Abstract
Resistance to glucocorticoids (GCs) is a major clinical problem in the treatment of acute lymphoblastic leukemia (ALL), but the underlying mechanisms are not well understood. Although mutations in the glucocorticoid receptor (GR) gene can give rise to therapy resistance in vitro, acquired somatic mutations in the GR are rarely encountered in patients. Here we report that the protein encoded by the BTG1 gene, which is frequently deleted in (pediatric) ALL, is a key determinant of GC responsiveness. Using RNA interference, we show that loss of BTG1 expression causes GC resistance both by decimating GR expression and by controlling GR-mediated transcription. Conversely, reexpression of BTG1 restores GC sensitivity by potentiating GC-induced GR expression, a phenomenon known as GR autoinduction. In addition, the arginine methyltransferase PRMT1, a BTG1-binding partner and transcriptional coactivator, is recruited to the GR gene promoter in a BTG1-dependent manner. These results implicate the BTG1/PRMT1 complex in GR-mediated gene expression and reveal that deregulation of a nuclear receptor coactivator complex can give rise to GC resistance. Further characterization of this complex as part of the GR regulatory circuitry could offer novel opportunities for improving the efficacy of GC-based therapies in ALL and other hematologic malignancies.
Introduction
Acute lymphoblastic leukemia (ALL) is the most common type of cancer in children. Although cure rates are exceeding 80%, therapy resistance and associated disease relapse remain a significant clinical problem. Pediatric ALL is a genetically heterogeneous disease characterized by chromosomal abnormalities, including translocations, aneuploidies, and deletions. Molecular analysis of the most common chromosomal alterations in ALL has allowed classification into different subtypes that are associated with distinct clinical outcome. In addition to these chromosomal abnormalities, prognosis is determined by blast count at diagnosis, lineage (B or T cell), and the initial response to therapy.1 Despite these advances in the molecular pathology of pediatric ALL, little is known about the mechanisms that contribute to relapse or therapy resistance in the patient.
Synthetic glucocorticoids (GCs) are essential drugs in the treatment of pediatric ALL, as they effectively induce apoptosis in leukemic blasts. GCs exert their effects by binding to the glucocorticoid receptor (GR), a ligand-activated receptor and member of the nuclear receptor family. Activation of the GR by GCs leads to recruitment of GR to large transcription regulatory complexes that modulate expression of GC response genes, which leads to the induction of apoptosis in lymphoblasts. Resistance to GCs is a major clinical problem predictive of poor therapy outcome.2 In most in vitro models, resistance to GCs appears to occur at the level of the receptor, either by mutations in the GR or by the inability of cells to up-regulate the GR in response to GC exposure, a phenomenon known as GR autoinduction.3 However, mutations in the GR are rarely seen in (relapsed) ALL,4 and it remains unclear to what extent differences in GR expression affect therapy response in the patient.5,6
Through genome-wide profiling using single nucleotide polymorphism (SNP)-based array comparative genomic hybridization, we and others recently identified several novel submicroscopic lesions that may either contribute to the development of ALL or determine the response to therapy.7,8 Focal microdeletions, often smaller than 0.5 Mb and containing single genes, were identified with a relatively high frequency. The genes most frequently affected by these microdeletions are implicated in B-cell differentiation, cell survival, or cell-cycle progression.7,8 A novel recurrent single-gene lesion observed in pediatric pre-B ALL involves the B-cell translocation 1 gene (BTG1).7-9 This gene was originally identified as a translocation partner of the c-MYC gene in a case of B-cell chronic lymphocytic leukemia and belongs to a family of antiproliferative genes that also includes BTG2, BTG3, TOB, and TOB2.10,11 Proteins encoded by members of this gene family have been implicated in the induction of growth arrest or apoptosis in a variety of cell systems.12 Overexpression of BTG1 was found to block proliferation during normal erythroid differentiation13 and to induce growth arrest in a B-cell lymphoma model.14 The BTG1 protein has no proven intrinsic enzymatic activity, but the presence of several protein interaction domains suggests that BTG1 functions either as an adaptor molecule or as a cofactor involved in transcriptional regulation. For instance, BTG1 was identified as coactivator of the homeobox transcription factor HoxB915 and the transcriptional regulator CAF1.16 Moreover, BTG1 was shown to stimulate the activity of several myogenic transcription factors as well as nuclear receptors during muscle cell differentiation.17
BTG1 interacts with and regulates the activity of the arginine methyltransferase PRMT1.11,18 Members of this enzyme family, including PRMT1, are considered global regulators of gene expression that act as transcriptional coregulators by arginine methylation of histone tails and critical transcriptional regulators.19 PRMT1 was shown to act as a histone H4 Arg-3–specific methyltransferase involved in transcriptional activation of several nuclear receptors.20-22 However, a role for either BTG1 or PRMT1 in GR activation has not been investigated. In the present study, we identify BTG1 as a critical determinant of GC-induced therapy responses in vitro, which acts by controlling GR autoinduction as well as by regulating GR-mediated gene expression. Moreover, we demonstrate that PRMT1 is recruited to the GR promoter in a BTG1-dependent manner.
Methods
Collection of leukemia samples and maintenance of ALL cell lines
Lymphoblast samples of patients treated according to the current clinical protocol were obtained with informed consent in accordance with the Declaration of Helsinki and with the approval and supervision of the Medical Ethics Committee of the Radboud University Nijmegen Medical Center. ALL-derived cell lines RS4;11, REH, and SUP-B15 were purchased from ATCC. Primary lymphoblasts, RS4;11, and REH cells were maintained in RPMI 1640 with L-glutamine (Invitrogen) supplemented with 10% fetal bovine serum (FBS; Greiner Bio-One) and 100 units/mL penicillin/streptomycin (Invitrogen), whereas SUP-B15 cells were maintained in Iscove modified Dulbecco medium with L-glutamine, 20% FBS, and 100 units/mL penicillin/streptomycin.
MLPA and sequence analyses
Nine Multiplex Ligation-dependent Probe Amplification (MLPA) probes covering the BTG1 gene (supplemental Table 1; supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) were combined in one MLPA assay along with 4 standard control probes in 3 different genes, ie, VIRP2, MRPL41, and KIAA0056, according to guidelines of MRC-Holland. Amplification products were quantified and identified by capillary electrophoresis on an ABI 3730 genetic analyzer (Applied Biosystems). Normalization of the data was performed by dividing the peak area of each probe by the average peak area of the control probes. This normalized peak pattern was divided by the average peak pattern of all the samples in the same experiment. The resulting values were approximately 1.0 for every wild-type peak, 0.5 for heterozygous deletions, and 1.5 for heterozygous duplications.
Lentiviral knockdown in ALL cell lines
shRNAs were introduced using the BLOCK-iT U6 RNAi Entry Vector in combination with the pLenti6/BLOCK-iT DEST vector according to the manufacturer's protocol (Invitrogen). Two shRNA sequences were directed against BTG1 mRNA (NM_001731.1): 5′-gctctctgtacatttgctagc-3′ (shRNA1) and 5′-gctgctggcagaacattataa-3′ (shRNA2). A nonfunctional shRNA (5′-gctacaagagaaaccaaatct-3′), originally designed to target myosin X, which is not expressed in lymphocytes, was used as a negative control. The individual vectors were cotransfected with packaging vectors into the 293FT cells according to the manufacturer's instructions. Virus was produced in Dulbecco modified Eagle medium containing Glutamax (Invitrogen) supplemented with 10% FBS and 100 units/mL penicillin/streptomycin and collected from tissue culture supernatant 48 hours after transfection. Tissue-culture plates were coated with retronectin (Takara), and virus supernatant was cocultured with ALL cell lines for 8 hours. Transduced cells were selected with 4 μg/mL blasticidin (Invitrogen).
Retroviral transduction and Western blot analysis
A BTG1 expression construct was generated by polymerase chain reaction (PCR; forward primer: 5′-gcgaattcgaccatggcttacccatacgatgttcc-agattacgctcatcccttctacacccggg-3′ (HA tag in bold); reverse: 5′-gcctcgagttaacctgatacagtcatcatattg-3′), cloned into pLZRS-Neo, and verified by sequencing. pLZRS-HA-BTG1 and pLib-T7-MSC2-IRESpuro-hGR (obtained from Prof Dr R. Kofler, Innsbruck, Austria) were transfected into Phoenix amphotropic producer cells to generate retrovirus-containing supernatants. Cells were transduced in retronectin-coated tissue-culture dishes, and transduced cells were selected using 400 μg/mL G418 (Invitrogen) and 1 μg/mL puromycin. Antibodies directed against the GR (Santa Cruz Biotechnology), α-tubulin (Sigma-Aldrich), and the HA epitope, clone 3F10 (Roche Diagnostics) were used in immunoblot assays using standard procedures.
Cytotoxicity assays
Cell viability was determined using the CellTiter 96 Aqueous One Solution Cell Proliferation Assay (MTS; Promega). A total of 1 × 105 cells were seeded in a 96-well culture plate and treated with increasing amounts of prednisolone (5 × 10−3 to 5 × 103 μg/mL; Organon) or dexamethasone (5 × 10−4 to 5 × 102 μg/mL; Centrafarm) for 72 hours (RS4;11 cells) or 48 hours (REH cells). Apoptosis induction was measured using the annexin V–PE Apoptosis Detection Kit I (BD Biosciences) according to the manufacturer's protocol and analyzed on a FACSCalibur flow cytometer (BD Biosciences).
Microarray analysis and quantitative real-time RT-PCR
Total RNA was isolated using the RNeasy Mini kit (QIAGEN) according to the manufacturer's instructions, and RNA quality was tested using the Agilent 2100 Bioanalyzer. A total of 1 μg of total RNA from each sample was used to perform hybridizations on the GeneChip Human Exon 1.0 ST Array (Affymetrix) in duplicate according to the Affymetrix GeneChip Expression Analysis Technical Manual. Probe set intensities were summarized into transcript intensities by taking the mean of corresponding probe sets. Mean expression of 2 independent experiments was calculated. Analysis of variance was applied comparing the log2 intensities of BTG1 shRNA1 knockdown cells to the empty vector-transduced cells. The resulting P values were corrected for multiple testing using the Benjamini-Hochberg False Discovery Rate. The data discussed in this publication have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus and are accessible through Gene Expression Omnibus Series accession number GSE18027. For reverse-transcribed (RT)–PCR, cDNA was generated with the iScript cDNA synthesis kit (Bio-Rad) and analyzed by quantitative real-time PCR using SYBR Green technology on a 7500 Fast Real-Time PCR System (Applied Biosystems). Expression levels were normalized against the housekeeping gene TATA box-binding protein (TBP). Primer sequences used in RT-PCR experiments are depicted in supplemental Table 2.
ChIP assays
Chromatin immunoprecipitations (ChIP) were performed as described elsewhere,23 using 2 μg of anti-PRMT1 ab3768 (Abcam) or 07-404 (Upstate Biotechnology) or anti-TBP (TBPCSH-100; Diagenode Epigenetics; positive control) antibody. Efficacy of both anti-PRMT1 antibodies in ChIP was compared on control RS4;11 cells. Because ab3768 gave the most reproducible results (supplemental Figure 2), it was used in all further experiments. Primer sequences used in real-time PCR experiments were: 5′-ccgtgcagcaccctttatagt-3′ forward, 5′-atggtgcgatattcggattg-3′reverse (BTG1), 5′-tcctccttgggaatgagaaa-3′ forward, 5′-atcaagcaggtgccagaaaa-3′ reverse (NR3C1). Primers were designed to detect the genomic area around the transcription initiation site of all genes. All ChIP experiments were performed at least 3 times, and the real-time PCR reactions were performed in duplicate.
Luciferase reporter assays
The mouse mammary tumor virus promoter cloned upstream of the luciferase gene (MMTV-Luc) contains multiple GC-responsive elements, located between positions −202 and −79 upstream of the RNA start site, and was originally described by Hollenberg and Evans.24 293FT cells were seeded in 24-well plates (2 × 105 cells/well). Six hours later, each well was transfected with 60 ng of MMTV-Luc, 20 ng pCMV-HA-GR, 2 ng of pRL-CMV (Promega), and increasing concentrations of pcDNA3.1-HA-BTG1 using linear polyethylenimine (Polysciences). The DNA amount was supplemented with pcDNA plasmid to a total of 300 ng. The next day, prednisolone (3.6 μg/mL) or medium control was added to the wells and 16 hours later cells were harvested and analyzed with a Dual-Luciferase Reporter Assay according to the supplier's instructions (Promega). Firefly luciferase values were normalized with Renilla luciferase values, which served as an internal control.
Results
BTG1 copy number loss occurs frequently in pediatric precursor-B ALL
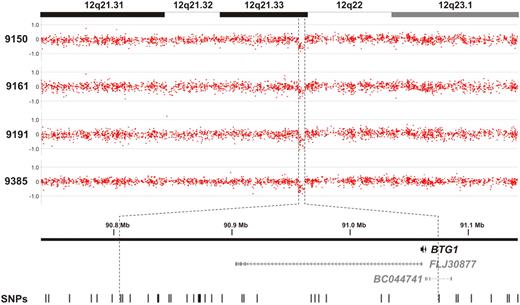
By high-resolution genomic profiling of childhood ALL, we and others recently identified a recurrent microdeletion on the q-arm of chromosome 12 affecting the BTG1 locus in approximately 10% of precursor-B (pre-B) ALL cases (Figure 1).7-9 In our study, copy number losses of the BTG1 gene were found in 4 of 33 cases examined. MLPA was used to confirm the presence of these microdeletions (supplemental Figure 1). Because these BTG1 microlesions appeared to be monoallelic, we performed exon sequencing to determine whether loss of function mutations in BTG1 could be observed in the other allele. Sequencing of the BTG1 gene was also performed in the remaining 29 cases to reveal the potential presence of smaller lesions or point mutations. However, in none of the cases showing a single-copy loss of BTG1 or in the other leukemia samples, additional mutations in BTG1 were found. The reported role of BTG1 as a regulator of differentiation, proliferation, and cell survival12 suggests that BTG1 haploinsufficiency contributes to ALL development, preferentially through focal microdeletions. However, it remains to be established how BTG1 contributes to ALL development.
BTG1 copy number loss is observed in approximately 10% of pediatric pre-B ALL cases. Genomic profiles of the chromosome 12q21.31-23.1 segment in 4 pre-B ALL cases. Indicated is a region less than 300 kb in size containing the BTG1 gene, with the positions of the 2 hypothetical proteins FLJ30877 and BC044741 and the SNPs represented on the array.
BTG1 copy number loss is observed in approximately 10% of pediatric pre-B ALL cases. Genomic profiles of the chromosome 12q21.31-23.1 segment in 4 pre-B ALL cases. Indicated is a region less than 300 kb in size containing the BTG1 gene, with the positions of the 2 hypothetical proteins FLJ30877 and BC044741 and the SNPs represented on the array.
shRNA-mediated knockdown of BTG1 leads to GC resistance
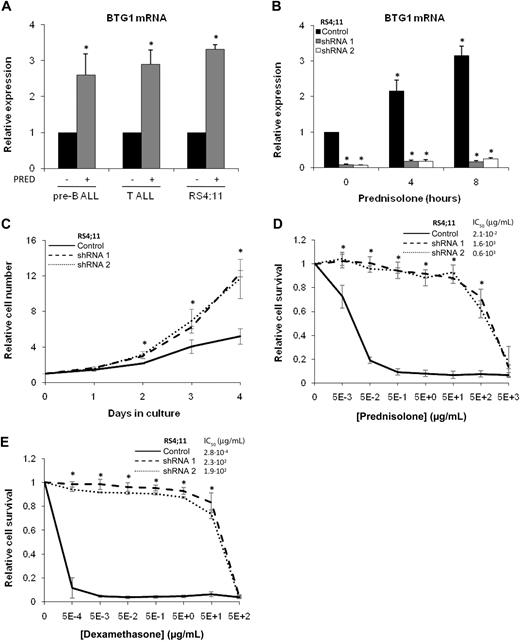
BTG1 expression and function have been linked to the inhibition of cell growth and the induction of apoptosis in a variety of model systems.12 Moreover, with one exception,25 several studies have identified BTG1 as one of many genes up-regulated in clinical isolates of ALL cells in response to synthetic GCs.26-28 Similarly, we detected a 3-fold induction of BTG1 mRNA expression after exposure of primary pre-B ALL cells, primary T-ALL cells, or the pre-B ALL cell line RS4;11 to 1 mg/mL prednisolone for 8 hours (Figure 2A). Next, we determined to what extent BTG1 expression could be involved in the induction of GC-induced cell death. Using a lentiviral vector, we expressed shRNAs targeting different areas of the BTG1 gene in RS4;11 pre-B ALL cells. With either of these shRNA constructs, BTG1 mRNA expression was reduced to less than 10% compared with cells transduced to express an irrelevant shRNA control (Figure 2B). Prednisolone treatment had only a minor effect on BTG1 levels, which remained low in BTG1 knockdown cells (Figure 2B).
BTG1 expression is essential for the induction of GC-mediated cell death. (A) Quantitative RT-PCR analysis of BTG1 in total mRNA of primary pre-B and T-ALL and RS4;11 cell line after exposure to 1 mg/mL prednisolone dissolved in medium or medium alone for 8 hours. Expression values were normalized for TBP. Titration of the prednisolone concentration revealed that efficient induction of BTG1 mRNA, after 8-hour exposure to prednisolone, could be observed at concentrations as low as 5.0 × 10−1 μg/mL (supplemental Figure 3B). (B) Quantitative PCR analysis of BTG1mRNA in RS4;11 cell lines transduced with control or either of 2 independent shRNAs targeting different regions of the BTG1 gene (shRNA1 and shRNA2). Cells were exposed to 1 mg/mL prednisolone for up to 8 hours. (C) The relative increase in cell number of the RS4;11 control, BTG1 shRNA 1, and BTG1 shRNA 2 cell lines was determined during 4 days of culture. (D-E) Determination of relative viability of RS4;11 control, BTG1 shRNA 1, and BTG1 shRNA 2 cell lines as determined by an MTS-based cytotoxicity assay after 3 days of exposure to increasing concentrations of prednisolone (D) or dexamethasone (E). Relative survival is determined as relative compared with the medium control. Data are mean ± SEM of triplicate experiments. Statistical significance was tested with 2-sided Student t test. (A) *Significant change compared with control cells (P < .05). (B) *Significant change compared with mock-treated control cells (P < .05). (C-E) *Significant change in both shRNA 1 and 2 compared with control cells that had received similar treatment (P < .05).
BTG1 expression is essential for the induction of GC-mediated cell death. (A) Quantitative RT-PCR analysis of BTG1 in total mRNA of primary pre-B and T-ALL and RS4;11 cell line after exposure to 1 mg/mL prednisolone dissolved in medium or medium alone for 8 hours. Expression values were normalized for TBP. Titration of the prednisolone concentration revealed that efficient induction of BTG1 mRNA, after 8-hour exposure to prednisolone, could be observed at concentrations as low as 5.0 × 10−1 μg/mL (supplemental Figure 3B). (B) Quantitative PCR analysis of BTG1mRNA in RS4;11 cell lines transduced with control or either of 2 independent shRNAs targeting different regions of the BTG1 gene (shRNA1 and shRNA2). Cells were exposed to 1 mg/mL prednisolone for up to 8 hours. (C) The relative increase in cell number of the RS4;11 control, BTG1 shRNA 1, and BTG1 shRNA 2 cell lines was determined during 4 days of culture. (D-E) Determination of relative viability of RS4;11 control, BTG1 shRNA 1, and BTG1 shRNA 2 cell lines as determined by an MTS-based cytotoxicity assay after 3 days of exposure to increasing concentrations of prednisolone (D) or dexamethasone (E). Relative survival is determined as relative compared with the medium control. Data are mean ± SEM of triplicate experiments. Statistical significance was tested with 2-sided Student t test. (A) *Significant change compared with control cells (P < .05). (B) *Significant change compared with mock-treated control cells (P < .05). (C-E) *Significant change in both shRNA 1 and 2 compared with control cells that had received similar treatment (P < .05).
The BTG1 knockdown cells showed a marked increase in growth rate compared with shRNA control cells (Figure 2C), consistent with the observation that overexpression of BTG1 leads to a cessation of growth in other cell types.12 Subsequently, a role for BTG1 in GC-induced apoptosis was examined by exposing control shRNA cells and BTG1 knockdown cells to increasing concentrations of prednisolone and assessing in vitro toxicity after 3 days of exposure using an MTS cell viability assay. Whereas vector control-transfected RS4;11 cells were highly sensitive to prednisolone exposure, with a 50% inhibitory concentration (IC50) of 2.1 × 10−2 μg/mL prednisolone at 72 hours, BTG1 knockdown cells had become largely refractory (IC50 of 1.6 × 103 and 0.6 × 103 μg/mL for shRNA1 and shRNA2, respectively) to the cytotoxic effects of this drug (Figure 2D). During these 3 days of exposure to prednisolone, BTG1 mRNA levels continued to increase in the control cells; whereas in the BTG1 knockdown cells, the expression level remained low (supplemental Figure 3A). Similar to prednisolone, BTG1 knockdown cells also acquired resistance to the synthetic GC dexamethasone (IC50 of 2.8 × 10−4, 2.3 × 102, and 1.9 × 102 μg/mL for control shRNA, shRNA1, and shRNA2, respectively; Figure 2E). The protective effect of BTG1 knockdown on GC-induced cytotoxicity was confirmed using a flow cytometry–based apoptosis assay, which measures exposure of annexin-V and 7-amino-actinomycin D (supplemental Figure 4A-B). These results demonstrate that loss of BTG1 expression is sufficient to induce GC resistance in vitro.
Loss of BTG1 expression effectively interferes with GR autoinduction and GC-induced gene expression
Having identified BTG1 as a critical determinant of GC-induced apoptosis, microarray expression analysis was used to compare GC-regulated transcript levels in RS4;11 control shRNA cells versus BTG1 knockdown cells. In the (GC-sensitive) RS4;11 control shRNA cells, we found that 951 genes were up- or down-regulated at least 2-fold after exposure to prednisolone (Figure 3A; supplemental Table 3). In contrast, in the BTG1 knockdown cells, only 38 genes were differentially expressed in response to prednisolone (Figure 3A; supplemental Table 4). Of the top 200 ranked genes most strongly up- or down-regulated by prednisolone in control cells, the majority had become unresponsive to prednisolone in the BTG1 knockdown cells (Figure 3B; supplemental Table 3). Expression of 3 representative genes induced by GC exposure was confirmed by quantitative RT-PCR (Figure 3C). Notably, one of the genes strongly affected by BTG1 knockdown was the NR3C1 gene, which encodes the GR. Expression of the GR was reduced more than 10-fold in the BTG1 knockdown cells relative to the RS4;11 control cells, as determined by quantitative PCR (Figure 3C) and confirmed at the protein level by Western blotting (Figure 3D). These findings suggested that the inability of the BTG1 knockdown cells to respond to GCs could be the result of severely reduced expression of the GR. To address whether and how BTG1 would also affect expression of other GR-regulated genes, we complemented expression of the GR in the BTG1 knockdown cells by retroviral transduction of a GR transgene. GR expression levels obtained in these cells were than those in the control RS4;11 cells, as determined by Western blotting (Figure 3E).
BTG1 knockdown in RS4;11 cells impairs GC-induced gene expression because of loss of GR expression. (A) Venn diagram summarizing microarray expression analysis in prednisolone-treated (8-hour) control RS4;11 cells and BTG1 knockdown (shRNA1) RS4;11 cells. The number of genes with a significant (2-fold change) in expression is indicated. (B) Heat map of the 200 genes with highest differential expression after prednisolone exposure in control cells. For comparison, expression changes in BTG1 shRNA 1 cells are shown in the right column. (C) mRNA expression of the GR and the GC-regulated genes SGK1 and VDR were analyzed by quantitative RT-PCR and normalized for TBP after exposure to 1 mg/mL prednisolone for up to 8 hours in RS4;11 control and BTG1 shRNA1 cells. (D) Immunoblot showing GR expression in control RS4;11 cells and in BTG1 knockdown cells in response to prednisolone treatment for up to 8 hours. (E) Immunoblot showing GR expression in RS4;11 cells transduced with a control shRNA, BTG1 shRNA1, or BTG1 shRNA1 with a GR overexpression construct. (F) MTS assay determining relative viability of RS4;11 cells transduced with a control shRNA, BTG1 shRNA1, or BTG1 shRNA1 with a GR overexpression construct after prednisolone treatment. Error bars represent the SEM of triplicate experiments. Statistical significance (P < .05) was assessed with the Student t test compared with untreated control cells.
BTG1 knockdown in RS4;11 cells impairs GC-induced gene expression because of loss of GR expression. (A) Venn diagram summarizing microarray expression analysis in prednisolone-treated (8-hour) control RS4;11 cells and BTG1 knockdown (shRNA1) RS4;11 cells. The number of genes with a significant (2-fold change) in expression is indicated. (B) Heat map of the 200 genes with highest differential expression after prednisolone exposure in control cells. For comparison, expression changes in BTG1 shRNA 1 cells are shown in the right column. (C) mRNA expression of the GR and the GC-regulated genes SGK1 and VDR were analyzed by quantitative RT-PCR and normalized for TBP after exposure to 1 mg/mL prednisolone for up to 8 hours in RS4;11 control and BTG1 shRNA1 cells. (D) Immunoblot showing GR expression in control RS4;11 cells and in BTG1 knockdown cells in response to prednisolone treatment for up to 8 hours. (E) Immunoblot showing GR expression in RS4;11 cells transduced with a control shRNA, BTG1 shRNA1, or BTG1 shRNA1 with a GR overexpression construct. (F) MTS assay determining relative viability of RS4;11 cells transduced with a control shRNA, BTG1 shRNA1, or BTG1 shRNA1 with a GR overexpression construct after prednisolone treatment. Error bars represent the SEM of triplicate experiments. Statistical significance (P < .05) was assessed with the Student t test compared with untreated control cells.
Even though GR expression was restored to endogenous levels in these cells, expression of GR-target genes in response to GCs could not be induced (supplemental Figure 5), although no reversal of prednisolone resistance was seen (Figure 3F). These results demonstrate that GR expression per se is not sufficient to restore therapy responses but requires the presence of additional regulatory protein complexes. We similarly tried to restore expression of BTG1 in the shRNA1 knockdown cells, by retroviral transduction of an HA-BTG1 transgene (resistant to shRNA1). However, despite multiple attempts, we observed that enforced expression of HA-BTG1 in these cells was not tolerated.
BTG1 and GR synergize to reverse therapy responses in GC resistant REH pre-B ALL cells
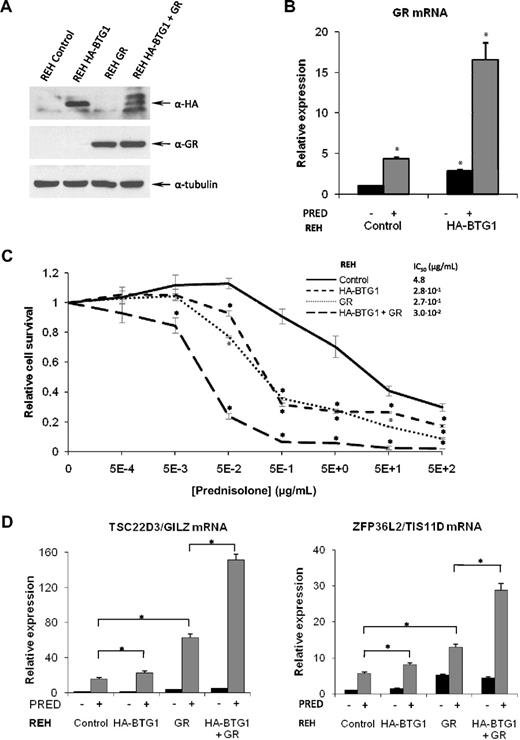
To further define a role for BTG1 in controlling GC responsiveness, we made use of the GC-resistant REH pre-B ALL cell line, which carries a single copy deletion of the BTG1 gene.29 Expression of the BTG1 mRNA could still be detected in these cells, albeit at a reduced level compared with RS4;11 cells (supplemental Figure 6A). In contrast, expression of the GR was almost undetectable in REH cells (supplemental Figure 6B), suggesting that also in these cells GC resistance may be the result of lack of GR expression. To determine whether BTG1 haploinsufficiency in REH cells was directly linked to reduced GR mRNA levels, we complemented BTG1 expression by retroviral transduction of an HA-BTG1 transgene. In contrast to RS4;11 cells, expression of the HA-BTG1 transgene was readily detected in these cells (Figure 4A). Indeed, expression of (HA-tagged) BTG1 resulted in a marked up-regulation of the GR mRNA, as determined by quantitative PCR, particularly after exposure to prednisolone (Figure 4B). This phenomenon of GC-induced GR expression (GR autoinduction) is considered to contribute to the apoptosis-inducing effects of GCs in leukemic blasts.5,30 Despite increased GR autoinduction, expression of GR mRNA in the HA-BTG1–expressing REH cells still remained approximately 20-fold lower compared with the (GC-sensitive) RS4;11 cells (supplemental Figure 6C). As a consequence, we were not able to detect the GR protein in these cells (Figure 4A; results not shown). In agreement with these observations, only a partial reversal of GC resistance was observed; IC50 value was reduced from 4.8 μg/mL in control cells to 2.8 × 10−1 μg/mL in HA-BTG1 cells (Figure 4C). Similarly, raising GR expression levels by retroviral transduction of a GR transgene was not sufficient to fully restore GC responses in the REH cells (IC50 was reduced to 2.7 × 10−1 μg/mL). However, when expression of HA-BTG1 and the GR was combined, a further 10-fold increase in sensitivity to GCs was observed (IC50 reduced to 3.0 × 10−2 μg/mL; Figure 4C), consistent with a role for BTG1 as a limiting cofactor in GR-mediated gene regulation. In REH cells expressing HA-BTG1, this recombinant protein migrated as a single band of approximately 20 kDa. When expression of HA-BTG1 was combined with that of the GR, several discrete products representing HA-BTG1 were observed (Figure 4A), suggesting that, under these conditions, the BTG1 protein is subject to modification. The nature of this modification is currently under investigation.
A BTG1 transgene rescues GR expression and GC therapy response in REH pre-B ALL cells. (A) Western blot showing protein expression of HA-BTG1 and GR protein obtained after retroviral transduction of REH cells with the vectors indicated at the top. (B) Expression of GR mRNA analyzed by quantitative RT-PCR and normalized for TBP in REH control vector versus REH HA-BTG1–transduced cells, after exposure to prednisolone or mock treatment. (C) Determination of relative viability of REH control vector-transduced cells compared with REH cells transduced with HA-BTG1, GR, or HA-BTG1 and GR retroviral constructs using an MTS-based cytotoxicity assay after exposure to prednisolone for 3 days. (D) Quantitative RT-PCR of GR target genes TSC22D3/GILZ and ZFP36L2/TIS11D in REH cells after exposure to 1 mg/mL prednisolone or mock treatment for 8 hours. Error bars represent the SEM of triplicate experiments. (C) *Statistical significance (P < .05) was assessed with the Student t test compared with control vector-transduced cells that received the same treatment. (B,D) *Statistical significance (P < .05) was assessed with the Student t test compared with control vector-transduced cells that were mock treated.
A BTG1 transgene rescues GR expression and GC therapy response in REH pre-B ALL cells. (A) Western blot showing protein expression of HA-BTG1 and GR protein obtained after retroviral transduction of REH cells with the vectors indicated at the top. (B) Expression of GR mRNA analyzed by quantitative RT-PCR and normalized for TBP in REH control vector versus REH HA-BTG1–transduced cells, after exposure to prednisolone or mock treatment. (C) Determination of relative viability of REH control vector-transduced cells compared with REH cells transduced with HA-BTG1, GR, or HA-BTG1 and GR retroviral constructs using an MTS-based cytotoxicity assay after exposure to prednisolone for 3 days. (D) Quantitative RT-PCR of GR target genes TSC22D3/GILZ and ZFP36L2/TIS11D in REH cells after exposure to 1 mg/mL prednisolone or mock treatment for 8 hours. Error bars represent the SEM of triplicate experiments. (C) *Statistical significance (P < .05) was assessed with the Student t test compared with control vector-transduced cells that received the same treatment. (B,D) *Statistical significance (P < .05) was assessed with the Student t test compared with control vector-transduced cells that were mock treated.
To further explore a functional interaction between BTG1 and the GR, we analyzed expression of the GR-target genes TSC22D3/GILZ and ZFP36L2/TIS11D, both of which have been linked to the antiproliferative effects of GCs in leukemic cells.26,31 Despite the low GR expression in parental REH cells, a moderate induction of these genes was observed in response to prednisolone, which was further enhanced in the presence of the GR transgene (Figure 4D). Expression of HA-BTG1 only slightly increased GC-induced expression of TSC22D3/GILZ and ZFP36L2/TIS11D; however, combined expression of HA-BTG1 and the GR resulted in a significantly larger induction (Figure 4D). Together, these results strongly suggest that BTG1 and the GR synergize in the induction of GC-mediated gene expression and apoptosis.
PRMT1 is recruited to GC-responsive promoter elements
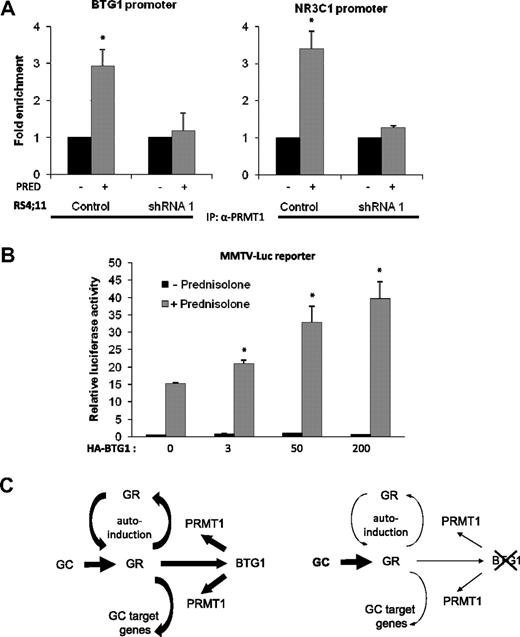
Several studies have demonstrated that BTG1 associates with and regulates the activity of PRMT1.11,18 Moreover, this arginine methyltransferase was shown to regulate transcription mediated by several nuclear receptors, but a functional interaction between PRMT1 and the GR has not been established. Therefore, we examined by ChIP analysis whether BTG1, in complex with PRMT1, may control GR autoinduction. To this end, 2 independent PRMT1 antibodies were tested (supplemental Figure 2). By doing so, a specific association between PRMT1 and the promoter regions of BTG1 and NR3C1 (GR) could be observed in response to GC exposure (Figure 5A; supplemental Figure 2). These results establish PRMT1 as a novel GR coactivator. Importantly, this association was no longer present in the BTG1 knockdown cells (Figure 5A). As the loss of BTG1 expression in the knockdown cells is accompanied by a loss of GR expression, the lack of association between PRMT1 and these promoters could be the result of either reduced BTG1 expression or reduced GR expression, or a combination of the two.
Involvement of BTG1 and PRMT1 in GC-dependent target gene regulation. (A) Association of PRMT1 with the promoter regions of BTG1 and GR in response to GC exposure is dependent on BTG1 expression. Quantitative ChIP analysis of PRMT1 binding to promoter sequences of BTG1 and NR3C1 in RS4;11 control vector-transduced cells versus BTG1-shRNA 1 cells treated with either 1 mg/mL prednisolone or mock treated for 8 hours. Error bars represent the SEM of 2 independent experiments. *Statistical significance (P < .05) was assessed with the Student t test compared with mock-treated cells. (B) Luciferase reporter assays were performed using MMTV-Luc in the presence of 20 ng of exogenous GR and increasing concentrations of HA-BTG1 (3-200 ng), with or without prednisolone treatment. Luciferase activity was normalized to Renilla values. Error bars represent the SEM of 2 independent experiments. *Statistical significance (P < .05) was assessed with Student t test compared with cells without HA-BTG1 expression. (C) Schematic representation explaining how BTG1 regulates expression of the GR and that of GR-target genes.
Involvement of BTG1 and PRMT1 in GC-dependent target gene regulation. (A) Association of PRMT1 with the promoter regions of BTG1 and GR in response to GC exposure is dependent on BTG1 expression. Quantitative ChIP analysis of PRMT1 binding to promoter sequences of BTG1 and NR3C1 in RS4;11 control vector-transduced cells versus BTG1-shRNA 1 cells treated with either 1 mg/mL prednisolone or mock treated for 8 hours. Error bars represent the SEM of 2 independent experiments. *Statistical significance (P < .05) was assessed with the Student t test compared with mock-treated cells. (B) Luciferase reporter assays were performed using MMTV-Luc in the presence of 20 ng of exogenous GR and increasing concentrations of HA-BTG1 (3-200 ng), with or without prednisolone treatment. Luciferase activity was normalized to Renilla values. Error bars represent the SEM of 2 independent experiments. *Statistical significance (P < .05) was assessed with Student t test compared with cells without HA-BTG1 expression. (C) Schematic representation explaining how BTG1 regulates expression of the GR and that of GR-target genes.
BTG1 directly affects GR-mediated gene expression
To more directly implicate BTG1 in GR-mediated transcriptional regulation, we studied the effects of BTG1 on a MMTV-LUC reporter plasmid by transient transfection of 293FT cells. This GC-responsive reporter plasmid, which contains the mouse mammary tumor (MMTV) promoter cloned upstream of the luciferase gene, harbors multiple GC-responsive elements.24 When this reporter was transfected in combination with low amounts of recombinant GR, we observed a robust (15-fold) activation of this promoter in response to prednisolone (Figure 5B). Importantly, titration of increasing amounts of HA-BTG1 resulted in a dose-dependent increase in GR-induced transcriptional activation (Figure 5B). Based on these findings, we propose that BTG1, possibly in association with the arginine methyltransferase PRMT1, promotes GR-mediated gene expression, whereas loss of BTG1 function contributes to GC resistance. This type of therapy resistance appears to involve both defective GR autoregulation as well as reduced activation of GR target genes (Figure 5C).
Discussion
Although GCs are essential drugs in the treatment of ALL and other lymphoid malignancies, the molecular mechanisms that lead to GC resistance in the patient remain unclear.2,5 It is evident, mostly from in vitro studies, that GC resistance can occur at the level of the GR, either by loss-of-function mutations in the GR32-34 or by defective GR autoinduction.32 However, acquired somatic mutations in the GR are rarely found in the patient and not related to GC resistance in vivo.35 Moreover, in a recent study comparing GC-resistant with GC-sensitive ALL, no correlation between GR expression in patient samples and therapy response was seen.6 These studies indicate that GR expression alone does not accurately predict therapy response but rather that GC sensitivity is highly dependent on cellular context, such as the metabolic state of the cell36 or the presence of specific genetic changes.
In this study, we identify BTG1, a gene recurrently deleted in pre-B ALL, as a novel determinant of GC-induced apoptosis that acts by regulating GR-mediated gene expression. Reduced expression of BTG1, either induced by shRNA-mediated knockdown or by (single copy) deletion of the BTG1 gene, as observed in the REH cells, renders pre-B ALL cells refractory to the apoptosis-inducing effects of GCs. This acquired GC resistance appears to be, at least in part, the result of defective GR autoinduction. In RS4;11 cells, shRNA-mediated knockdown of BTG1 leads to a more than 10-fold reduction in GR expression, whereas restoration of BTG1 expression in BTG1-haploinsufficient REH cells promotes GR autoinduction. However, the effects of BTG1 on GR-mediated gene expression and associated therapy responses cannot be explained simply by increased GR expression levels. Restoring GR expression in RS4;11 BTG1 knockdown cells could not restore therapy responses in these cells. Similarly, in the REH cells, which show reduced expression of BTG1 coincident with a single copy loss of BTG1, retroviral transduction of the GR only partially rescued GR-mediated gene transcription and associated therapy response. This suggests that, in both leukemia cell lines, expression of BTG1 might be a rate-limiting factor. Indeed, combined expression of BTG1 and the GR further enhanced GC-induced gene expression in the REH cells and consequently the induction of apoptosis. Similar to GR, elevating BTG1 expression in the REH cells was not sufficient to fully restore therapy response. Apparently, GR expression levels need to exceed a certain threshold before proper activation by BTG1 can take place. As BTG1 expression also increases in response to GC stimulation and GR autoinduction is dependent on BTG1, we conclude that BTG1 and the GR are part of a potent feed-forward autoregulatory loop that enhances the apoptosis-inducing effects of GCs in ALL cells.
Similar to other nuclear receptors, GR-mediated transcription involves the sequential recruitment of transcriptional coregulators into multiprotein complexes. Components of these complexes include acetyltransferases and methyltransferases that modify transcription factors present in the complex and bring about specific combinations of histone modifications at these promoter sites.37,38 In this context, the arginine methyltransferase PRMT1 has been identified as a coactivator of the nuclear receptors HNF4,20,39 ER,40 and FXR.21 Activation of the nuclear receptor HNF4-α by PRMT1 involves a 2-step process. Initially, PRMT1 methylates the receptor, thereby facilitating binding to the target promoter element, after which PRMT1 can bind to the receptor complex and locally methylate histone tails to further enhance transcription.20 Hence, PRMT1 functions as a global regulator of gene expression, and it was shown that disruption of normal PRMT1 function can contribute to leukemia formation.41 As BTG1 was found to interact with PRMT1,42 we postulated that the effects of BTG1 knockdown on GR-mediated gene expression might be the result of a failure to recruit PRMT1 to GC-responsive promoter elements. Consistent with such a model, ChIP assays using PRMT1 antibodies revealed that on GC exposure PRMT1 associates with the promoter regions of both BTG1 and the GR (NR3C1) in a BTG1-dependent manner. As ChIP grade antibodies for BTG1 are not available, we could not establish a direct association of BTG1 with these promoter elements by this method. Instead, luciferase reporter assays using the GC-responsive MMTV promoter were used to demonstrate that expression of recombinant HA-BTG1 enhances prednisolone-induced activation of this promoter in a dose-dependent manner. Because BTG1 and PRMT1 appear to cooperate in GC-mediated gene expression, shRNA-mediated knockdown of PRMT1 was expected to similarly affect GC responsiveness. Lentiviral knockdown of PRMT1 in RS4;11 cells, however, induced massive cell death within days after transduction (results not shown), which probably reflects other important nonredundant functions of PRMT1. The latter may be explained by the fact that PRMT1 is responsible for approximately 80% of all arginine methylation activity within the cell.43
Although our leukemia cell models provide valuable insights into how BTG1 and GR synergize in the control of GR-mediated gene expression, these results cannot be directly translated to the clinical situation. For instance, a near-complete loss of GR expression, as found in the REH cells, is not observed in primary ALL. Hence, it remains to be established to what extent BTG1 copy number losses contribute to therapy resistance in the patient. Assessment of our small patient cohort (33 cases) revealed that a single copy loss of BTG1 at the time of diagnosis was not necessarily associated with a poor therapy response. Indeed, in none of the patients was a poor therapy response to initial GC therapy observed. We therefore suspect that the effect of BTG1 copy number loss on GC responsiveness is highly dependent on the genetic and cellular context in which the lesion occurs. For instance, BTG2, which shows overlapping expression patterns and functions with BTG1,14 may compensate for the effects of BTG1 copy number loss. Consistent with such a compensatory role, GC-sensitive SUP-B15 pre-B ALL cells, which exhibit a BTG1 copy number loss, showed high expression of BTG2, particularly after stimulation with prednisolone; whereas in GC-resistant REH cells, which also lack one copy of the BTG1 gene, expression of BTG2 was undetectable (supplemental Figure 6D). A first clue that deletions in BTG1 may influence therapy outcome was recently provided by Mullighan et al.44 They showed that (therapy-induced) selection of malignant subclones, often already present at the time of diagnosis, may result in a therapy-resistant relapse. In one of the cases examined, a BTG1 lesion not present at diagnosis appeared in the relapse.44 In addition, 4 other relapse samples were found to contain single copy deletions of the NR3C1 gene, which were not present at diagnosis. These results suggest that the GC therapy may have contributed to the selective outgrowth of these clones.
In the present study, we show that loss of BTG1 expression, either induced by deletion or by shRNA-mediated knockdown, is sufficient to induce GC resistance in the defined context of 2 model cell lines. It will be important to determine to what extent BTG1 or other members of the BTG/TOB family modulate GC-induced therapy responses in the patient. As GC-based therapies are used in a variety of medical conditions, a detailed understanding of these novel regulators of GR signaling may provide more rational approaches to overcoming GC resistance.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Prof Dr R. Kofler for providing the GR retroviral expression vector and Prof Dr G. Adema for the CMV-GR expression vector.
This work was supported by the Dutch Cancer Society (grant KUN2005-3660; F.N.v.L. and P.M.H.) and Kinderen Kankervrij (F.N.v.L, P.M.H., and R.P.K.).
Authorship
Contribution: J.C.v.G., L.v.E., M.L., and E.T. performed and interpreted most cell biologic experiments; R.P.K., E.W., S.V.v.R., and C.G. performed and analyzed SNP arrays, expression arrays, and MLPA; J.C.v.G., R.P.K., B.S., A.G.v.K., P.M.H., and F.N.v.L. conceived experiments; J.C.v.G. and F.N.v.L. wrote the manuscript; and all authors reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Frank N. van Leeuwen, Radboud University Nijmegen Medical Centre, Laboratory of Pediatric Oncology, PO Box 9101, 6500 HB Nijmegen, The Netherlands; e-mail: fn.vanleeuwen@cukz.umcn.nl.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal