage of VWF by ADAMTS13 by 75% and 100%, respectively (Figure 7). These data demonstrate that although there may be VWF binding sites present within the TSP1 repeats and the CUB domains, the cooperative activity among these domains appears to be crucial for productive binding and efficient cleavage of VWF under flow.
Inhibition of VWF proteolysis by ADAMTS13 under flow by the C-terminal fragments of ADAMTS13. (A) The C-terminal fragments block cleavage of VWF by ADAMTS13. Purified VWF (18.75 μg/mL or 50 nM) was incubated 10 mM EDTA (control) or 0 to 150 nM of CUB, T2-8, T5-8, T5-8CUB, and T2-8CUB (lanes 2-6) for 60 minutes. ADAMTS13 (50 nM) was then added into the reaction mixture in presence of 50 mM HEPES buffer containing 0.25% BSA, 5 mM CaCl2, and 0.25 mM ZnCl2 (total volume, 20 μL) in a 0.2 mL PCR tube with dome caps. The reaction mixture was subjected to vortexing at a fixed rotation rate of about 2500 rpm (set “8”) for 3 minutes on a mini vortexer. The cleavage of VWF was determined by Western blot with anti-VWF IgG, peroxidase conjugated anti–rabbit IgG and ECL reagents (arrowheads indicate the dimers of 176 kDa). (B) Quantitation of chemiluminescent signal. The signal on X-ray film within the 30 seconds to 1 minute was quantified by densitometry using NIH ImageJ software. The relative proteolytic activity of ADAMTS13 (%) after being inhibited by various C-terminal fragments was plotted against the concentrations of C-terminal fragments of ADAMTS13 added into the reaction.
Inhibition of VWF proteolysis by ADAMTS13 under flow by the C-terminal fragments of ADAMTS13. (A) The C-terminal fragments block cleavage of VWF by ADAMTS13. Purified VWF (18.75 μg/mL or 50 nM) was incubated 10 mM EDTA (control) or 0 to 150 nM of CUB, T2-8, T5-8, T5-8CUB, and T2-8CUB (lanes 2-6) for 60 minutes. ADAMTS13 (50 nM) was then added into the reaction mixture in presence of 50 mM HEPES buffer containing 0.25% BSA, 5 mM CaCl2, and 0.25 mM ZnCl2 (total volume, 20 μL) in a 0.2 mL PCR tube with dome caps. The reaction mixture was subjected to vortexing at a fixed rotation rate of about 2500 rpm (set “8”) for 3 minutes on a mini vortexer. The cleavage of VWF was determined by Western blot with anti-VWF IgG, peroxidase conjugated anti–rabbit IgG and ECL reagents (arrowheads indicate the dimers of 176 kDa). (B) Quantitation of chemiluminescent signal. The signal on X-ray film within the 30 seconds to 1 minute was quantified by densitometry using NIH ImageJ software. The relative proteolytic activity of ADAMTS13 (%) after being inhibited by various C-terminal fragments was plotted against the concentrations of C-terminal fragments of ADAMTS13 added into the reaction.
Discussion
Present study demonstrates that multimeric VWF can be readily cleaved by recombinant or plasma ADAMTS13 within 3 minutes under mechanic-induced shear stress on a mini vortexer. The cleavage is specific at the Tyr1605-Met1606 bond as shown by the presence of dimers of 176 kDa. The VWF proteolysis is rotation speed– (Figure 2A) and ADAMTS13 concentration–dependent (Figure 2B). Addition of EDTA (10 mM) or omission of ADAMTS13 enzyme into the reaction abrogates cleavage of VWF (Figure 2C), confirming the specific cleavage of VWF by ADAMTS13, not simply by the mechanic-induced shear stress. VWF can also be cleaved by normal human plasma but not by TTP-patient plasma in presence of heparin (Figure 2B), suggesting that the simple flow-based assay may be applicable to determine plasma ADAMTS13 activity in patients with congenital and acquired TTP.
ADAMTS13 does not bind or cleave native VWF in absence of flow shear stress or denaturing regents. However, how much shear stress is required for ADAMTS13 to interact with VWF remains unclear. An early study has shown that a 1500 s−1 shear rate may be required to detect VWF proteolysis by plasma ADAMTS13 enzyme.35 Yet, in a mouse model, thrombi are formed in the venules of the mesentery (shear rate of about 200 to 250 s−1) in adamts13−/− mice after topical fusion of calcium ionophore A23187 but not in adamts13+ / + mice or in adamts13−/− mice supplemented with recombinant ADAMTS13 protein via tail vein injection,36 suggesting that ADAMTS13 and VWF interaction may occur at low shear stress. Consistent with this hypothesis, our data show that the cleavage of VWF is detectable at low vortexing-rotation speed (about 640 rpm); the cleavage product accumulates in a rotation speed–dependent manner and reaches the plateau at the rotation rates between about 2500 rpm and about 3200 rpm (with an estimated shear rate of about 12 000 s−1) (Figure 2A). On a BIAcore system, the multimeric plasma VWF binds ADAMTS13 at injection rate of 10 μL/min (shear rates about 500 s−1), but the affinity is not enhanced with increasing injection rates up to 100 μL/min (with shear rate of about 5000 s−1; Figure 3A). These data indicate that ADAMTS13 may be physiologically important in preventing thrombus formation in both arterioles and venules.
Although the N-terminal domains of ADAMTS13 appear to be sufficient to bind and cleave VWF under denatured and static condition,22–24;29 the C-terminal domains are clearly required for recognition of VWF under flow. A removal of the CUB domains (delCUB) or more (MDTCS) significantly impairs ADMATS13's ability to cleave VWF under vortex-induced mechanic shear stress (Figure 2C). Yet, these C-terminal truncated variants at the concentrations tested are able to cleave guanidine-HCl–denatured VWF (Figure 2D) or GST-VWF73 (Figure 2E,F) or FRETS-VWF73 (data not shown) with more or similar efficacy compared with full-length ADAMTS13. Analysis on the BIAcore system has also shown that full-length ADAMTS13 binds VWF in high affinity (KD of about 50 nM). The removal of the CUB domains results in about 5-fold decrease in the binding affinity (Table 1; Figure 3), and further removal of the TSP1 2-8 repeats almost completely abolishes its ability to bind VWF under flow. Again, predenatured VWF is able to bind ADAMTS13 substantially, with a KD of about 330 nM, comparable with that of the construct delCUB (Table 1). These data indicate that the C-terminal TSP1 repeats and CUB domains participating in substrate recognition under flow and the predenaturization of VWF exposes additional cryptic sites that are otherwise not available under flow alone.
To determine whether the CUB domains are sufficient to bind VWF under flow or whether the other adjacent structure is required for binding, we performed the direct binding and competition inhibition assays with various purified C-terminal fragments of ADAMTS13. We show that the isolated CUB domains are not able to bind VWF under flow detectably on BIAcore system (Figure 6A) or microtiter plate (data not shown). Neither does the CUB fragment, nor does the TSP1 2-8 or T5-8 fragment, inhibit the cleavage of VWF by recombinant ADAMTS13 in a flow-based assay (Figure 7). However, addition of the TSP1 5-8 repeats to the CUB domains (construct T5-8CUB) restores the binding affinity toward VWF under flow (KD of about 212) (Figure 6) and their inhibitory potency toward cleavage of VWF by ADAMTS13 (Figure 7). Further addition of the TSP1 2-4 repeats (construct T2-8CUB) increases the affinity by about 1.5-fold (Figure 6) and their inhibitory potency (Figure 7), suggesting that the cooperative activity between the distal TSP1 repeats and the CUB domains is critical for productive recognition of VWF under flow. The cooperative binding of the TSP1 and the CUB domains to VWF may trigger the flow-induced VWF conformational change and expose its other cryptic binding sites for the N-terminal domains (such as Cys-rich and spacer domains) of ADAMTS13, resulting in cleavage of the Tyr1605-Met1606 bond in the VWF-A2 subunit. Alternatively, the CUB domains may be required to present or orientate the TSP1 repeats for high-affinity interaction with the unfolded VWF by flow shear stress.
Our data are consistent with some but not all of observations made by others using a parallel-flow chamber assay.27,28 For example, the short construct MDTCS was shown to be more active in removing the stringlike structure on the endothelial surface.27 In addition, the recombinant fragments consisting of the first CUB domain or both CUB domains but not second CUB domain immobilized on the microtiter plate or microbeads were able to bind VWF in solution or on the endothelial cells.37 The peptides derived from the CUB domains inhibit the cleavage of stringlike structure by plasma or recombinant ADAMTS13.37 The discrepancy between our results and the data published so far may be caused by the different assays used. Our activity assay directly detects the accumulation of the specific cleavage product (dimers of 176 kDa) by Western blot; it is highly sensitive and reproducible. Our binding assay on BIAcore system detects ADAMTS13 and VWF interaction under flow in real time and in a purified system without additional detection steps that may disrupt equilibrium binding. In contrast, the parallel-flow chamber assay detects the disappearance of the platelet-VWF strings and is only an indirect estimate of the breaking down of VWF from endothelial cell surface,38,39 which is highly complex and involves live endothelial cells, labeled or unlabeled platelets, histamine stimulation, and VWF–endothelial cell interactions.27,28,38,39 This makes the data interpretation less certain and quantitative. However, it might be possible that certain proteins or nonprotein cofactors in plasma or on the surface of endothelial cells or platelets rescue the defect in proteolytic activity of the C-terminal truncated ADAMTS13 variants. For example, addition of heparin or binding of platelet glycoprotein 1b to VWF moderately increased the proteolytic cleavage of VWF by ADAMTS13 under static and denatured condition.40 However, such cofactors that may enhance VWF proteolysis by ADAMTS13 under flow are yet to be identified.
In summary, we demonstrate that multimeric VWF can be readily cleaved by full-length recombinant and plasma-derived ADAMTS13 but not by the C-terminal truncated variants under the vortex-induced mechanic shear stress. The interaction between VWF and ADAMTS13 under flow is a high-affinity one. The cooperative activity between the middle C-terminal TSP1 repeats and the distal C-terminal CUB domains of ADAMTS13 appears to be crucial for productive recognition and cleavage of VWF under flow.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by grants from the National Institutes of Health (R01HL079027 and P50 HL081012 [X.L.Z.] and R01 HL 078726 [B.S.S.]).
We thank Dr Sriram Krishnaswamy at the Children's Hospital of Philadelphia and Dr Scott Diamond at Institute for Medicine and Engineering, University of Pennsylvania, for their helpful advise on kinetic data analysis and flow-based assays.
National Institutes of Health
Authorship
Contribution: P.Z. designed and performed experiments and analyzed the data; W.P., A.H.R., and B.S.S. provided technical assistance and experimental design and analysis of the BIAcore data, and revised the manuscript; and X.L.Z. designed research, analyzed data, and wrote and revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: X. Long Zheng, Department of Pathology and Laboratory Medicine, the Children's Hospital of Philadelphia and the University of Pennsylvania, 34th St and Civic Center Blvd, ARC 816G, Philadelphia, PA 19104; e-mail: zheng@email.chop.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal