Abstract
Previous European studies suggest NOD2/CARD15 and interleukin-23 receptor (IL-23R) donor or recipient variants are associated with adverse clinical outcomes in allogeneic hematopoietic stem cell transplantation. We reexamined these findings as well as the role of another inflammatory bowel disease (IBD) susceptibility gene (immunity-related GTPase family, M [IRGM]) on transplantation outcomes in 390 US patients and their matched unrelated donors, accrued between 1995 and 2004. Patients received T-replete grafts with mostly myeloablative conditioning regimens. Multivariate analyses were performed for overall survival, disease-free survival, transplantation-related mortality, relapse, and acute and chronic graft-versus-host disease. Of 390 pairs, NOD2/CARD15 variant single nucleotide polymorphisms (SNPs) were found in 14% of donors and 17% of recipients. In 3% both donor and recipient had a mutant SNP. Thirteen percent of donors and 16% of recipients had variant IL23R SNPs, with 3% having both donor and recipient variants. Twenty-three percent of both donors and recipients had variant IRGM SNPs. None of the 3 IBD-associated alleles showed a statistically significant association with any adverse clinical outcomes. Our results do not support an association between the 3 IBD-associated SNPs and adverse outcomes after matched unrelated donor hematopoietic cell transplantations in US patients.
Introduction
Allogeneic hematopoietic stem cell transplantation (HSCT) is an important and potentially curative intervention in the management of a range of hematologic malignancies, but the efficacy and outcome of this treatment is frequently limited by its inherent immunologic sequelae. Among the dominant complications of HSCT, graft-versus-host disease (GVHD) remains a main cause of morbidity and mortality, and considerable attention has focused on strategies to limit its development while not completely eliminating the beneficial graft-versus-leukemia effect.1 These strategies have included allelic human leukocyte antigen (HLA) matching, appropriate donor selection, T-cell depletion, and other prophylactic immunosuppressive regimens.
Earlier observations showed extensive gastrointestinal mucosal damage in patients with GVHD, raising the suggestion that resultant bacterial translocation might play a role in both the initiation and maintenance of a systemic inflammatory response as a result of the conditioning regimen and which could potentially represent a key inciting event in the pathogenesis of GVHD.2 These observations, coupled with an extensive body of literature that points to the role of bacterial flora in the cause of idiopathic inflammatory bowel disease (IBD), have together reinforced the concept that luminal bacterial communities may represent key elements in the host response to injury.3 Indeed, genomewide association studies have identified more than 30 distinct loci linked with susceptibility to Crohn disease, including genes involved in innate host defense.4-7 Accordingly, an emerging consensus suggests that GVHD and IBD, both immune-mediated inflammatory conditions, may share overlapping pathways in their pathogenesis.
Among the many candidate IBD-associated genes, NOD2/CARD15 was initially identified as a susceptibility gene in Crohn disease with a frequency of 7.7% to 9.1% in the healthy US population.8 NOD2/CARD15 encodes a cytosolic protein that recognizes a bacterial cell wall component, muramyl dipeptide, and results in downstream activation of innate pathways of host defense. Three informative single nucleotide polymorphisms (SNPs) in NOD2/CARD15 have been identified, including Leu1007fsinsC, a cytosine insertion that produces a 33 amino acid protein truncation, and 2 missense mutations Arg702Trp and Gly908Arg (SNPs 8, 12, and 13, respectively).9,10
The importance of these allelic variants in the pathogenesis of GVHD was suggested by Holler et al,11 who initially demonstrated that SNPs 8, 12, and 13 of the NOD2/CARD15 gene in either donor or recipient (from a European cohort) was associated with increased incidence and severity of acute GVHD, increased transplantation-related mortality (TRM), and reduced survival in HLA-matched sibling transplantations. These effects were most pronounced in pairs with the risk variant in both donor and recipient.11 A follow-up study by the same group showed an increased risk of severe acute GVHD, but not mortality in unrelated donor (URD) transplantations. In addition, when broken down by individual polymorphism, SNP 13 was the only significant risk factor.12 Although these findings suggested the importance of NOD2/CARD15 variants in GVHD, other studies have shown conflicting results that failed to confirm these associations.13-15
Accordingly, the aim of the current study was to evaluate the effect of NOD2/CARD15 and 2 other IBD-associated SNPs on the outcomes of HSCT in a US population. We selected IRGM (immunity-related GTPase family, M), a gene product involved in autophagy and intracellular pathogen clearance, because variations in or near the IRGM locus have been associated with increased susceptibility to Crohn disease,16 and IL23R (interleukin-23 receptor) because variants appear to have a protective role in the development of IBD.17 In addition, recent findings in adults18 and, more recently, in children19 indicated a protective effect of the variant IL23R in donors from the development of GVHD after HSCT. Nevertheless, our findings fail to show a significant effect (at the P < .01 level) of variants in these SNPs and the outcomes of HSCT.
Methods
Patient populations and treatment regimen
Pretransplantation samples from 390 patients and their 10 of 10 HLA-A–, -B–, -C–, -DRB1, and -DQB1–matched unrelated donors from 85 different centers within the United States were provided by the National Marrow Donor Program Research Repository (NMDP). Patients received T-replete transplants for early stage acute myeloid leukemia, acute lymphoblastic leukemia, chronic myeloid leukemia, myelodysplastic syndrome accrued between 1995 and 2004. The majority (98%) underwent a myeloablative conditioning regimen (Table 1). GVHD prophylaxis used combinations of cyclosporine, tacrolimus, and methotrexate as detailed in Table 1. Gut decontamination was not routinely performed, but the prevalence of this practice varies widely and was not systematically recorded in the data collected. Patients and donors were of predominately European American descent, 92.2% and 94.5%, respectively, with 6.7% racial/ethnic minorities (Table 1). Median time of follow-up was 77 months (range, 12-149 months). Approval was obtained from the Washington University Human Studies Committee, and all patients and donors gave their informed and written consent to the NMDP for genetic analysis of their biologic samples in accordance with the Declaration of Helsinki.
Genotyping of SNPS
DNA was extracted from frozen whole blood samples collected from donors and patients before transplantation with the use of QIAGEN Blood Kit with Proteinase K. Genomic DNA was isolated with the use of a QIAmp DNA Blood Mini Kit (QIAGEN) following the manufacturer's instructions. Control samples of each genotype, which were confirmed by sequencing, were included in each run. DNA was genotyped for 3 NOD2/CARD15 SNPs (rs2066844, rs2066845, and rs2066847), 2 IL23R SNPs (rs11209026 and rs11465804), and 2 IRGM SNPs (rs4958847 and rs13361189) with the use of Sequenom and/or Taqman SNP Genotyping Assay (PE Applied Biosystems). Each reaction was performed in a 96-well plate. The optimized reaction mix of a final volume of 50 μL consisted of Universal Master Mix and the DNA template. The sequence of the primers used is available on request.
Data analysis and statistical considerations
Persons heterozygous for at least 1 of the 3 NOD2/CARD15 SNPs analyzed (SNPs 8, 12, and 13) were considered variant, whereas persons lacking these SNPs were considered wild type. Clinical variables analyzed include cytomegalovirus status, disease, graft source, GVHD prophylaxis, Karnofsky score, donor and patient ages, donor/recipient sex mismatch, myeloablative versus nonmyeloablative conditioning, and year of transplantation. Diagnosis for acute or chronic GVHD was based on clinical signs, and/or histopathologic findings in biopsies from skin, liver, and the gastrointestinal tract. Grading of acute GVHD was performed and based on the Glucksberg criteria.20 Multivariate analyses were performed with Cox proportional hazards regression analysis for overall (OS) and disease-free survival, TRM, relapse, and acute and chronic GVHD, adjusting for the previously mentioned clinical variables. Because of the multiple testing performed within the multivariate analysis, a P value of less than .01 is considered significant. SAS software Version 9.1 (SAS Institute) was used in all analyses.
Results
Patient characteristics
The distribution by age, sex, and disease is shown in Table 1. Most (74%) patients receiving transplants were between the ages of 20 and 50 years, with the majority (63%) being performed for chronic myeloid leukemia. The majority of patients underwent bone marrow grafting (81%), and 98% of subjects underwent a myeloablative conditioning regimen before transplantation (Table 1).
Characteristics of patients with AML, ALL, CML, or MDS in early disease stage receiving either a BM or PBSC first transplant between 1995 and 2004 with a myeloablative conditioning regimen and having an allele-level matched donor at HLA-A, -B, -C, -DRB1, and -DQB1 from the National Marrow Donor Program
| Variable . | No. evaluated . | Value . |
|---|---|---|
| No. of patients | 390 | |
| No. of centers | 85 | |
| Median age, y, (range) | 390 | 37 (< 1 to 59) |
| Age at transplantation, n (%) | 390 | |
| 0-9 y | 8 (2) | |
| 10-19 y | 44 (11) | |
| 20-29 y | 69 (18) | |
| 30-39 y | 107 (27) | |
| 40-49 y | 113 (29) | |
| 50 y and older | 49 (13) | |
| Male sex, n (%) | 390 | 211 (54) |
| Karnofsky before transplantation > 90, n (%) | 382 | 329 (86) |
| Disease at transplantation, n (%) | 390 | |
| AML | 64 (16) | |
| ALL | 51 (13) | |
| CML | 244 (63) | |
| MDS | 31 (8) | |
| Graft type, n (%) | 390 | |
| Bone marrow | 317 (81) | |
| Peripheral blood | 73 (19) | |
| Conditioning regimen, n (%) | 390 | |
| Myeloablative | 384 (98) | |
| TBI/Cy | 287 (74) | |
| Bu/Cy | 75 (19) | |
| TBI | 11 (3) | |
| Melphalan | 4 (1) | |
| Bu (reduced-intensity)/Cy | 4 (1) | |
| Bu | 3 (1) | |
| Nonmyeloablative | 6 (2) | |
| Cy/Flu | ||
| GVHD prophylaxis, n (%) | 390 | |
| Tacrolimus ± other | 124 (32) | |
| Cyclosporine + methotrexate ± other | 256 (66) | |
| Cyclosporine ± other (no methotrexate) | 10 (3) | |
| Donor/recipient sex match, n (%) | 390 | |
| Male to male | 161 (41) | |
| Male to female | 96 (25) | |
| Female to male | 50 (13) | |
| Female to female | 83 (21) | |
| Donor/recipient cytomegalovirus match, n (%) | 390 | |
| Negative/negative | 157 (40) | |
| Negative/positive | 97 (25) | |
| Positive/negative | 53 (14) | |
| Positive/positive | 72 (18) | |
| Unknown | 11 (3) | |
| Median donor age, y (range) | 390 | 36 (19-59) |
| Donor age, n (%) | 390 | |
| Younger than 20 y | 2 (1) | |
| 20-29 y | 107 (27) | |
| 30-39 y | 146 (37) | |
| 40-49, y | 104 (27) | |
| 50 y and older | 31 (8) | |
| Year of transplantation, n (%) | 390 | |
| 1995 | 33 (8) | |
| 1996 | 31 (8) | |
| 1997 | 47 (12) | |
| 1998 | 53 (14) | |
| 1999 | 45 (12) | |
| 2000 | 60 (15) | |
| 2001 | 39 (10) | |
| 2002 | 29 (7) | |
| 2003 | 27 (7) | |
| 2004 | 26 (7) | |
| Median follow-up of survivors, mo (range) | 77 (12-149) |
| Variable . | No. evaluated . | Value . |
|---|---|---|
| No. of patients | 390 | |
| No. of centers | 85 | |
| Median age, y, (range) | 390 | 37 (< 1 to 59) |
| Age at transplantation, n (%) | 390 | |
| 0-9 y | 8 (2) | |
| 10-19 y | 44 (11) | |
| 20-29 y | 69 (18) | |
| 30-39 y | 107 (27) | |
| 40-49 y | 113 (29) | |
| 50 y and older | 49 (13) | |
| Male sex, n (%) | 390 | 211 (54) |
| Karnofsky before transplantation > 90, n (%) | 382 | 329 (86) |
| Disease at transplantation, n (%) | 390 | |
| AML | 64 (16) | |
| ALL | 51 (13) | |
| CML | 244 (63) | |
| MDS | 31 (8) | |
| Graft type, n (%) | 390 | |
| Bone marrow | 317 (81) | |
| Peripheral blood | 73 (19) | |
| Conditioning regimen, n (%) | 390 | |
| Myeloablative | 384 (98) | |
| TBI/Cy | 287 (74) | |
| Bu/Cy | 75 (19) | |
| TBI | 11 (3) | |
| Melphalan | 4 (1) | |
| Bu (reduced-intensity)/Cy | 4 (1) | |
| Bu | 3 (1) | |
| Nonmyeloablative | 6 (2) | |
| Cy/Flu | ||
| GVHD prophylaxis, n (%) | 390 | |
| Tacrolimus ± other | 124 (32) | |
| Cyclosporine + methotrexate ± other | 256 (66) | |
| Cyclosporine ± other (no methotrexate) | 10 (3) | |
| Donor/recipient sex match, n (%) | 390 | |
| Male to male | 161 (41) | |
| Male to female | 96 (25) | |
| Female to male | 50 (13) | |
| Female to female | 83 (21) | |
| Donor/recipient cytomegalovirus match, n (%) | 390 | |
| Negative/negative | 157 (40) | |
| Negative/positive | 97 (25) | |
| Positive/negative | 53 (14) | |
| Positive/positive | 72 (18) | |
| Unknown | 11 (3) | |
| Median donor age, y (range) | 390 | 36 (19-59) |
| Donor age, n (%) | 390 | |
| Younger than 20 y | 2 (1) | |
| 20-29 y | 107 (27) | |
| 30-39 y | 146 (37) | |
| 40-49, y | 104 (27) | |
| 50 y and older | 31 (8) | |
| Year of transplantation, n (%) | 390 | |
| 1995 | 33 (8) | |
| 1996 | 31 (8) | |
| 1997 | 47 (12) | |
| 1998 | 53 (14) | |
| 1999 | 45 (12) | |
| 2000 | 60 (15) | |
| 2001 | 39 (10) | |
| 2002 | 29 (7) | |
| 2003 | 27 (7) | |
| 2004 | 26 (7) | |
| Median follow-up of survivors, mo (range) | 77 (12-149) |
HLA indicates human leukocyte antigen; AML, acute myeloid leukemia; ALL, acute lymphoblastic leukemia; CML, chronic myeloid leukemia; BM, bone marrow; PBSC, peripheral blood stem cell; MDS, myelodysplastic syndrome; TBI, total body irradiation; Cy, cyclophosphamide; Bu, busulfan; Flu, fludarabine; and GVHD, graft-versus-host disease.
Frequency distribution of NOD2/CARD15, IL-23R, and IRGM variants
NOD2/CARD15 variant SNPs were found in 56 donors (14%) and 67 recipients (17%), with an overall frequency of 16% in this cohort (Figure 1). In 44 pairs (11%) the mutation was detected in the donor only, in 55 pairs (14%) the mutation occurred in the recipient only, and in 12 pairs (3%) both donor and recipient had a mutant SNP. The small number of donor/recipient pairs that both had a variant NOD2/CARD15 SNP precludes further analysis to determine statistical significance. Frequencies for individual mutated SNPs were 3% for donors and 3% for recipients for SNP 8, 5% of donors and 5% of recipients for SNP 12, and 6.4% for donors and 9.5% for recipients for SNP 13 (Table 2).
Incidence of CARD15, IL-23R, and IRGM SNPs in donors, recipients, or both
| Variable . | No. evaluated . | n (%) . |
|---|---|---|
| CARD15 | 390 | |
| Donor negative/recipient negative | 279 (72) | |
| Donor positive/recipient negative | 44 (11) | |
| Donor negative/recipient positive | 55 (14) | |
| Donor positive/recipient positive | 12 (3) | |
| IL-23R | 353 | |
| Donor negative/recipient negative | 263 (75) | |
| Donor positive/recipient negative | 35 (10) | |
| Donor negative/recipient positive | 44 (12) | |
| Donor positive/recipient positive | 11 (3) | |
| IRGM | 385 | |
| Donor negative/recipient negative | 229 (60) | |
| Donor positive/recipient negative | 66 (17) | |
| Donor negative/recipient positive | 66 (17) | |
| Donor positive/recipient positive | 24 (6) |
| Variable . | No. evaluated . | n (%) . |
|---|---|---|
| CARD15 | 390 | |
| Donor negative/recipient negative | 279 (72) | |
| Donor positive/recipient negative | 44 (11) | |
| Donor negative/recipient positive | 55 (14) | |
| Donor positive/recipient positive | 12 (3) | |
| IL-23R | 353 | |
| Donor negative/recipient negative | 263 (75) | |
| Donor positive/recipient negative | 35 (10) | |
| Donor negative/recipient positive | 44 (12) | |
| Donor positive/recipient positive | 11 (3) | |
| IRGM | 385 | |
| Donor negative/recipient negative | 229 (60) | |
| Donor positive/recipient negative | 66 (17) | |
| Donor negative/recipient positive | 66 (17) | |
| Donor positive/recipient positive | 24 (6) |
SNP indicates single nucleotide polymorphism.
For IL23R, 46 (13%) of donors and 55 (16%) of recipients had variant SNPs (n = 353 pairs analyzed; Figure 1). In 35 pairs (10%) the mutation occurred in the donor only, in 44 pairs (12%) the mutation occurred in the recipient only, and in 11 pairs (3%) both donor and recipient had variant SNPs (Table 2).
For IRGM, variant SNPs were seen in 90 donors (23%) and 90 recipients (23%; n = 385 pairs analyzed; Figure 1). In 66 pairs (17%) the mutation occurred in the donor only, in 66 pairs (17%) the mutation occurred in the recipient only, and in 24 pairs (6%) both donor and recipient had a mutant SNP (Table 2).
Incidence of clinical end points
The overall incidence of the end points analyzed is shown in Table 3. The overall incidence of 100-day severe acute GVHD (grade III-IV) was 21% (17%-25%) and 1-year chronic GVHD was 53% (48%-58%). Gastrointestinal GVHD rates at 100 days for stages II to IV was 14% (11%-18%) and for stages III to IV was 8% (5%-11%). These rates and those of other major outcomes were similar to previously published results in a US cohort.21 In comparison, the overall incidence of severe acute GVHD (grade III-IV) for matched URD transplants cited in the study of Holler et al12 was higher at 28%. Rates of gastrointestinal GVHD (grade II-IV) were calculated to be higher in the mixed cohort of unrelated and related donor transplants in the study of Holler et al11 (41 of 169 donor/patient pairs or 24.5%). In general, the incidence of chronic GVHD in European centers has been reported at 27% to 57%.22,23
Overall incidence of evaluated clinical outcomes (n = 390)
| Clinical outcome . | % (95% CI) . |
|---|---|
| 100 days (acute) | |
| Grade III-IV GVHD | 21 (17-25) |
| 100-day GI GVHD | |
| Grade II-IV | 14 (11-18) |
| Grade III-IV | 8 (5-11) |
| 1-year chronic GVHD | 53 (48-58) |
| Rate of relapse | |
| 100 d | 2 (1-4) |
| 6 mo | 5 (3-8) |
| 1 y | 10 (7-13) |
| Overall survival | |
| 100 d | 85 (82-89) |
| 6 mo | 77 (73-81) |
| 1 y | 67 (62-72) |
| Disease-free survival | |
| 100 d | 84 (80-88) |
| 6 mo | 74 (70-78)) |
| 1 y | 63 (58-68) |
| Clinical outcome . | % (95% CI) . |
|---|---|
| 100 days (acute) | |
| Grade III-IV GVHD | 21 (17-25) |
| 100-day GI GVHD | |
| Grade II-IV | 14 (11-18) |
| Grade III-IV | 8 (5-11) |
| 1-year chronic GVHD | 53 (48-58) |
| Rate of relapse | |
| 100 d | 2 (1-4) |
| 6 mo | 5 (3-8) |
| 1 y | 10 (7-13) |
| Overall survival | |
| 100 d | 85 (82-89) |
| 6 mo | 77 (73-81) |
| 1 y | 67 (62-72) |
| Disease-free survival | |
| 100 d | 84 (80-88) |
| 6 mo | 74 (70-78)) |
| 1 y | 63 (58-68) |
CI indicates confidence interval; GI, gastrointestinal; and GVHD, graft-versus-host disease.
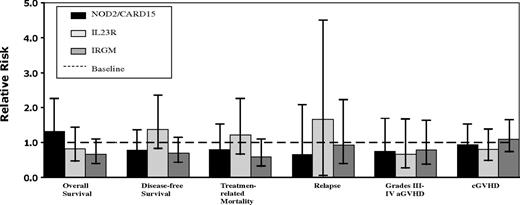
None of the 3 IBD-associated alleles showed evidence of a statistically significant association with any of the clinical outcomes at P less than .01 (Figure 2). In particular, there was no association between the 3 NOD2/CARD15 SNPs (8, 12, or 13) and the incidence of acute or chronic GVHD or TRM (Tables 4, 5). For pairs with any variant NOD2/CARD15 SNP the relative risk (RR) of developing acute GVHD (grade II-IV) was 0.99 (95% CI, 0.68-1.42; P = .93), severe acute (grade III-IV) GVHD was 0.79 (95% CI, 0.43-1.46; P = .45), chronic GVHD was 1.02 (95% CI, 0.70-1.48; P = .93), and TRM was 0.86 (95% CI, 0.53-1.39; P = .54). Extending this analysis to include acute GVHD grades II to IV, the data show a RR in pairs with any variant IL23R SNP was 0.74 (95% CI, 0.49-1.13; P = .16), severe acute GVHD was 0.66 (95% CI, 0.33-1.34; P = .25), chronic GVHD was 0.84 (95% CI, 0.56-1.25; P = .39), and TRM was 1.24 (95% CI, 0.78-1.98; P = .36). For pairs with any variant IRGM SNP the RR of developing acute GVHD (grade II-IV) was 0.97 (95% CI, 0.70-1.34; P = .85), severe acute GVHD was 0.82 (95% CI, 0.47-1.42; P = .48), chronic GVHD was 1.14 (95% CI, 0.83-1.56; P = .42), and TRM was 0.59 (95% CI, 0.37-0.95; P = .03). Even combining all 3 IBD-associated alleles into 1 “combination variable” (ie, whereby any mutation in any 1 of the 3 alleles would be considered variant) failed to show an association with outcome.
Adjusted relative risks of SNP-positive versus SNP-negative variant status in patients who have undergone allogeneic stem cell transplantation. aGVHD indicates acute graft-versus-host disease.
Adjusted relative risks of SNP-positive versus SNP-negative variant status in patients who have undergone allogeneic stem cell transplantation. aGVHD indicates acute graft-versus-host disease.
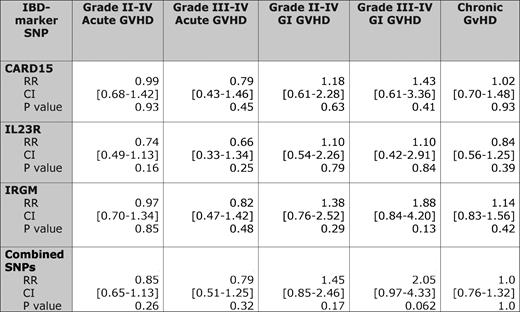
SNP variant status and relative risks of graft-versus-host disease compared with wild-type pairs
Discussion
Among the target organs for GVHD, tissue injury is most pronounced throughout the gastrointestinal tract, which functions as a reservoir for bacterial communities. These resident bacterial flora interact with the host to generate luminal bacterial products, such as lipopolysaccharide, that then translocate across the bowel and perpetuate both a local and systemic inflammatory response.2 The role of luminal bacterial contents and host interactions in mediating intestinal injury is further underscored by findings in both animal models as well as earlier clinical studies that each suggested that gut decontamination was associated with decreased incidence of GVHD.13,24,25 The further demonstration that polymorphic variants (including NOD2/CARD15) known to modulate the innate immune response may be informative predictors of adverse outcomes in GVHD prompted us to reexamine this association in a US population with standardized transplantation protocols. This was an important objective in view of distinctive practice patterns, including the use of T-cell depletion, conditioning regimen, gut decontamination, and HLA matching, that make those earlier findings from European centers difficult to extrapolate to a US population. Gut decontamination, for example, is not routine practice in the US, but its use does vary widely among various transplantation centers. Our findings, by contrast with those earlier reports, suggest that NOD2/CARD15 variants in either the donor or recipient are not significantly associated with clinical outcomes after HSCT in matched URD transplantations. We also concluded that IL-23R donor genotype, despite previous reports of a protective effect on the occurrence of acute GVHD19 and improved death rates in remission,18 had no effect on outcome. We also included another IBD-associated gene, IRGM, which has heretofore not been reported in the context of HSCT outcomes. This gene also did not show an association with any clinically significant outcome. These findings bear further discussion in light of conflicting reports involving the association of NOD2/CARD15 and IL-23R SNPs and HSCT outcomes in different populations.
The initial findings of Holler et al11 suggested a pathogenic role of NOD2/CARD15 variants in the outcomes of HSCT and were based on observations in 169 recipients and their matched donors from 2 European centers. Those investigators reported that NOD2/CARD15 mutations in either the donor or recipient increased the incidence of TRM and acute GVHD. Furthermore, pairs in which NOD2/CARD15 risk alleles were present in both donor and recipient experienced a 1-year TRM of 83% compared with 20% in wild-type pairs (P < .001). Our study, a retrospective cohort, did not contain sufficient numbers of pairs in which both the donor and recipient had a NOD2/CARD15 mutation and was thus underpowered to perform this particular analysis. A follow-up study by Holler et al24 expanded the original cohort of sibling donor transplantations (n = 78) with an additional 225 donor/recipient pairs from 4 additional European centers. The association of NOD2 mutations and adverse outcomes was again confirmed, and the findings suggested that gut decontamination attenuated the effect of NOD2 mutations on TRM.24 A further study yet again expanded the study cohort and pooled all the related (n = 358) and unrelated (n = 342) donor recipient pairs to show that there was a weak association with NOD2/CARD15 variants and GVHD, and there was no overall effect on TRM and OS.12
The implications of those initial reports, namely that NOD2/CARD15 genotyping might have a role in preemptive stratification of risk profiles in patients undergoing HSCT, prompted studies at other centers to validate those findings.14,26 van der Velden et al26 used partially T-depleted grafts in sibling donor transplantations and found a significant effect of NOD2/CARD15 polymorphisms on severe acute GVHD and TRM rates when both the donor and recipient had a mutation. A separate study in the United Kingdom that involved 196 URD transplantations for acute leukemia, most of which were T-depleted grafts, also confirmed an increased risk of relapse and overall mortality in recipients with NOD2/CARD15 mutations.14 Although these findings support at least some of the earlier conclusions linking NOD2/CARD15 polymorphisms to worse outcomes after transplantation, other work has reported less consistent findings. Studies have shown, for example, that donor NOD2/CARD15 mutations led to an unexpected reduction in acute GVHD, whereas both donor and recipient NOD2/CARD15 mutations led to the highest incidence of severe GVHD and gut GVHD.13 Granell et al22 found an increased risk of relapse and death in patients with NOD2/CARD15 variants undergoing T cell–depleted sibling transplantations for acute leukemia with those outcomes being independent of the development of acute or chronic GVHD. Yet another study, involving 198 unrelated and sibling transplantations, showed no association between NOD2/CARD15 variants and negative outcomes after HSCT.15 Clearly, subtle differences in practice patterns in these various centers may account for the variance in the findings, but the findings suggest at the very least that there is uncertainty about the association between NOD2/CARD15 mutations and the outcomes of HSCT.
In attempting to reconcile our findings with these earlier studies, we can point to some important differences between our study and previous reports. First, the current study group had a lower incidence of NOD2/CARD15 variants, which may have led to our cohort being underpowered to detect a difference in outcome. Among the reasons for the lower mutant frequencies are that our patients were drawn from a more ethnically diverse population, which included 6.7% racial/ethnic minorities. Previous epidemiologic studies have shown that mutations in the NOD2/CARD15 gene are extremely rare outside the white population.8 Second, our observed rates of GVHD were also lower than earlier studies, which may reflect our exclusion of any donor-recipient mismatched pairs and higher proportion of bone marrow as the stem cell source as opposed to peripheral blood stem cells. Although peripheral blood stem cells are used more commonly today, the inclusion period of this study (1995-2004) dates back to when bone marrow was still the more common source of stem cells and the number of bone marrow transplantations in our cohort exceeded peripheral blood stem cell transplantations until 2003. Future studies will be required to address the outcomes with the use of predominantly or exclusively peripheral blood stem cells. Finally, it is probable that NOD2/CARD15 effects may be more pronounced if donor T cells are strongly suppressed as originally suggested12 and recently reported.26
Note that the use of T-replete grafts, unrelated donors, and lack of gut decontamination as used in the current study are factors that one would typically associate with a higher incidence of GVHD. In particular, given the emerging role of luminal flora in the pathogenesis of IBD,27 one would have predicted a priori that the lack of gut decontamination would, if anything, exacerbate the incidence of GVHD. However, this was not the case. It is not routine practice in US transplantation centers to use gut decontamination as was performed in most of the aforementioned studies from European centers, but the exact frequency is unknown and not recorded in the NMDP database.
In designing this study, we selected several IBD markers with a plausible role in GVHD. The findings in regard to IRGM, which has not previously been studied in relation to HSCT outcomes, have yielded provocative but as yet inconclusive results. Our findings suggest that IRGM variant status did show a trend toward improved OS, disease-free survival, and TRM. Nevertheless, because of the multiple variables examined, we selected a P value of less than .01 to meet statistical significance, and we conclude that further study will be required to confirm and extend these associations. Several other IBD-associated genes have recently been elucidated, and these may also merit examination in the future to determine whether there is a role for polymorphic variants in the development of GVHD. In this context, it is worth emphasizing that the use of non-HLA genes in association studies of transplantation outcome should be interpreted with caution and, in the context of biologic plausibility, to justify further analysis of a particular inflammatory cytokine. By way of example, mouse genetic studies may serve as an invaluable tool in this regard because the findings from NOD2/CARD15 knockout mice point to a key role in innate intestinal immunity,28 and other recent findings that suggest that genetic deletion of the p19 dimer of IL23 may be protective against GVHD.29
In conclusion, because of the lack of any clear association between the studied variant SNPs and outcomes after HSCT, our data do not support the routine use of NOD2/CARD15, IL-23R, or IRGM polymorphism screening in the patient-donor selection process for HSCT. Further studies are needed to determine whether these polymorphisms would have an effect in sibling donor transplantations, a population not included in this study, or in exclusively peripheral blood stem cell–based transplantations in the United States.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Anne Bowcock for helpful discussion and assistance with the Sequenom assays and Dr Effie Petersdorf for discussions and insights about the role of NOD2/CARD15 mutants.
This work was supported in part by grants (Digestive Disease Research Core Center DK-52574, HL-38180, and DK-56260; N.O.D.). The Center for International Blood and Marrow Transplant Research (CIBMTR) is supported by the National Cancer Institute (NCI; Public Health Service Grant/Cooperative Agreement U24-CA76518) the National Heart, Lung, and Blood Institute (NHLBI), and the National Institute of Allergy and Infectious Diseases (NIAID); the NHLBI and NCI (grant/cooperative agreement 5U01HL069294); Health Resources and Services Administration (HRSA/DHHS; a contract HHSH234200637015C); the Office of Naval Research (grants N00014-06-1-0704 and N00014-08-1-0058); and grants from AABB; Aetna; American Society for Blood and Marrow Transplantation; Amgen Inc; Anonymous donation to the Medical College of Wisconsin; Astellas Pharma US Inc; Baxter International Inc; Bayer HealthCare Pharmaceuticals; Be the Match Foundation; Biogen IDEC; BioMarin Pharmaceutical Inc; Biovitrum AB; BloodCenter of Wisconsin; Blue Cross and Blue Shield Association; Bone Marrow Foundation; Canadian Blood and Marrow Transplant Group; CaridianBCT; Celgene Corporation; CellGenix GmbH; Centers for Disease Control and Prevention; Children's Leukemia Research Association; ClinImmune Labs; CTI Clinical Trial and Consulting Services; Cubist Pharmaceuticals; Cylex Inc; CytoTherm; DOR BioPharma Inc; Dynal Biotech, an Invitrogen Company; Eisai Inc; Enzon Pharmaceuticals Inc; European Group for Blood and Marrow Transplantation; Gamida Cell Ltd; GE Healthcare; Genentech Inc; Genzyme Corporation; Histogenetics Inc; HKS Medical Information Systems; Hospira Inc; Infectious Diseases Society of America; Kiadis Pharma; Kirin Brewery Co Ltd; The Leukemia & Lymphoma Society; Merck & Company; The Medical College of Wisconsin; MGI Pharma Inc; Michigan Community Blood Centers; Millennium Pharmaceuticals Inc; Miller Pharmacal Group; Milliman USA Inc; Miltenyi Biotec Inc; National Marrow Donor Program; Nature Publishing Group; New York Blood Center; Novartis Oncology; Oncology Nursing Society; Osiris Therapeutics Inc; Otsuka America Pharmaceutical Inc; Pall Life Sciences; PDL BioPharma Inc; Pfizer Inc; Pharmion Corporation; Saladax Biomedical Inc; Schering Corporation; Society for Healthcare Epidemiology of America; StemCyte Inc; StemSoft Software Inc; Sysmex America Inc; Teva Pharmaceutical Industries; THERAKOS Inc; Thermogenesis Corporation; Vidacare Corporation; Vion Pharmaceuticals Inc; ViraCor Laboratories; ViroPharma Inc; and Wellpoint Inc.
The views expressed in this article do not reflect the official policy or position of the National Institute of Health, the Department of the Navy, the Department of Defense, or any other agency of the US Government.
Authorship
Contribution: Y.N. designed the research plan and wrote the paper; A.A.-L. and E.G. performed the sequencing; E.L. assisted with design of assays and provided data interpretation; M.H. contributed to editing the paper, analyzed data, and performed statistical analysis; T.W. performed statistical analysis; S.S. and S.J.L. provided editorial assistance; and N.O.D. devised the project concept and wrote and edited the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nicholas O. Davidson, Division of Gastroenterology, Washington University School of Medicine, 660 S Euclid Ave, Campus Box 8124, St Louis, MO 63110; e-mail: nod@wustl.edu.