Abstract
Mutations in the nucleophosmin 1 (NPM1) gene are the most frequent genetic aberrations of acute myeloid leukemia (AML) and define a clinically distinct subset of AML. A monoclonal antibody (T26) was raised against a 19-amino acid polypeptide containing the unique C-terminus of the type A NPM1 mutant protein. T26 recognized 10 of the 21 known NPM1 mutants, including the A, B, and D types, which cover approximately 95% of all cases, and did not cross-react with wild-type NPM1 or unrelated cellular proteins. It performed efficiently with different detection technologies, including immunofluorescence, immunohistochemistry, and flow cytometry. Within a series of consecutive de novo AML patients, 44 of 110 (40%) and 15 of 39 (38%) cases scored positive using the T26 antibody in immunofluorescence and flow cytometry assays, respectively. T26-positive cases were found to be all carrying mutations of NPM1 exclusively, as determined by molecular analysis. T26 is the first antibody that specifically recognizes a leukemia-associated mutant protein. Immunofluorescence or flow cytometry using T26 may thus become a new tool for a rapid, simple, and cost-effective molecular diagnosis of AMLs.
Introduction
Nucleophosmin 1 (NPM1) is a multifunctional phosphoprotein that plays multiple roles in DNA repair and cell proliferation. Although NPM1 has a predominantly nucleolar localization, it shuttles continuously between the cytoplasm and the nucleus, a characteristic that is critical for its function as a molecular chaperone (reviewed by Okuwaki1 ). Somatic mutations in exon 12 of the NPM1 gene are the most frequent genetic abnormality in adult acute myeloid leukemia (AML), found in approximately 35% of all cases and in up to 60% of patients with normal karyotype AML.2 Mutations usually consist of a tetranucleotide duplication at the 3′ end of the gene and are highly heterogeneous, with mutation A composing 75% to 80% of all cases, mutation B approximately 10%, mutation D approximately 5%, and all others being very rare (reviewed by Falini et al3 ). Despite genetic heterogeneity, all of the aforementioned mutations generate a nuclear export signal leading to the aberrant localization of the NPM1 protein in the cytoplasm, a characteristic that underlies the designation of this disease subtype as AML with cytoplasmic NPM1 (NPMc+ AML).3
NPMc+ AML possesses distinctive biologic and clinical features and is included as a provisional entity in the 2008 World Health Organization classification of tumors of hematopoietic and lymphoid tissues.4 NPM1 mutations are mutually exclusive of recurrent chromosomal translocations; they are frequently associated with internal tandem duplications (ITDs) in the FMS-like tyrosine kinase 3 (FLT3) gene and are characterized by unique gene expression5 and microRNA6 signature profiles. Clinically, mutations of NPM1 have an important prognostic significance: in the absence of a coexisting FLT3-ITD mutation, NPMc+ AML is associated with a favorable response to chemotherapy and improved outcome.7 Moreover, NPM1 mutations represent a suitable marker for minimal residual disease (MRD) detection in NPMc+ AML.8,9 Thus, the identification of NPM1 mutations is recommended as an essential part of the diagnostic screening of AML to improve the prognostic stratification potentially allowing for a better assessment of response to therapy through MRD monitoring.7
Currently available methods to detect NPM1 mutations include DNA sequencing, real-time quantitative polymerase chain reaction (PCR), denaturing high-performance liquid chromatography, capillary electrophoresis, locked nucleic acid–mediated PCR clamping, and allele-specific reverse-transcribed PCR assays.10-18 Although highly specific, these methods are expensive and laborious. Alternatively, NPM1 mutations can be identified indirectly, using antibodies that recognize epitopes that are common for both the wild-type (wt) and mutant NPM1 proteins and allow recognition of the cytoplasmic dislocation of NPM1.19 However, as the subcellular localization of NPM1 protein is not reliably detected in smears and cytospins,9,20 this approach is only recommended for bone marrow trephine biopsy samples and requires assistance of expert pathologists for the interpretation of slides.19
We report here the production and characterization of a monoclonal antibody (mAb) against the NPM1 mutant A (NPM1mutA) protein, which allows for a simple and inexpensive identification of the most frequent types of NPM1 mutants. The T26 mAb performs efficiently in both immunofluorescence (IF) and flow cytometry and thus can be used for the diagnosis and follow-up of NPMc+ AMLs.
Methods
Production of the anti-NPM1mutA antibody
A 19-amino acid polypeptide (CQEAIQDLCLAVEEVSLRK) containing the C-terminal end of the NPM1mutA protein and spanning its de novo sequence was synthesized by Primm, conjugated to the keyhole limpet hemocyanin protein, and used to immunize 4 5-week-old female B6C3F1 mice (5 injections at 2-week intervals, with a final boost after 4 weeks). Blood samples were obtained from the tail of the immunized mice and tested for titers against unconjugated peptide by enzyme-linked immunosorbent assay (ELISA). The spleen from the mouse that showed the highest titer was removed, and splenocytes were fused with the mouse myeloma cell line SP2/0. The screening of culture supernatants by ELISA against unconjugated peptide led to the identification of 6 positive clones. The 6 clones were therefore tested by Western blotting (WB) and IF to uncover the one with best performance. The selected clone (T26, producing IgG2k) was subcloned, adapted to growth in serum-free conditions, and seeded in a stationary bioreactor for large-scale antibody production. IgG fractions were affinity-purified using HiTrap MabSelect SuRe (GE Healthcare) protein A-derived ligand. ELISA, WB, and immunoprecipitation experiments were performed according to standard protocols. Mice have been kept and treated according to Italian Law (DL 116/92 and following additions), which enforce the EU 86/609 European Directive.
Epitope mapping
Overlapping peptides (10 amino acids in length with an offset of one amino acid) that cover the entire sequence of the immunogen (JPT Peptide Technologies) were covalently bound to cellulose membrane and tested for their binding of the antibody. Epitope mapping was refined using a series of non-A NPM1 mutants in a format of 13-amino acid–long peptides. Dot-blot hybridization experiments were performed according to the manufacturer's instructions.
Cell lines
Antibody specificity was assessed using a panel of cell lines, including OCI-AML3, OCI-AML5, PLB, U937 leukemia cell lines, and the NPMnull murine embryonic fibroblasts (NPMnull-MEFs). OCI-AML3 cells, which express the NPM1mutA protein, were transduced with a lentivirus (pSicoPuroR) expressing an anti-NPM1mutA -specific short hairpin RNAs (shRNAs; target sequence 5′-gatctctgtctggcagtgg-3′). NPM-null MEFs were engineered to express green fluorescent protein (GFP)–tagged versions of NPM1 or NPM1mutA (NPM1-GFP or NPM1mutA-GFP) by retroviral transduction (cloned in frame with the GFP protein into the PINCO vector).
Patients
In this study, we included 118 hematologic malignancies (110 de novo AMLs, 3 acute lymphoblastic leukemias, 2 myeloproliferative syndromes, and 3 chronic lymphocytic leukemias) referred to the Hematology Department of the Tor Vergata University Hospital (Rome, Italy) during 2008 and 2009. All cases were analyzed by morphology, cytochemistry, immunophenotyping, and cytogenetics. We obtained approval from the Internal Review Board for the research described in this article. Bone marrow/peripheral blood samples were acquired with patients' informed consent in accordance with the Declaration of Helsinki.
IF
Cytospin preparations or bone marrow/peripheral blood smears were fixed in 4% paraformaldehyde for 10 minutes at room temperature, washed in phosphate-buffered saline (PBS), permeabilized with 0.1% Triton X 100/0.2% bovine serum albumin/PBS for 10 minutes, and blocked with 2% bovine serum albumin/PBS for 30 minutes. Slides were incubated with the T26 antibody (1:200 in 2% bovine serum albumin/PBS; ∼ 10 ng/slide) for 1 hour at room temperature and, after one PBS wash, with a Cy3-conjugated goat antimouse immunoglobulin (1:500, Jackson ImmunoResearch) for 1 hour, counterstained with 4′,6-diamidino-2-phenylindole (DAPI), mounted with mowiol, and analyzed with an Olympus BX61 fluorescent microscope using UPlanApo 100×/1.35 Oil objective and equipped with CoolSNAP EZ camera (Photometrics). Single color images were taken with the Metapmorph 7.5.6.0 software (Molecular Devices); overlap of different fluorescence channels and pseudocolor assignment was performed with ImageJ 1.42q (Wayne Rasband, National Institutes of Health). For each patient, we prepared 2 control slides: one stained with DAPI alone and another with DAPI and the secondary antibody, respectively, to evaluate autofluorescence or aspecific binding. Two independent observers unaware of the results of the molecular tests scored each slide.
Flow cytometry
For flow cytometric analyses, cells from 100 μL of whole blood or bone marrow were fixed and permeabilized using the IntraPrep Kit (Beckman Coulter), incubated with 20 μL of the T26 antibody (1:200) for 20 minutes in the dark, blocked with 10 μL of purified IgG1 for 5 minutes, incubated with 10 μL of a fluorescein isothiocyanate-conjugated secondary antibody (goat anti–mouse, Dako Denmark) for 20 minutes, then stained with the appropriate surface antibody (anti–CD45-peridinin chlorophyll protein, anti–CD34-phycoerythrin, anti–CD33-allophycocyanin; BD Biosciences), and analyzed on the FACSCanto I (Becton Dickinson). The surface staining was performed after the intracytoplasmic indirect staining to avoid the cross-reaction of the secondary antibody anti–mouse IgG with the antibodies directed against the surface CD markers. To test the potential use of the T26 antibody for MRD assessment, we performed an assay in which wt NPM1 HL60 cells were mixed with a decreasing number of NPM1mutA-positive OCI-AML3 cells until reaching the dilution of 1 positive cell/105 negative cells. A percentage of 0.0014 positive cells, similar to the expected percentage of 0.001, was obtained acquiring more than 700 000 events (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Immunohistochemistry
Three-micron-thick bone marrow sections were pretreated with an antigen retrieval solution (0.01M ethylenediaminetetraacetic acid buffer, pH 8) for 3 4-minute cycles at 99°C in a microwave oven and then incubated with the T26 antibody at a working dilution of 1:3000 in Tris-buffered saline for 1 hour at room temperature. Detection steps were performed using the Dako EnVision Plus-HRP kit (Dako Denmark) according to the manufacturer's instructions. Peroxidase activity was revealed with 3–3-diaminobenzidine-copper sulfate (Sigma-Aldrich) to obtain a brown-black end product.
Results
The T26 mAb specifically recognizes the mutated NPM1 protein
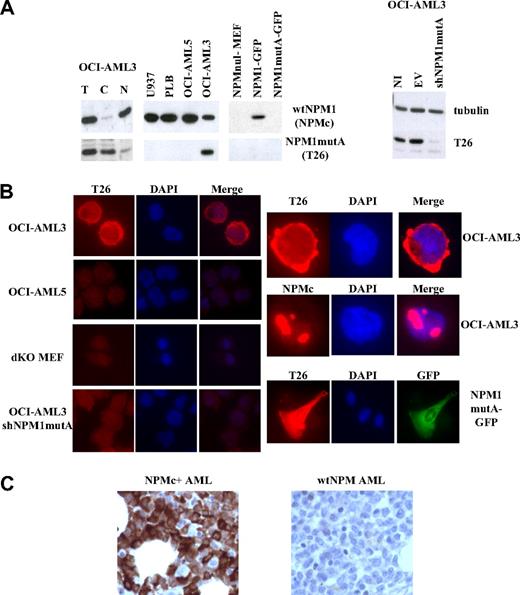
Anti–mutant-NPM1 mAbs were raised by conventional techniques against a 19-amino acid peptide composing the NPM1mutA specific sequence (“Production of the anti-NPM1mutA antibody”). Of the 6 clones found to be positive in the ELISA screen of culture supernatants, the T26 antibody was selected based on its excellent performance in IF. After large-scale production and immunopurification, the specificity of the T26 antibody was assessed by WB and IF using a series of human and murine cell lines: (1) OCI-AML3, a patient-derived cell line carrying the mutation A of NPM1, which expresses both wt and mutant NPM1; (2) an OCI-AML3-derivative with expression of mutant NPM1 (OCI-AML3-shNPM1mutA) silenced by lentivirally mediated expression of NPM1mutA-specific shRNAs; (3) myeloid leukemia cell lines expressing wt NPM1 only (OCI-AML5, U937, and PLB); and (4) NPM-null mouse embryo fibroblasts (NPMnull-MEF) engineered to express wt or mutated (mutA) NPM1 fused to the green fluorescent protein (NPM1-GFP or NPM1mutA-GFP). The T26 mAb recognized an approximately 37-kDa immunoreactive polypetide in the OCI-AML3 cells, which was mainly localized in the cytoplasm (WB of total lysates and nucleus/cytoplasm fractions in Figure 1A left panel). Conversely, T26 failed to recognize such a protein in the PLB, OCI-AML5, U937, or NPM1mutA RNA-interfered OCI-AML3 cells (Figure 1A). All these cell lines, however, expressed wt NPM1 (as revealed by their reactivity with NPMc, a mAb recognizing a C-terminal epitope of wt NPM1 that is lost in the NPM1mut21 ; Figure 1A), suggesting that T26 recognizes mutant NPM1 and does not cross-react with the wt NPM1 protein. Accordingly, T26 recognized NPM1mutA-GFP when exogenously expressed, but not NPM1-GFP, in the context of NPM-null murine fibroblasts (Figure 1A-B).
Validation of the T26 mAb. (A) Western blotting analysis of cells expressing or not the NPM1mutA protein. Lysates from the indicated cell lines were analyzed using mAbs specific to NPM1mutA (T26), wt NPM1 (NPMc), or tubulin β (Santa Cruz Clone H-235). (Left panel) Total lysates (T) were fractionated into cytoplasmic (C) or nuclear (N) fractions. (Right panel) Lysates were prepared from not infected OCI-AML3 cells (NI) or the same cells after infection with lentiviral vector expressing (shNPM1mutA) or not (empty vector; EV) shRNAs against NPM1mutA. (B) IF analysis of the indicated cell lines with the specified antibodies (T26 or NPMc). Staining with T26 or NPMc, DAPI staining, and merged channels (Merge) are shown. Original magnifications: left panels, ×600; right panels, ×1000. (C) IHC of bone marrow trephine biopsy sections from NPMc+ AML (left) or wt NPM AML (right) patients.
Validation of the T26 mAb. (A) Western blotting analysis of cells expressing or not the NPM1mutA protein. Lysates from the indicated cell lines were analyzed using mAbs specific to NPM1mutA (T26), wt NPM1 (NPMc), or tubulin β (Santa Cruz Clone H-235). (Left panel) Total lysates (T) were fractionated into cytoplasmic (C) or nuclear (N) fractions. (Right panel) Lysates were prepared from not infected OCI-AML3 cells (NI) or the same cells after infection with lentiviral vector expressing (shNPM1mutA) or not (empty vector; EV) shRNAs against NPM1mutA. (B) IF analysis of the indicated cell lines with the specified antibodies (T26 or NPMc). Staining with T26 or NPMc, DAPI staining, and merged channels (Merge) are shown. Original magnifications: left panels, ×600; right panels, ×1000. (C) IHC of bone marrow trephine biopsy sections from NPMc+ AML (left) or wt NPM AML (right) patients.
IF analysis showed a predominantly cytoplasmic localization pattern of the T26 mAb in the OCI-AML3 cells, with virtually no staining in the control OCI-AML5, OCI-AML3 knocked-down for NPM1mutA, or NPM-null cells (Figure 1B). The T26 antibody did not cross-react with wt NPM1, as revealed by the different localization patterns obtained with T26 (cytoplasmic) and NPMc (nucleolar) antibodies in the OCI-AML3 cell line. Notably, the T26 staining of cells expressing NPM1mutA-GPF was identical to that of the GFP-fusion protein (Figure 1B). Finally, the T26 mAb specifically stained NPM1mut+ cells in sections of paraffin-embedded bone marrow biopsies (Figure 1C).
Taken together, these data demonstrate that the T26 mAb recognizes the mutated NPM1 protein in WB, IF, and immunohistochemistry (IHC) assays, and that it does not cross-react with wt NPM1.
T26 recognizes the most frequent NPM1 mutations
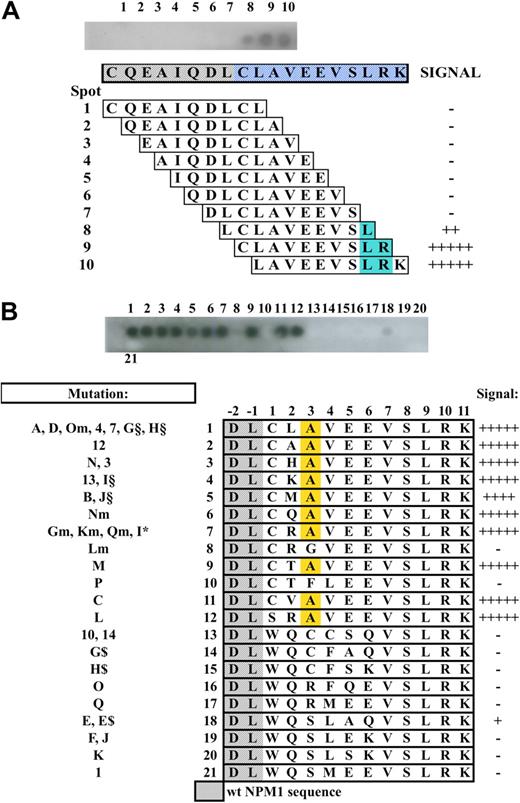
We then tested the specificity of the T26 antibody against the mutation A of NPM1, compared with the other NPM1 mutations. Thirty-five different types of mutations of the NPM1 gene have been described in NPMc+ AML patients, which result in 21 distinct NPM1 mutant proteins (reviewed by Falini et al3 ). All mutant proteins contain a novel C-terminally located sequence of 11 amino acids, of which the last 5 (VLSRK) are common to all mutants. The A, D, Om, 4, 7, G, and H mutations generate the same mutant sequence (CLAVEEVLSRK), which is the most frequently found in NPMc+ AML (95%).3 The 19-amino acid peptide used to generate the T26 mAb contains this sequence and 8 amino acids from wt NPM1 (CQEAIQDLCLAVEEVLSRK; wt NPM1 amino acid are indicated in italics). Epitope mapping by overlapping-peptide scan (using 10 amino acid peptides with an off-set of one amino acid) showed that the LSR 3-amino acid sequence is critical for optimal T26-mAb binding (Figure 2A), thus confirming that the T26 specificity is restricted to the NPM1-mutated sequence.
Epitope mapping and non-A mutant reactivity. (A) Top blot: Dot-blot hybridization of the T26 mAb onto overlapping 10-amino acid peptides (with an offset of one amino acid) spanning the used immunogen. Lower diagram: Schematic representation of the corresponding peptide sequence and T26 mAb binding. Blue represents the common part of the peptides recognized by the antibody. (B) Non-A mutant reactivity. Top blot shows dot-blot hybridization of the T26 mAb onto 21 13-amino-acid peptides containing the unique sequence of the known non-A NPM1 mutants. (Bottom diagram) Schematic representation of the corresponding peptide sequence and T26 mAb binding. Yellow represents the alanine residue at position +3.
Epitope mapping and non-A mutant reactivity. (A) Top blot: Dot-blot hybridization of the T26 mAb onto overlapping 10-amino acid peptides (with an offset of one amino acid) spanning the used immunogen. Lower diagram: Schematic representation of the corresponding peptide sequence and T26 mAb binding. Blue represents the common part of the peptides recognized by the antibody. (B) Non-A mutant reactivity. Top blot shows dot-blot hybridization of the T26 mAb onto 21 13-amino-acid peptides containing the unique sequence of the known non-A NPM1 mutants. (Bottom diagram) Schematic representation of the corresponding peptide sequence and T26 mAb binding. Yellow represents the alanine residue at position +3.
Because the LSR sequence is retained in all NPM1 mutant proteins so far identified, we investigated the ability of T26 to interact with NPM1 mutants other than type A. To test this hypothesis, we performed a dot-blot experiment applying the T26 antibody to a series of 13-amino acid peptides representing all of the known NPM1 mutants, covalently bound to a cellulose membrane (Figure 2B). The results revealed that the T26 mAb reacted with all of the NPM1 mutant sequences containing an alanine residue in position +3 of the 13-amino acid peptides, regardless of the amino acid present at the other variable positions (+1, +2, +4, +5, and +6). Therefore, these data demonstrate that the T26 mAb recognizes in vitro all of the NPM1 mutant proteins matching the xxAxxxVLSRK sequence, which is encoded by the majority of the characterized NPM1 mutations (A, B, C, M, N, Gm, Km, Lm, Nm, Qm, 3, 12, 13, I§, J§, I*, E, E$, and L; ∼ 95%).
The T26 mAb predicts the presence of NPM1 mutations in AML samples
We next validated the T26 mAb as a diagnostic tool for NPMc+ AMLs. To this end, we analyzed bone marrow or peripheral blood samples of 110 consecutive de novo AML and 9 non-AML patients by IF, using the T26 mAb. The same samples were also analyzed in parallel by capillary electrophoresis and allele-specific oligonucleotide (ASO) reverse-transcribed PCR, to identify and characterize NPM1 mutations. Molecular analyses revealed the presence of NPM1 mutations in 44 AML cases. Of them, 38 were of type A, 2 type B, 2 type D, one type I, and one type M (Table 1). IF analysis showed the presence of T26-positive cells in all 44 NPM1-mutated AMLs, regardless of the type of mutation, and in none of the AMLs with wt NPM1 or the non-AMLs. Table 1 and Figure 3A show representative results of 2 positive AMLs carrying type A or B mutation, respectively, and of a negative AML case. Thus, the T26 mAb recognizes AML patients carrying type A NPM1 mutation, as well as other more rare types of NPM1 mutations, as predicted by the in vitro reactivity analysis.
Summary of phenotypic and molecular analyses of AMLc+ samples
| Case no. . | Patient ID . | Blast (FACS), % . | T26+ IF, % . | T26+ (FACS), % . | Mutation . | ||
|---|---|---|---|---|---|---|---|
| Capillary electrophoresis . | ASO-PCR . | Type . | |||||
| 1 | BM 450/08 | 65 | 67 | ND | + | + | A |
| 2 | 9520 | 40 | 35 | ND | + | + | A |
| 3 | 10654 | 72 | 53 | ND | + | + | A |
| 4 | BM 309/08 | 39 | 40 | 95 | + | + | A |
| 5 | 10450 | 82 | 78 | ND | + | + | A |
| 6 | BM 219/08 | 94 | 90 | ND | + | + | A |
| 7 | 10002 | 80 | 84 | ND | + | + | A |
| 8 | BM 627/08 | 79 | 70 | 86 | + | + | A |
| 9 | BM 647/08 | 95 | 60 | 60 | + | + | A |
| 10 | BM 972/08 | 35 | 40 | 99 | + | + | A |
| 11 | BM 227/08 | 64 | 60 | ND | + | + | A |
| 12 | BM 1373/08 | 80 | 30 | ND | + | + | A |
| 13 | 10261 | 70 | 66 | ND | + | + | A |
| 14 | 9700 | 87 | 30 | ND | + | + | A |
| 15 | 9836 | 91 | 90 | ND | + | + | A |
| 16 | BM 929/08 | 75 | 63 | 75 | + | + | A |
| 17 | BM 1620/08 | 70 | 74 | 21 | + | + | A |
| 18 | BM 1637/08 | 77 | 71 | 13 | + | + | A |
| 19 | 10471 | 90 | 88 | ND | + | + | A |
| 20 | BM 1033/08 | 48 | 35 | 85 | + | + | A |
| 21 | BM 831/08 | 52 | 45 | 88 | + | + | A |
| 22 | 9898 | 86 | 60 | ND | + | + | A |
| 23 | 9727 | 83 | 80 | ND | + | + | A |
| 24 | BM 565/08 | 88 | 85 | ND | + | + | A |
| 25 | BM 1557/08 | 65 | 88 | 73 | + | + | A |
| 26 | 10540 | 90 | 80 | ND | + | + | A |
| 27 | BM 1298/08 | 70 | 60 | 35 | + | + | A |
| 28 | BM 240/08 | 80 | 80 | ND | + | + | A |
| 29 | 9908 | 38 | 45 | ND | + | + | A |
| 30 | BM 812/08 | 74 | 65 | ND | + | + | A |
| 31 | BM 1157/08 | 92 | 94 | 54 | + | + | A |
| 32 | BM 10/09 | 42 | 68 | ND | + | + | A |
| 33 | BM 366/08 | 80 | 83 | ND | + | + | A |
| 34 | BM 1478/08 | 57 | 70 | 66 | + | + | A |
| 35 | BM 1068/08 | 65 | 62 | ND | + | + | A |
| 36 | BM 204/09 | 80 | 70 | ND | + | + | A |
| 37 | 10571 | 90 | 90 | ND | + | + | A |
| 38 | BM 1479/08 | 78 | 95 | 80 | + | + | A |
| 39 | BM 242/09 | 40 | 38 | ND | + | − | B |
| 40 | 10400 | 68 | 60 | ND | + | − | B |
| 41 | 10361 | 90 | 80 | ND | + | − | D |
| 42 | BM 68/08 | 95 | 92 | ND | + | − | D |
| 43 | BM 1452/08 | 47 | 68 | 72 | + | − | I |
| 44 | BM 222/09 | 90 | 45 | ND | + | − | M |
| Case no. . | Patient ID . | Blast (FACS), % . | T26+ IF, % . | T26+ (FACS), % . | Mutation . | ||
|---|---|---|---|---|---|---|---|
| Capillary electrophoresis . | ASO-PCR . | Type . | |||||
| 1 | BM 450/08 | 65 | 67 | ND | + | + | A |
| 2 | 9520 | 40 | 35 | ND | + | + | A |
| 3 | 10654 | 72 | 53 | ND | + | + | A |
| 4 | BM 309/08 | 39 | 40 | 95 | + | + | A |
| 5 | 10450 | 82 | 78 | ND | + | + | A |
| 6 | BM 219/08 | 94 | 90 | ND | + | + | A |
| 7 | 10002 | 80 | 84 | ND | + | + | A |
| 8 | BM 627/08 | 79 | 70 | 86 | + | + | A |
| 9 | BM 647/08 | 95 | 60 | 60 | + | + | A |
| 10 | BM 972/08 | 35 | 40 | 99 | + | + | A |
| 11 | BM 227/08 | 64 | 60 | ND | + | + | A |
| 12 | BM 1373/08 | 80 | 30 | ND | + | + | A |
| 13 | 10261 | 70 | 66 | ND | + | + | A |
| 14 | 9700 | 87 | 30 | ND | + | + | A |
| 15 | 9836 | 91 | 90 | ND | + | + | A |
| 16 | BM 929/08 | 75 | 63 | 75 | + | + | A |
| 17 | BM 1620/08 | 70 | 74 | 21 | + | + | A |
| 18 | BM 1637/08 | 77 | 71 | 13 | + | + | A |
| 19 | 10471 | 90 | 88 | ND | + | + | A |
| 20 | BM 1033/08 | 48 | 35 | 85 | + | + | A |
| 21 | BM 831/08 | 52 | 45 | 88 | + | + | A |
| 22 | 9898 | 86 | 60 | ND | + | + | A |
| 23 | 9727 | 83 | 80 | ND | + | + | A |
| 24 | BM 565/08 | 88 | 85 | ND | + | + | A |
| 25 | BM 1557/08 | 65 | 88 | 73 | + | + | A |
| 26 | 10540 | 90 | 80 | ND | + | + | A |
| 27 | BM 1298/08 | 70 | 60 | 35 | + | + | A |
| 28 | BM 240/08 | 80 | 80 | ND | + | + | A |
| 29 | 9908 | 38 | 45 | ND | + | + | A |
| 30 | BM 812/08 | 74 | 65 | ND | + | + | A |
| 31 | BM 1157/08 | 92 | 94 | 54 | + | + | A |
| 32 | BM 10/09 | 42 | 68 | ND | + | + | A |
| 33 | BM 366/08 | 80 | 83 | ND | + | + | A |
| 34 | BM 1478/08 | 57 | 70 | 66 | + | + | A |
| 35 | BM 1068/08 | 65 | 62 | ND | + | + | A |
| 36 | BM 204/09 | 80 | 70 | ND | + | + | A |
| 37 | 10571 | 90 | 90 | ND | + | + | A |
| 38 | BM 1479/08 | 78 | 95 | 80 | + | + | A |
| 39 | BM 242/09 | 40 | 38 | ND | + | − | B |
| 40 | 10400 | 68 | 60 | ND | + | − | B |
| 41 | 10361 | 90 | 80 | ND | + | − | D |
| 42 | BM 68/08 | 95 | 92 | ND | + | − | D |
| 43 | BM 1452/08 | 47 | 68 | 72 | + | − | I |
| 44 | BM 222/09 | 90 | 45 | ND | + | − | M |
The percentage of blasts was assessed by flow cytometry using logical gating based on the physical parameters (FSC/SSC) and on surface antigen expression (CD45, CD33, and CD34 when expressed), IF, or FACS after T26 mAb staining, capillary electrophoresis, or ASO-PCR (specific for the mutation A).
ASO indicates allele-specific oligonucleotide; +, positive for the presence of mutations of the NPM1 gene; −, negative for the presence of mutations of the NPM1 gene. The type of mutation is reported according to the currently used nomenclature3; and ND, not done.
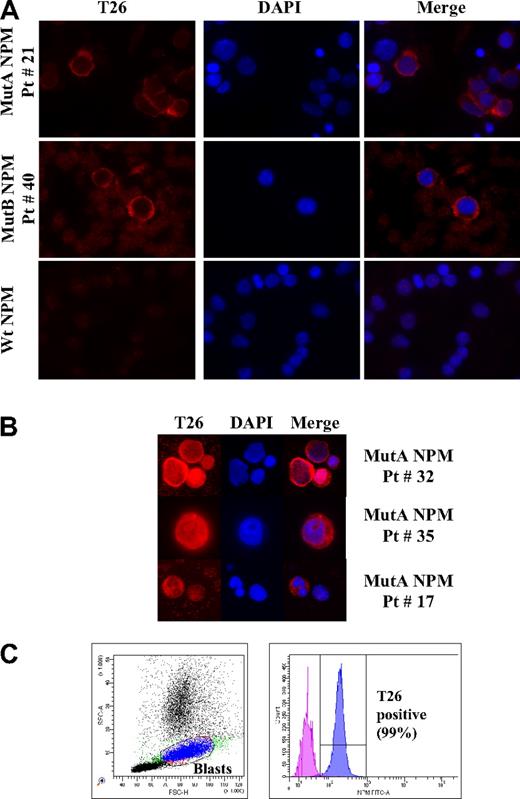
IF and flow cytometric analyses of AML patient samples using the T26 mAb. (A) Representative IF images of bone marrow cells from type A (top panel), type B (middle panel), NPMc+ AML or wt NPM1 AML (bottom panel) patients, as indicated. Patient identification numbers are indicated as in Table 1. (B) Representative IF images of NPMc+ blasts with nuclear + cytoplasmic (top panels) or nucleolar + cytoplasmic (middle panels), and cytoplasmic T26 staining in NPMc+ leukemic cells with segmented nuclei (bottom panels). T26 staining, DAPI staining, and merged channels (Merge) are shown. (C) Representative example of flow cytometric analysis of a bone marrow sample from an NPMc+ AML patient (Pt # 10). Dot-plot showing the blast gate set on forward scatter/side scatter (left) and the histogram displaying the fluorescence of cells within the blast gate (right). Patients' lymphocytes present in the sample were used as a negative control.
IF and flow cytometric analyses of AML patient samples using the T26 mAb. (A) Representative IF images of bone marrow cells from type A (top panel), type B (middle panel), NPMc+ AML or wt NPM1 AML (bottom panel) patients, as indicated. Patient identification numbers are indicated as in Table 1. (B) Representative IF images of NPMc+ blasts with nuclear + cytoplasmic (top panels) or nucleolar + cytoplasmic (middle panels), and cytoplasmic T26 staining in NPMc+ leukemic cells with segmented nuclei (bottom panels). T26 staining, DAPI staining, and merged channels (Merge) are shown. (C) Representative example of flow cytometric analysis of a bone marrow sample from an NPMc+ AML patient (Pt # 10). Dot-plot showing the blast gate set on forward scatter/side scatter (left) and the histogram displaying the fluorescence of cells within the blast gate (right). Patients' lymphocytes present in the sample were used as a negative control.
In the majority of patient samples (∼ 90%), all T26-positive cells showed a strong staining pattern concentrated in the cytoplasm (Figure 3A for representative results). In approximately 10% of patient samples, a small fraction of the T26-positive cells also displayed nuclear-diffuse (∼ 10% of cells) or nucleolar (∼ 2%) staining (Figure 3B).
The frequency of T26-positive cells varied in the different samples from 30% to 95% and paralleled, in the majority of cases (33 cases; 75%), the estimated frequency of leukemic blasts (Table 1). However, in 5 cases (11%), the number of T26-positive cells exceeded significantly (13%-23%) that of blasts, with some T26-positive cells showing segmented nuclei (Figure 3B). In another 6 cases (14%), the fraction of stained cells was inferior to the blast number (12%-50%, Table 1).
Finally, we tested the performance and utility of the T26 mAb in the diagnosis of NPMc+ AML by flow cytometry. To this end, we analyzed 39 AML samples by fluorescence-activated cell sorter (FACS) analysis (including 15 NPMc+ AML cases). Results showed positivity for the T26 mAb staining in all of the 15 NPMc+ AML samples, although it was consistently absent in the 24 AMLs carrying wt NPM1 (Table 1; Figure 3C). The blast population was identified by logic gating based on the physical parameters (forward scatter/side scatter) and on surface antigen expression selected from the immunophenotypic panel used at diagnosis (CD45, CD33, and CD34 when expressed). The percentage of positive cells within the blast gate varied between 13% and 99%.
Discussion
We report here a mAb (T26) that recognizes the mutant NPM1 protein without cross-reacting with the wt NPM1. This is the result of the property of T26 to interact with a specific epitope (AVEEVLSR) included in the de novo CLAVEEVLSRK sequence generated by the NPM1 (type A) mutation and located at the C-terminus of the mutant NPM1 protein. Furthermore, the T26 mAb did not show significant cross-reactivity with other cellular proteins, as revealed by staining of NPMc+ cells after the silencing of the mutated NPM1-allele. As a consequence of these 2 properties, the T26 mAb specifically recognizes only the cells carrying a mutated NPM1 allele. To our knowledge, T26 represents the first antibody that specifically detects a leukemia-associated mutant.
T26 was generated using a peptide immunogen containing the CLAVEEVLSRK sequence encoded by the NPM1 type A mutation. However, T26 also recognized all the NPM1 mutants that introduce an alanine residue at position +3 of the CLAVEEVLSRK sequence, regardless of the identity of the other variant positions (+1, +2, +4, +5, and +6). The alanine residue at position +3 is generated by different nucleotide insertions at positions 959 or 960 of the NPM1 RNA (corresponding, respectively, to mutations L or B, C, M, N, Gm, Km, Lm, Nm, Qm, 3, 12, 13, I§, J§, I*, E, and E$). In summary, T26 recognized 20 of the 35 known NPM1 mutations, including A, B, and D mutations, which cover approximately 95% of all known NPM1 mutations, thus supporting its use as a diagnostic tool for NPMc+ AMLs.
Our data indicate that the T26 mAb performs efficiently with different detection technologies, including immunoprecipitation (not shown), ELISA, WB, IF, IHC, and flow cytometry. Of these, IF and FACS analyses are of particular relevance for diagnostic purposes because of their reduced costs, execution time, and technical difficulty, compared with IHC or molecular tests. We propose, therefore, to add the T26 mAb to the standard panel of antibodies commonly used for IF or FACS characterization of AML immunophenotype at diagnosis. By this approach, the NPM1 mutation status of the patient and the degree of leukemia-cell infiltration can be rapidly determined at the time of diagnosis without the need for more laborious molecular analyses. However, although sequencing for multiple mutations is not yet part of routine diagnostics, we think that the rapid developments in DNA sequencing technology, and the increasing number of mutations identified in AML may lead in the near future to an integrated diagnostic workup for this disease. This may include mAb-based technologies for most homogeneous alterations (eg, NPM1 mutA) and sequencing technologies for highly variable lesions, such as FLT3-ITD.
Smears and/or cytospin preparations, which are usually used for the execution of IF staining, are not currently considered suitable for the diagnosis of NPMc+ AML (using an N-terminal anti-NPM1 antibody that recognizes both wt and mutated NPM1). This is based on reports showing artifactual diffusion of the wt NPM1 protein to the cytoplasm during slide preparation and sample fixation.9,20 In our hands, however, N- or C-terminal anti-NPM1 antibodies recognize nucleolar NPM1 in smears and/or cytospin preparations of both wt and NPMc+ AML samples (supplemental Figure 2). Nevertheless, this limit is overcome, in principle, using the T26 antibody because it does not cross-react with the wt NPM1.
A limit of the IF technology that we encountered in the present study is represented by rare (highly) autofluorescent cells that may be detected in the analysis of bone marrow or peripheral blood smears. These cells are large and show a characteristic granular pattern of fluorescence in all fluorescence channels (supplemental Figure 3). However, if appropriate controls (DAPI only and secondary/DAPI staining) are included, they can be easily identified and excluded from the counting of T26-positive cells.
Furthermore, the use of T26 in combination with antibodies specific for wt NPM1 may prove useful to investigate the effects of the NPM1 mutation on the wt NPM1 protein. It has been reported that the NPM1 mutants delocalize the wt protein to the cytoplasm because of their ability to form stable complexes.22 We observed, however, that a sizable fraction of wt and mutated NPM1 proteins remain in separate compartments in the NPMc+ AML blasts (some of the mutated protein remains in the nucleus and nucleolus, whereas a large fraction of the wt NPM1 remains in the nucleolus; Figure 1B; supplemental Figure 4).
Finally, in patients who test positive at diagnosis, the T26 mAb staining may also provide a reliable tool to assess MRD after treatment or disease evolution at relapse. Notably, serial dilution experiments and FACS analysis showed that the T26 antibody can detect as few as 0.001% positive cells (supplemental Figure 1), a range of sensitivity that may prove informative for residual disease or imminent relapse.23 As an example of how the T26 mAb may help identify leukemic cells regardless of their changing phenotype, we have longitudinally analyzed the immunophenotype (including mutated NPM1) of a NPMc+ AML patient at the time of diagnosis and at relapse and found that the same leukemia clone converted from CD34-positive to CD34-negative (supplemental Figure 4).
More in general, the availability of a mAb that unambiguously recognizes leukemic cells will allow high-sensitivity tracking of the leukemic clone during the natural history of the disease.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Annunziata Venuto and Serena Laterza for technical support and the medical staff of the Department of Biopathology, University Tor Vergata, Rome for excellent support, collection of patient samples, and information; and Drs Emanuela Colombo and Myriam Alcalay for critical reading of the manuscript.
This work was supported by AIRC, European Union (FP7-Genica), and the Vollaro Foundation.
Authorship
Contribution: A.M.G. designed and performed research, analyzed data, and wrote the paper; S.L., M.I.C., T.O., and F.B. performed research and analyzed data; C.M., M.C., G.O., G.P., and M.D. performed research; M.C. collected data; E.A. and A.V. interpreted data; A.d.M. designed research and interpreted data; and F.L.-C. and P.G.P. designed research, interpreted data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pier Giuseppe Pelicci, European Institute of Oncology at the IFOM-IEO Campus, Via Adamello, 16, 20139 Milan, Italy; e-mail: piergiuseppe.pelicci@ifom-ieo-campus.it.
References
Author notes
F.L.-C. and P.G.P. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal