Abstract
The JAK2V617F mutation does not elucidate the phenotypic variability observed in myeloproliferative neoplasm (MPN) families. A putative tumor suppressor gene, TET2, was recently implicated in MPN and myelodysplastic syndromes through the identification of acquired mutations affecting hematopoietic stem cells. The present study analyzed the TET2 gene in 61 MPN cases from 42 families. Fifteen distinct mutations were identified in 12 (20%) JAK2V617F-positive or -negative patients. In a patient with 2 TET2 mutations, the analysis of 5 blood samples at different phases of her disease showed the sequential occurrence of JAK2V617F and TET2 mutations concomitantly to the disease evolution. Analysis of familial segregation confirmed that TET2 mutations were not inherited but somatically acquired. TET2 mutations were mainly observed (10 of 12) in patients with primary myelofibrosis or patients with polycythemia vera or essential thrombocythemia who secondarily evolved toward myelofibrosis or acute myeloid leukemia.
Introduction
Families of myeloproliferative neoplasms (MPNs) are characterized by a clinical and genetic heterogeneity. First, within MPN families, distinct clinical entities are observed, the 3 main ones being polycythemia vera (PV), essential thrombocythemia (ET), and primary myelofibrosis (PMF).1,2 Second, disease evolution can be highly variable within families presenting with the same type of MPN.2,3 The acquired JAK2V617F mutation present in most PV and half of ET and PMF sporadic or familial cases does not totally explain the phenotypic variability.2-7 The existence of additional molecular events, either germline or acquired, may explain the MPN predisposition and the distinct phenotypes observed within MPN families.8-10
Recently, acquired mutations of the Ten-Eleven Translocation 2 gene, (TET2),were reported in approximately 12% of sporadic MPN (PV, ET, and PMF) as well as in myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML).11 By in vitro clonal assays, Delhommeau et al showed that TET2 defects target both multipotent and committed progenitors and were associated with a selective advantage of early hematopoiesis.11 These recent findings led us to analyze TET2 in familial MPN cases to determine whether TET2 could be a predisposition factor, to estimate the prevalence of acquired TET2 events in MPN cases, and to describe the clinical profile of patients with TET2 mutations.
Methods
Families with at least 2 affected patients with MPN were collected through a national network.2 The diagnoses of MPN were reviewed based on the 2008 World Health Organization criteria.12 The study was approved by the French Comité de Protection des Personnes. All participants gave their written informed consent in accordance with the Declaration of Helsinki.
We analyzed 61 patients from 42 MPN families, 40 European and 2 African (F3 and F4). Thirty-three patients displayed a simple phenotype consisting of PV (14), ET (12), or PMF (7) with no observed hematologic evolution of the disease after a mean follow-up period of 12 years. Twenty-eight other patients had experienced an evolution in their MPD phenotype with a mean follow-up of 14.3 years: PV evolving into myelofibrosis (post-PV MF, 6) and/or into AML (12), ET evolving into MF (4) and/or AML (5), or PMF turning into AML (1).
TET2 molecular analysis
Amplifications of TET2 (NM_001127208.1) were performed either on genomic DNA extracted from mononuclear blood cells and from buccal swabs or directly on hematopoietic colonies after heating at 95°C for 10 minutes. Purified polymerase chain reaction (PCR) products were sequenced using the BigDye Terminator chemistry (Applied Biosystems) and run on an ABI3100 capillary sequencer.
The search for large genomic rearrangement was performed by quantitative multiplex PCR of short fluorescent fragments and applied to patients homozygous for all TET2 polymorphisms to exclude hemizygosity of the region. Primer pairs were designed for exons 3, 6, and 11. PCR products were separated on an ABI3100 capillary sequencer. Analysis using GeneMapper, Version 4.0 (Applied Biosystems), is based on the comparison of the peak heights generated from the tested DNA sample and the control DNA. A heterozygous exon deletion will lead to a 2-fold reduction of the height of the corresponding peak. Primer sequences for TET2 sequencing and quantitative multiplex PCR of short fluorescent fragments are shown in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Cell culture
Peripheral blood granulocytes were isolated using standard protocols. Thawed cells were sorted on the CD34 and CD38 antigens using a FACSDiva cell sorter. CD34+CD38+ cells were seeded in methylcellulose medium supplemented with erythropoietin, interleukin-3, and stem cell factor. Hematopoietic colonies were picked at day 14.
Results and discussion
Sixty-one patients were screened for mutations in the 6009-bp coding sequence of the TET2 gene, composed of 11 exons spanning 130 kb. Fifteen distinct molecular defects, including 11 (73%) truncating mutations, 3 (20%) missense mutations, and one whole gene deletion, were identified (Table 1). They were spread throughout the gene. The 11 truncating mutations consisted of 3 nonsense mutations, 6 out-of-frame insertions/deletions, and 2 splice site mutations. The 3 missense mutations affected amino acids that were conserved in at least one of the TET2 paralogs, TET1 and TET3, and in TET2 orthologs.13,14 Two, p.Leu1340Pro and p.His1868Arg, were located in the 2 highly conserved TET2 functional domains (1134-1444 and 1842-1921) of the Drosophila ortholog.13 Furthermore, all 3 missense mutations were absent from 165 control individuals of ethnically matched populations, leading us to consider them as putative mutations. The proportion and the type of TET2 mutations identified in familial MPNs were similar to the ones found in sporadic MPN cases.11,15 TET2 mutations have also been reported in other myeloid malignancies.11,15-19
Clinical profile of the 12 patients with TET2 mutations
| . | P1 (F1) . | P2 (F2) . | P3 (F2) . | P4 (F3) . | P5 (F3) . | P6 (F4) . | P7 (F4) . | P8 . | P9 . | P10 . | P11 . | P12 . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex | Female | Female | Female | Female | Male | Female | Female | Female | Male | Female | Male | Male |
| Diagnosis | PV | PV | PV | ET > PV | ET | ET | ET | ET | PMF | PMF | PMF | PV |
| Age at diagnosis, y | 75.3 | 62.1 | 58.4 | 37.5 | 34.8 | 47 | 48.6 | 45.3 | 78 | 77.2 | 64.3 | 54.6 |
| Overall duration of the disease, y | 5.9 | 16.3 | 16.4 | 18.1 | 7.3 | 1.2 | 6.2 | 29.2 | 3.5 | 4.3 | 3.6 | 13.2 |
| Outcome | Alive | Alive | Alive | Deceased | Deceased | Deceased | Alive | Alive | Deceased | Deceased | Deceased | Alive |
| Complications | ||||||||||||
| Disease duration at first complication, y | 4.7 | 13 | — | 15.7 | 7.1 | 1.0 | — | 16.0 | — | — | — | 9.4 |
| Myelofibrosis, yes/no | Yes | Yes | No | Yes | Yes | No | No | Yes | — | — | — | Yes |
| Leukemic transformation, yes/no | No | No | No | Yes | Yes | Yes | No | No | — | — | — | No |
| JAK2V617F | ||||||||||||
| Disease duration at first observance, y | 4.6 | 14.4 | 14.5 | 8.4 | 3.6 | 1.0 | 5.7 | 20.3 | 0.2 | 2.9 | 2.7 | 3.4 |
| Allele burden, % | 95 | 65 | 50 | 25 | 0 | 0 | 40 | 90 | 35 | 35 | 65 | 80 |
| TET2 mutations | ||||||||||||
| Disease duration at first observance, y | 4.6 | 14.4 | 14.5 | 8.4/15.1* | 3.6 | 1.0 | 5.7 | 20.3 | 0.2 | 2.9 | 2.7 | 3.4 |
| TET2 allele burden, % | 50 | 25 | 20 | 5 / 40† | 40 | 50 / 35 | 50 | 45 / 60 | 45 | 45 | 20 | |
| Location | Exon 11 | Intron 7 | Exon 3 | Exon 3 | Exon 3 | Exon 3 | Intron 4 | All exons | Exons 3/11 | Exon 3 | Exon 8 | Exon 11 |
| Nucleotide change | c.5695delC | c.3954+2T>A | c.3138delT | c.1648C>T/c.2570delA | c.2058A>T | c.1955delA/c.2490dupA | c.3500+3A>C | c.1_6009del | c.1720C>T/c.4999_5014del16 | c.694C>T | c.4019T>C | c.5603A>G |
| Protein change | p.Leu1899fs | p. ? | p.Thr1047fs | p.Arg550X/Asn857fs | p.Arg686Ser | p.Gln652fs/p.Gln831fs | p. ? | p. ? | p.Gln574X/p.Leu1667fs | p.Gln232X | p.Leu1340Pro | p.His1868Arg |
| . | P1 (F1) . | P2 (F2) . | P3 (F2) . | P4 (F3) . | P5 (F3) . | P6 (F4) . | P7 (F4) . | P8 . | P9 . | P10 . | P11 . | P12 . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex | Female | Female | Female | Female | Male | Female | Female | Female | Male | Female | Male | Male |
| Diagnosis | PV | PV | PV | ET > PV | ET | ET | ET | ET | PMF | PMF | PMF | PV |
| Age at diagnosis, y | 75.3 | 62.1 | 58.4 | 37.5 | 34.8 | 47 | 48.6 | 45.3 | 78 | 77.2 | 64.3 | 54.6 |
| Overall duration of the disease, y | 5.9 | 16.3 | 16.4 | 18.1 | 7.3 | 1.2 | 6.2 | 29.2 | 3.5 | 4.3 | 3.6 | 13.2 |
| Outcome | Alive | Alive | Alive | Deceased | Deceased | Deceased | Alive | Alive | Deceased | Deceased | Deceased | Alive |
| Complications | ||||||||||||
| Disease duration at first complication, y | 4.7 | 13 | — | 15.7 | 7.1 | 1.0 | — | 16.0 | — | — | — | 9.4 |
| Myelofibrosis, yes/no | Yes | Yes | No | Yes | Yes | No | No | Yes | — | — | — | Yes |
| Leukemic transformation, yes/no | No | No | No | Yes | Yes | Yes | No | No | — | — | — | No |
| JAK2V617F | ||||||||||||
| Disease duration at first observance, y | 4.6 | 14.4 | 14.5 | 8.4 | 3.6 | 1.0 | 5.7 | 20.3 | 0.2 | 2.9 | 2.7 | 3.4 |
| Allele burden, % | 95 | 65 | 50 | 25 | 0 | 0 | 40 | 90 | 35 | 35 | 65 | 80 |
| TET2 mutations | ||||||||||||
| Disease duration at first observance, y | 4.6 | 14.4 | 14.5 | 8.4/15.1* | 3.6 | 1.0 | 5.7 | 20.3 | 0.2 | 2.9 | 2.7 | 3.4 |
| TET2 allele burden, % | 50 | 25 | 20 | 5 / 40† | 40 | 50 / 35 | 50 | 45 / 60 | 45 | 45 | 20 | |
| Location | Exon 11 | Intron 7 | Exon 3 | Exon 3 | Exon 3 | Exon 3 | Intron 4 | All exons | Exons 3/11 | Exon 3 | Exon 8 | Exon 11 |
| Nucleotide change | c.5695delC | c.3954+2T>A | c.3138delT | c.1648C>T/c.2570delA | c.2058A>T | c.1955delA/c.2490dupA | c.3500+3A>C | c.1_6009del | c.1720C>T/c.4999_5014del16 | c.694C>T | c.4019T>C | c.5603A>G |
| Protein change | p.Leu1899fs | p. ? | p.Thr1047fs | p.Arg550X/Asn857fs | p.Arg686Ser | p.Gln652fs/p.Gln831fs | p. ? | p. ? | p.Gln574X/p.Leu1667fs | p.Gln232X | p.Leu1340Pro | p.His1868Arg |
— indicates not applicable.
Disease duration at first observation for each mutation.
TET2 allele burden at first observation for each mutation.
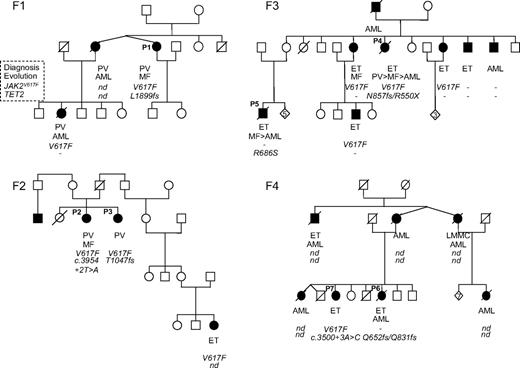
All mutations identified were each found in a single patient, and segregation with the phenotype could never be observed in the families (Figure 1). Moreover, in 3 families (F2, F3, and F4), different TET2 mutations were identified in affected members. Sequence analysis of buccal swabs available for patients P1 (F1) and P4 (F3) showed the absence of the TET2 mutations. Therefore, the analysis of the 42 families showed that TET2 is not a major predisposing gene factor in familial MPN and that identified TET2 defects were acquired events.
Pedigrees of 4 MPN families (F1, F2, F3, and F4) showing that TET2 mutations do not segregate with the MPN phenotype. Filled symbols represent patients; their clinical phenotype is indicated below. Under each symbol, the first top line represents the phenotype at the time of diagnosis; the second line, the evolution (blank where there is none); the third line, the JAK2V617F status (V617F when the mutation was found; – otherwise); and the fourth line, any TET2 mutation. Mutations are annotated in amino acid one-letter code. nd indicates not done.
Pedigrees of 4 MPN families (F1, F2, F3, and F4) showing that TET2 mutations do not segregate with the MPN phenotype. Filled symbols represent patients; their clinical phenotype is indicated below. Under each symbol, the first top line represents the phenotype at the time of diagnosis; the second line, the evolution (blank where there is none); the third line, the JAK2V617F status (V617F when the mutation was found; – otherwise); and the fourth line, any TET2 mutation. Mutations are annotated in amino acid one-letter code. nd indicates not done.
Two distinct TET2 mutations were found in 3 unrelated patients (P4, P6, and P9, Table 1). We picked 3 individual colonies derived from CD34+CD38+ cells from the blast phase of patient P4; and using allele-specific PCR, we found that each TET2 mutation affected a different allele of the same colony (data not shown). In addition, we showed in mononuclear cells from patient P6 that the 2 exon 3 mutations were not located in cis. These observations are in favor of a biallelic inactivation of TET2, in agreement with the hypothesis of a tumor suppressor role of TET2.11
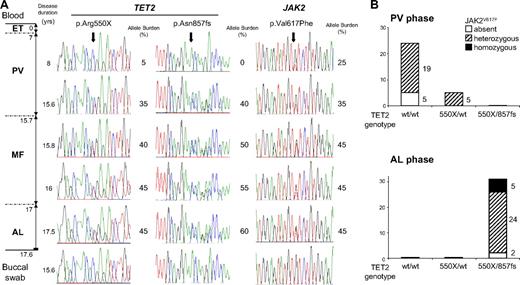
In patient P4 for whom 5 blood samples were available throughout the last 3 steps of her evolution, PV, MF, and AML (Figure 2A), sequence analysis showed that JAK2V617F and the TET2 p.Arg550X mutation were present at the PV stage and later on. The p.Asn857fs mutation only started being detectable in the second sample, 7 years later and 5 months before the diagnosis of MF. The sequential analysis showed that the burden of JAK2 and TET2 mutant alleles grew in time, concomitantly with the development of the disease. Blood progenitor cells were available at the PV and blast phases (Figure 2B). During the PV stage, endogenous erythroid colonies carried the p.Arg550X mutation (5 of 29) but p.Asn857fs was never observed and 19 of 29 clones were found JAK2V617F-positive in the absence of any TET2 mutation. These results suggest the initial occurrence of JAK2V617F followed by TET2 p.Arg550X and subsequently by the TET2 p.Asn857fs defect. After leukemic transformation, all colonies but 2 (29 of 31) carried JAK2V617F and both TET2 mutations. We genotyped the rs16922579 SNP located in intron 9 of JAK2 and heterozygous in patient P4. A homozygous G/G genotype was found in JAK2V617F-negative colonies, whereas G/A and A/A genotypes were observed in JAK2V617F heterozygous and homozygous colonies, respectively. These preliminary results suggest a loss of the JAK2V617F allele through mitotic recombination during disease progression. In contrast, in 2 other studies, mitotic recombination was excluded in 2 informative patients who transformed to AML and for whom all blast cells were JAK2V617F-negative.20,21
Sequential study of TET2 mutations and JAK2V617F in patient P4 (F3) in mononuclear cells and committed progenitors. (A) Sequence electrophorograms are shown for each TET2 mutation and for JAKV617F. (Left diagram) The different phases of the disease with their time lapse from diagnosis. (Right diagram) The disease duration (in years) at each sample date. Allele burdens of TET2 mutations and JAK2V617F, semiquantitatively estimated by sequencing as Bellanne-Chantelot et al,2 are indicated. (B) Histograms show for PV and acute leukemia (AL) phases the 3 distinct TET2 genotypes (each bar corresponding to a specific genotype, wt/wt, 550X/wt, 550X/857fs). The JAK2V617F mutation was absent, heterozygous, or homozygous. The number of genotyped clones is indicated for each group.
Sequential study of TET2 mutations and JAK2V617F in patient P4 (F3) in mononuclear cells and committed progenitors. (A) Sequence electrophorograms are shown for each TET2 mutation and for JAKV617F. (Left diagram) The different phases of the disease with their time lapse from diagnosis. (Right diagram) The disease duration (in years) at each sample date. Allele burdens of TET2 mutations and JAK2V617F, semiquantitatively estimated by sequencing as Bellanne-Chantelot et al,2 are indicated. (B) Histograms show for PV and acute leukemia (AL) phases the 3 distinct TET2 genotypes (each bar corresponding to a specific genotype, wt/wt, 550X/wt, 550X/857fs). The JAK2V617F mutation was absent, heterozygous, or homozygous. The number of genotyped clones is indicated for each group.
The TET2 defects with an allele burden semiquantitatively estimated by sequencing varied from 20% to 60% and were identified in 12 patients (12 of 61, 20%) diagnosed with PV (4 of 32), ET (5 of 21), and PMF (3 of 8; Table 1). All but 2 patients were positive for the JAK2V617F mutation. Altogether, 20% of the JAK2V617F-positive patients were found mutated for TET2 (10 of 49) and 17% of the JAK2V617F-negative patients (2 of 12). The 2 negative cases were ET patients who developed very active AML and died rapidly (P5 and P6, Table 1). In contrast to results recently reported, we did not observe an older age for patients with TET2 mutations (supplemental Table 1).15
Twenty-nine percent (10 of 35) of patients with PMF or with hematologic complications (post-PV/ET MF or AML) were found mutated in TET2 compared with 7.7% (2 of 26) of patients without any diagnosed hematologic complications after a mean disease duration of 12 years.
We first performed a univariate analysis to compare the incidence of transformation between the 2 groups (supplemental Table 1). The results indicated a trend toward hematologic complications in TET2-mutated patients, although this difference was not statistically significant (P = .055). This was confirmed by a multivariate analysis (supplemental Table 1). Similarly, a trend toward malignant proliferation was recently reported in MDS/MPN syndromes.18 Studies on larger cohorts of MPN cases would be required to estimate the prevalence of TET2 events in patients with more aggressive outcome.
Recent data showed that TET1 catalyzes conversion of 5-methylcytosine to 5-hydroxy-methylcytosine.22 TET gene family may play an important role in epigenetic regulation of stem cell functions. In addition, it was shown that deletion of TET2 induced a clonal advantage at the level of early hematopoietic progenitors.11 This may explain that patients with a defect in TET2 are prone to progress to MF. Moreover, TET2 defects may be a primary or secondary event, before or after JAK2V617F as reported for the 20q- deletion.21 In future studies, it will be important to understand whether different kinetics of occurrence of these 2 independent genetic defects have the same impact on the clinical course of the disease and are both associated with a clonal dominance at early hematopoietic stages.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Nicole Casadevall for helpful discussions, Myriam Labopin for statistical analysis, and the patients and their families for their participation in this study.
This work was supported by grants from the Institut National du Cancer (INCa; 2008), GIS-Maladies Rares, Project A0709ILS (F.D., C.B.-C.), Programme Hospitalier de Recherche Clinique (AOR07014; A.N., C.B.-C., G.L.), and Association pour la Recherche sur la Moelle Osseuse (A.N.). S.D. is a Ministère de la Recherche et de la Technologie recipient. I.P. is supported by the Cancéropole Ile de France. F.D. is the recipient of a European Hematology Association fellowship (2005/27), the Myeloproliferative Disorders Foundation (Investigator grant), and the Laurette Fugain Association and the Fondation de France.
Authorship
Contribution: C.B.-C., F.D., W.V., and A.N. drew the original study design; D.B., C.C., A. Delannoy, A. Devidas, M.G.-P., F.I., A.N., G.P., Y.P., W.V., and the French Group of Familial Myeloproliferative Disorders recruited the patients; A.N. recorded all clinical and hematologic data; C.S.-M., G.L., and C.J. performed molecular analyses; F.D., S.D., I.P., and O.B. assisted with interpretation of research; C.B.-C. and C.S.-M. analyzed data and wrote the manuscript; and A.N., F.D., S.D., and W.V. critically reviewed the manuscript. All authors contributed to the amendment of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A complete list of the French Group of Familial Myeloproliferative Disorders appears in the supplemental Appendix.
Correspondence: Christine Bellanné-Chantelot, Centre de Génétique Moléculaire et Chromosomique, Groupe Hospitalier Pitié Salpêtrière, 47-83 blvd de l'Hôpital, 75651 Paris Cedex 13, France; e-mail: christine.bellanne-chantelot@psl.aphp.fr.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal