Polycomb group (PcG) proteins are transcriptional repressors with a central role in the establishment and maintenance of gene expression patterns during development. We have investigated the role of polycomb repressive complexes (PRCs) in hematopoietic stem cells (HSCs) and progenitor populations. We show that mice with loss of function mutations in PRC2 components display enhanced HSC/progenitor population activity, whereas mutations that disrupt PRC1 or pleiohomeotic repressive complex are associated with HSC/progenitor cell defects. Because the hierarchical model of PRC action would predict synergistic effects of PRC1 and PRC2 mutation, these opposing effects suggest this model does not hold true in HSC/progenitor cells. To investigate the molecular targets of each complex in HSC/progenitor cells, we measured genome-wide expression changes associated with PRC deficiency, and identified transcriptional networks that are differentially regulated by PRC1 and PRC2. These studies provide new insights into the mechanistic interplay between distinct PRCs and have important implications for approaching PcG proteins as therapeutic targets.
Introduction
Polycomb group (PcG) proteins are epigenetic repressors important in the maintenance of transcriptional silencing. PcG proteins exist in 3 distinct complexes: polycomb repressive complexes (PRC)1, PRC2 and pleiohomeotic repressive complex (PhoRC). PRC2 has 4 constituent proteins: E(z), Su(z)12, Esc, and Pcl in flies and Ezh2, Suz12, Eed, and Phf1 in mammals (Figure 1A). Ezh2 is the enzymatic component of PRC2, catalyzing methylation of lysine 27 of histone 3 (H3; H3K27),1,,–4 whereas Suz12 and Eed are required for complete function of Ezh2 and stable formation of the complex.5,6 Phf1 influences the enzymatic specificity of Ezh2, promoting trimethylation of H3K27 (H3K27me3) in preference to dimethylation (H3K27me2).7 Dimethylation is broadly distributed throughout the genome and is thought to have a structural role, whereas H3K27me3 is enriched on promoters and associated with transcriptionally silent regions.8,,–11
There has been a vast expansion in the number of PRC1 components in mammals. PRC1 components include Bmi1 and Mel18 (orthologues of Drosophila Psc), Cbx2, Cbx4, and Cbx8 (orthologues of Drosophila Pc), Scmh1 and Scmh2 (orthologues of Drosophila Scm), Phc1/Rae28, Phc2 and Phc3 (orthologues of Drosophila Ph), and Ring1A and Ring1B (orthologues of Drosophila Ring; Figure 1A). Ring1A and Ring1B are the enzymatic components of PRC1 responsible for the mono-ubiquitination of histone 2A (H2A) at lysine 119 (H2AK119Ub),12 other components such as Bmi1 and Mel18 are able to stimulate this activity. H2AK119Ub is also associated with transcriptional repression.13,–15
PhoRC is composed of the DNA binding transcription factor Yy1 (Drosophila Pho) and Sfmbt1 (Figure 1A). PhoRC is not known to possess any enzymatic activity. Rather, Sfmbt binds methylated histone residues,16 and Yy1 is the only mammalian PcG protein with specific DNA binding activity,17 suggesting that PhoRC may direct PRC1 or PRC2.
Some years ago the chromodomain of Drosophila polycomb was shown to bind H3K27me3, leading to the hierarchical recruitment model (reviewed in Wang et al18 ), in which PRC2 trimethylates H3K27, leading to PRC1 recruitment and ubiquitination of H2AK119. Several lines of evidence support this prevailing model: (1) a high degree of overlap between sites bound by PRC1 and PRC2 in human and mouse cells9,10,19 ; (2) the failure of Drosophila E(z) mutants to recruit PRC120 ; and (3) a correlation between H3K27me3 levels and PRC1 recruitment and H2AK119Ub accumulation.21 A growing number of reports now suggest that PRC1 and PRC2 may not always act in this fashion: PRC1 binds nucleosomes that lack N-terminal tails in vitro22 ; PRC1 is recruited in PRC2-deficient mammalian cells23 ; and genome-wide chromatin immunoprecipitation studies show PRC2 bound to loci devoid of PRC119 and vice versa.15 These observations imply that the interaction between various PRCs is complex.
Genome-wide chromatin immunoprecipitation studies suggest PRCs play a role in embryonic stem (ES) cells by repressing key developmental transcription factors,8,9 although they also play roles in adult neural, mammary, and hematopoietic stem cells (HSCs). HSCs deficient in Bmi1, Mel18, or Phc1/Rae28 have defective repopulating capacity that is partially explained by de-repression of the Ink4a/Arf locus, which encodes the cyclin-dependent kinase p16Ink4a and tumor suppressor p19Arf.24,25 Deletion of the Ink4a/Arf locus only partially rescues the Bmi1−/− phenotype, pointing to a broader role for PRC1 in regulating HSC function. In contrast to the role of PRC1, the PRC2 component Suz12 restricts HSC activity; reduced levels of Suz12 in HSCs result in enhanced HSC activity,26 reminiscent of earlier studies using hypomorphic alleles of Eed.27 Although there is growing evidence that PRC1 and PRC2 have opposing activities in HSCs, the molecular bases for the differences remain undefined.
The study of PRC2 in HSCs has been limited by the embryonic lethality observed for null mutants. We have used the Mpl−/− genetic background, which provides a sensitized environment to detect small changes in HSC activity in viable heterozygous animals.28 c-Mpl is the receptor for thrombopoietin, a cytokine critical for both HSC function and platelet production.29 Our previous studies have shown that peripheral blood platelet count is a surrogate marker for stem cell activity in these mice.26,30 We reasoned that by combining the Mpl null-sensitized background and mutations in one or more of the PRCs, we would be able to systematically test whether the PRCs act in synergy, and thus test whether the hierarchical model for PRC action holds true in hematopoiesis.
Here we report that mutation of Ezh2, Eed, or Suz12 enhances HSC/progenitor activity, suggesting that PRC2 as a complex restricts HSC/progenitor activity. We establish genetically that PRC1 and PRC2 have opposing roles in HSC/progenitor populations, similar to what is seen in progenitors by others27 and contrary to the prediction of the hierarchical model of PRC action. We have also shown that PhoRC, like PRC1, enhances HSC/progenitor function. Finally, we investigated the gene expression signatures that account for the opposing role of the PRCs in HSC/progenitors, and have identified genes that are differentially regulated between PRC1 and PRC2. This set of genes encompasses the transcriptional networks downstream of HoxA9 and C/EBPα, both of which are central to normal development of the hematopoietic system and are implicated in disease.
Methods
Mouse strains
All mice were on an inbred C57BL/6 background, unless otherwise stated. Mice carrying the Eed3354 allele were purchased from Oak Ridge National Laboratories. The Eed3354 point mutation was detected using an allelic discrimination assay (supplemental Table 5; available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The Ezh2Del allele was generated from the Ezh2fl allele and genotyped as previously described.31 Animal studies were approved by The Walter and Eliza Hall Institute Animal Ethics Committee.
Hematologic analyses
Analyses were performed essentially as previously described.26 For bone marrow competitive transplantations, test CD45Ly5.2 bone marrow was mixed with an equal number or 10-fold excess of competitor CD45Ly5.1 bone marrow cells, and 2 × 106 nucleated cells injected into irradiated CD45Ly5.1/CD45Ly5.2 or CD45Ly5.1 recipients. For fetal liver lineage marker–negative (Lin−), Sca-1+, c-Kit+ (LSK) competitive transplantations, 4 to 5 livers per genotype were pooled before sorting. A total of 1000 test CD45Ly5.2 LSK cells, 1000 competitor CD45Ly5.1 LSK cells, and 200 000 CD45Ly5.1/CD45Ly5.2 filler bone marrow cells were injected into 4 lethally irradiated CD45Ly5.1/CD45Ly5.2 recipients, per sorted LSK sample. Test and competitor contribution was analyzed at 10 weeks after reconstitution for peripheral blood, and 12 to 16 weeks for other organs.
LSK analysis and fluorescence-activated cell sorting
Bone marrow LSK analysis was performed as previously described.26 Individual fetal livers were dissociated in isotonic buffer containing 5% (vol/vol) fetal calf serum (FCS; HyClone Laboratories). Samples for LSK enumeration were treated with red cell removal buffer, otherwise all cells were stained with rat monoclonal antibodies against Ter119, CD3, CD5, B220, Gr1, and CD8. Samples used for sorting were incubated with BioMag goat anti–rat IgG beads (QIAGEN), and lineage marker–positive cells removed using a Dynal magnet (Invitrogen). The lineage-depleted cells, or all cells in the case of LSK enumeration, were stained with fluorophore-conjugated anti–rat IgG antibody, then monoclonal antibodies to c-Kit, Sca-1, and CD45Ly5.1, and CD45Ly5.2 where applicable. Cells were flow sorted on a FACSDiva, FACSAria (BD Biosciences), or MoFlo (DakoCytomation).
shRNA-mediated viral knockdown and transplantation of fetal liver cells
Embryonic day 14.5 (E14.5) fetal livers were harvested and dissociated in isotonic buffer containing 5% FCS (HyClone Laboratories). Ter119-positive cells removed via magnetic bead depletion. Ter119-depleted cells were cultured in Iscove modified Dulbecco medium with 15% FCS and cytokines (10 ng/mL interleukin-6, 5 ng/mL Flt3 ligand, 50 ng/mL thrombopoietin, and 100 ng/stem cell factor) for 4 hours. Retroviral supernatants were applied to culture dishes treated with RetroNectin (Takara Biosciences). Supernatant was removed, and fetal liver cells cultured on the viral coated dishes in the presence of 4 μg/mL polybrene (Sigma-Aldrich). Fetal liver cells were washed off plates after 16 to 24 hours infection. The proportion of infected cells was determined by measuring green fluorescent protein (GFP), and approximately 3 to 5 × 105 viable nucleated cells were injected into each lethally irradiated recipient. The sequence of each short hairpin RNA (shRNA) is given in supplemental Table 5, and each was used in the LMS vector.
RNA extraction and quantitative polymerase chain reaction
RNA was extracted from 5 × 103 to 1 × 106 sorted cells using RNeasy micro or mini columns (QIAGEN). cDNA was synthesized using oligo(dT) primers and either Superscript III reverse transcriptase (Invitrogen) or Sensiscript reverse transcriptase (QIAGEN). Quantitative polymerase chain reactions (qPCRs) were set up with duplicate technical replicates, and the expression of mouse genes determined using TaqMan gene expression assays, as detailed in supplemental Figures 3, 4, 5, 6, and 7.
Microarray hybridization and differential expression
RNA extracted as described in “RNA extraction and quantitative polymerase chain reaction” from 40 000 to 70 000 sorted fetal liver LSK cells was labeled, amplified, and hybridized to Illumina MouseWG-6 Version 2.0 Expression BeadChips according to Illumina standard protocols at the Australian Genome Research Facility. Differential expression analysis was performed as described in supplemental Methods. Gene Expression Omnibus (National Center for Biotechnology Information) accession number GSE21404 is assigned.
Gene set testing
The overlap of the current microarray results with previous experiments was assessed using mean-rank gene set enrichment tests. A Wilcoxon signed-rank test was used to assess whether each external gene set was highly ranked when all genes on the microarrays were ordered from most to least differentially expressed using the current data.
Genuine association of expression profiles
A novel procedure, implemented in the genuine association of expression profiles (GENAS) function of the limma package (Bioconductor), was applied to estimate correlations between the expression profiles of different mouse mutants. Each mutant was compared with wild-type, producing a set of log2-fold changes. The GENAS analysis determines whether these fold changes are correlated between mutants, after allowing for estimation uncertainty. The method is a generalization of the limma empirical Bayes analysis32 in which the moderated t statistics are modeled using a multivariate t distribution. The biologic correlation between the mutants was estimated from the prior correlation of the log2-fold changes in the Bayesian hierarchical model. In this way, the method is able to separate true biologic correlation from technical correlation. The statistical significance of the correlation is assessed by a likelihood ratio test.
Romer gene set enrichment analysis
Gene set enrichment analysis (GSEA) is an approach that correlates a large database of coregulated gene sets with respect to a microarray dataset.33 Romer is a new GSEA method suitable for microarray linear model analyses including array weights and possibly a small number of replicates. Romer was applied using the curated (c2) gene sets from MSigDB (http://www.broadinstitute.org/gsea/). Each gene set was converted to mouse using the orthology mapping from The Jackson Laboratory. Romer can find gene sets associated with any contrast in a linear model. We looked for gene sets regulated in the same direction by Bmi1 and Suz12 by comparing the average of Bmi1−/− and Suz12Plt8/+ to wild-type. Gene sets regulated in opposite directions by Bmi1 and Suz12 were found by directly contrasting Bmi1−/− versus Suz12Plt8/+. The same steps were undertaken for Bmi1 and Yy1. In each case, sets with a P value less than .05 for up- or down-regulation were selected.
Results
All core PRC2 components restrict HSC/progenitor population activity
To study the role of PRC2 components (Figure 1A) in HSC/progenitor populations, we produced Mpl−/− animals heterozygous for null alleles of Eed (Eed3354)34 or Ezh2 (Ezh2Del),31 and compared peripheral blood counts to Mpl−/− animals heterozygous for a loss-of-function point mutant of Suz12 (Suz12Plt8).26 Heterozygosity for mutations in Suz12, Eed, or Ezh2 significantly ameliorated the Mpl−/− thrombocytopenia and significantly increased peripheral white blood cell counts (Figure 1B).
All core PRC2 components restrict HSC/progenitor activity. (A) The 3 PRCs, displaying mammalian components, Drosophila counterparts (shown in italics), and enzymatic activities. (B) Peripheral blood platelet (top panels) and white blood cell counts (bottom panels) at 7 weeks of age from mice of the given genotypes. The horizontal bar marks the mean for n = 20 to 25 samples per genotype. Eed3354 mice were on a mixed genetic background. (C) Bone marrow cells from 3 CD45Ly5.2 donor mice of the given genotype were mixed with Ezh2+/+Mpl−/−CD45Ly5.1 competitor cells at 1:1 (left) or 1:10 (right) ratios and used to reconstitute 3 lethally irradiated recipients per donor. Contribution of test cells is shown at 10 to 16 weeks after reconstitution. Blood indicates peripheral white blood cells; and Mϕ, macrophages. Statistical significance: *P < .05, **P < .01, ***P < .001, corrected for multiple testing; error bars indicate SEM.
All core PRC2 components restrict HSC/progenitor activity. (A) The 3 PRCs, displaying mammalian components, Drosophila counterparts (shown in italics), and enzymatic activities. (B) Peripheral blood platelet (top panels) and white blood cell counts (bottom panels) at 7 weeks of age from mice of the given genotypes. The horizontal bar marks the mean for n = 20 to 25 samples per genotype. Eed3354 mice were on a mixed genetic background. (C) Bone marrow cells from 3 CD45Ly5.2 donor mice of the given genotype were mixed with Ezh2+/+Mpl−/−CD45Ly5.1 competitor cells at 1:1 (left) or 1:10 (right) ratios and used to reconstitute 3 lethally irradiated recipients per donor. Contribution of test cells is shown at 10 to 16 weeks after reconstitution. Blood indicates peripheral white blood cells; and Mϕ, macrophages. Statistical significance: *P < .05, **P < .01, ***P < .001, corrected for multiple testing; error bars indicate SEM.
We tested the functional activity of long-term repopulating stem cells from Ezh2Del/+Mpl−/− mice by competitive reconstitution of lethally irradiated recipients. Transplantation of equal numbers or a 10 to 1 excess of Ezh2+/+ bone marrow over Ezh2Del/+ bone marrow resulted in an increased representation of the Ezh2Del/+ cells compared with Ezh2+/+ across all cell lineages and hematopoietic organs (Figure 1C). Our results suggest that PRC2 restricts HSC/progenitor activity.
PRC1 and PRC2 have opposing roles in the HSC/progenitor compartment
We tested the interaction between PRC1 and PRC2 in HSC/progenitors by examining the number of platelets in Mpl−/−Suz12Plt8/+ heterozygous mice, which were also heterozygous for mutations in the PRC1 genes Bmi1, Mel18, Phc1/Rae28, Phc2, Ring1A, Ring1B, and Cbx2.35,,,,–40 Heterozygosity for Bmi1 exacerbated Mpl−/− thrombocytopenia, whereas heterozygosity for Suz12 increased peripheral platelet and white blood cell counts (P < .001; Figure 2A). Compound heterozygotes for Bmi1 and Suz12 had peripheral platelet counts intermediate between each of the single mutants, in keeping with opposing functions of PRC1 and PRC2 (Figure 2A). Consistent results were seen in mice heterozygous or homozygous for each of the other PRC1 mutant alleles; where effects on platelet number were found, they were in the opposite direction to those of the Suz12Plt8 allele (supplemental Figure 1).
PRC1 and PRC2 do not synergize in HSC/progenitor function. (A) Peripheral blood platelet (top panel) and white blood cell counts (bottom panel) at 7 weeks of age from mice of the given genotypes, all additionally Mpl−/−. The horizontal bar marks the mean for n = 40 samples per genotype. (B) Bone marrow cells from 6 to 9 Mpl−/−CD45Ly5.2 donor mice of the given genotype were mixed 1:1 with wild-type Mpl−/−CD45Ly5.1 competitor cells and used to reconstitute 3 lethally irradiated recipients per donor. Contribution of test cells to each hematopoietic organ is shown 10 to 16 weeks after reconstitution. Blood indicates peripheral white blood cells. Statistical significance *P < .05, **P < .01, ***P < .001, corrected for multiple testing; error bars indicate SEM.
PRC1 and PRC2 do not synergize in HSC/progenitor function. (A) Peripheral blood platelet (top panel) and white blood cell counts (bottom panel) at 7 weeks of age from mice of the given genotypes, all additionally Mpl−/−. The horizontal bar marks the mean for n = 40 samples per genotype. (B) Bone marrow cells from 6 to 9 Mpl−/−CD45Ly5.2 donor mice of the given genotype were mixed 1:1 with wild-type Mpl−/−CD45Ly5.1 competitor cells and used to reconstitute 3 lethally irradiated recipients per donor. Contribution of test cells to each hematopoietic organ is shown 10 to 16 weeks after reconstitution. Blood indicates peripheral white blood cells. Statistical significance *P < .05, **P < .01, ***P < .001, corrected for multiple testing; error bars indicate SEM.
The long-term repopulating capacity of stem cells from the Bmi1 and Suz12 compound heterozygous animals was tested using 1:1 competitive transplantations of bone marrow cells into lethally irradiated recipients (Figure 2B). Bmi1 and Suz12 compound heterozygous animals show a level of contribution to recipients similar to wild-type, and significantly lower than the elevated contribution seen for Suz12 single heterozygotes (P < .01; Figure 2B). These results show that Bmi1 and Suz12 have opposing activities in HSC/progenitor function.
Yy1 as part of PhoRC enhances HSC/progenitor activity
Yy1 may be able to localize PRC1 or PRC2 through its DNA binding capacity, so we investigated whether PhoRC also has a role in HSC/progenitor function by using a null allele of Yy1.41 Because Yy1−/− mice die peri-implantation, we produced Mpl−/−Yy1+/− mice (Figure 3A) and found a significantly exacerbated thrombocytopenia and leukopenia (P < .001; Figure 3A). Subtle, but similar, defects were seen in Mpl+/+Yy1+/− mice (supplemental Figure 2). In the same experiment, we tested whether Yy1 and Suz12 cooperate in hematopoiesis. We found that compound heterozygotes for Yy1 and Suz12 displayed intermediate peripheral platelet and white blood cell counts on the Mpl−/− background (Figure 3A). Heterozygosity for Yy1 produced a PRC1-like phenotype, rather than a PRC2-like phenotype.
Yy1 does not synergize with PRC2 and has a role enhancing HSC/progenitor activity. (A) Peripheral blood platelet (top panel) and white blood cell counts (bottom panel) at 7 weeks of age from Mpl−/− mice of the given genotypes. The horizontal bar shows the mean for n = 11 to 40 samples. (B) Fetal liver cellularity (left), proportion (middle), and number (right) of LSK cells per E14.5 fetal liver of the given genotype. Data are from one litter of 4 Yy1+/+ and 5 Yy1+/− embryos, representative of data from 3 independent litters. (C) Fetal liver LSK cells from 4 to 5 individual CD45Ly5.2 donor embryos of the given genotype were mixed with an equal number of wild-type CD45Ly5.1 competitor fetal liver LSK cells (n = 2 samples) and used to reconstitute 4 lethally irradiated recipients per donor sample. Contribution of test cells to each hematopoietic organ and LSK cells is shown for 12 to 16 weeks after reconstitution. Blood indicates peripheral white blood cells. (D) E14.5 fetal liver cells were infected with retrovirus carrying shRNAs against Yy1 or a nonsilencing control (Nonsil) and used to reconstitute lethally irradiated recipients. The proportion of cells infected with each shRNA (GFP+) was compared at input and at 6 to 12 months after reconstitution in spleen, thymus, bone marrow, and the LSK cell compartment. The effect of the Nonsil shRNA in each of 3 experiments was normalized to 1. Statistical significance *P < .05, **P < .01, ***P < .001, corrected for multiple testing; error bars indicate SEM.
Yy1 does not synergize with PRC2 and has a role enhancing HSC/progenitor activity. (A) Peripheral blood platelet (top panel) and white blood cell counts (bottom panel) at 7 weeks of age from Mpl−/− mice of the given genotypes. The horizontal bar shows the mean for n = 11 to 40 samples. (B) Fetal liver cellularity (left), proportion (middle), and number (right) of LSK cells per E14.5 fetal liver of the given genotype. Data are from one litter of 4 Yy1+/+ and 5 Yy1+/− embryos, representative of data from 3 independent litters. (C) Fetal liver LSK cells from 4 to 5 individual CD45Ly5.2 donor embryos of the given genotype were mixed with an equal number of wild-type CD45Ly5.1 competitor fetal liver LSK cells (n = 2 samples) and used to reconstitute 4 lethally irradiated recipients per donor sample. Contribution of test cells to each hematopoietic organ and LSK cells is shown for 12 to 16 weeks after reconstitution. Blood indicates peripheral white blood cells. (D) E14.5 fetal liver cells were infected with retrovirus carrying shRNAs against Yy1 or a nonsilencing control (Nonsil) and used to reconstitute lethally irradiated recipients. The proportion of cells infected with each shRNA (GFP+) was compared at input and at 6 to 12 months after reconstitution in spleen, thymus, bone marrow, and the LSK cell compartment. The effect of the Nonsil shRNA in each of 3 experiments was normalized to 1. Statistical significance *P < .05, **P < .01, ***P < .001, corrected for multiple testing; error bars indicate SEM.
Approximately 1 in 6 Yy1+/− embryos on a mixed genetic background do not survive to weaning, attributable to developmental retardation and neural defects.41 On an inbred C57BL/6 background, we found that 30% of Yy1+/− animals die before weaning. The combination of Mpl deficiency and the C57BL/6 inbred background led to an even greater mortality of Yy1+/− animals, with 60% of Mpl−/−Yy1+/− animals dying before weaning (P < .001; Figure 3A). Thus, Yy1 and Mpl interact genetically, consistent with a role for both Yy1 and Mpl in HSCs.
As we observed, significantly reduced numbers of viable Yy1+/− adults, we used fetal livers from E14.5 embryos for further studies. We analyzed the number of HSC/progenitors using LSK staining. Heterozygosity for Yy1 in Mpl+/+ E14.5 fetal livers resulted in a significant reduction in the proportion and absolute number of LSK cells, without any decrease in fetal liver cellularity (P < .01; Figure 3B). Competitive transplantation of equal numbers of purified Mpl+/+Yy1+/− and Mpl+/+Yy1+/+ fetal liver LSK cells into lethally irradiated recipients (Figure 3C) suggested that the Yy1+/− cells are compromised in their ability to contribute to maintenance of the stem cell compartment and to blood cell formation (Figure 3C and data not shown).
To confirm the role of Yy1 in HSC/progenitors, we used shRNA-mediated viral knockdown of Yy1 in fetal liver cells, followed by transplantation into lethally irradiated recipients. At 6 to 12 months after transplantation, the proportion of Yy1 knockdown cells decreased significantly compared with the input cells and the nonsilencing control, across all cell lineages including LSK cells, and all hematopoietic organs (Figure 3D). Consistent with the phenotype of Suz12 mutant mice, Suz12 knockdown cells displayed an increased contribution across all cell types and hematopoietic organs (data not shown), as shown previously.26 Splenic B cells were isolated from recipient animals and knockdown of Yy1 and Suz12 confirmed in vivo in the shRNA-containing cells (GFP+, supplemental Figure 3). These results confirm that Yy1 enhances HSC/progenitor activity and number, independent of its role in embryonic development.
Identification of gene sets differentially regulated by PRC1, PRC2, and PhoRC
Identification of genes differentially regulated by the PRCs in hematopoietic cells was important to pinpoint the molecular bases for their opposing roles. The Ink4a/Arf locus is a recognized target of PRC1 in hematopoietic cells, and repressed by all 3 PRCs in fibroblasts. Using shRNA-mediated viral knockdown of Yy1, Bmi1, and Suz12, we investigated whether the Ink4a/Arf locus is repressed by each complex in Gata-1 null megakaryocyte erythroid (G1ME) cells.42 Using an in vitro competition assay between infected (GFP+) and uninfected (GFP−) cells, we found a mild selection against Suz12 shRNA-infected cells and a strong selection against cells infected with shRNAs directed against Bmi1 or Yy1 (supplemental Figure 4A).
Infected and uninfected cells were sorted on days 4 and 7 postinfection and analyzed by qPCR for knockdown of Bmi1, Yy1, and Suz12, and for expression of Cdkn2a (p16Ink4a, supplemental Figure 4B). Each hairpin produced greater than 80% knockdown of the target (supplemental Figure 4B). We did not observe any difference in Cdkn2a expression between cells uninfected and infected with the Nonsil or Suz12 shRNAs, but found significant up-regulation of Cdkn2a expression in cells infected with one Yy1 shRNA and the Bmi1 shRNA, and a small but not statistically significant increase in a different Yy1 shRNA (supplemental Figure 4B). Given the potency of p16Ink4A, increases in Cdkn2a need only be modest to slow growth of the cell and bring about a halt in cell-cycle progression. Although Suz12 knockdown did not alter Cdkn2a expression in G1ME cells (supplemental Figure 4B), knockdown was effective because we observe changes in 3 other genes we previously found to be sensitive to Suz12 dose in LSK or Lin− c-Kit+ cells (supplemental Figure 5).26 This suggests that the Ink4a/Arf locus is differentially sensitive to the amount of Suz12, Bmi1, and Yy1 in the G1ME hematopoietic cell line.
To test whether changes in G1ME cells also occurred in LSK cells, we analyzed Cdkn2a expression by qPCR in fetal liver LSK cells sorted from individual E14.5 embryos (supplemental Figure 6A). Cdkn2a was undetectable in wild-type fetal liver LSKs, more than 1000 times lower than the housekeeping gene Hprt1. Cdkn2a was detectable in fetal liver LSK cells from some Bmi1+/− embryos, and in all Bmi1−/− embryos tested, whereas it was never detected in Yy1+/− or Suz12Plt8/+ embryos (supplemental Figure 6A). We saw up-regulation of Cdkn2a in G1ME cells depleted of Yy1, but not in LSK cells heterozygous for Yy1, suggesting that although the locus is sensitive to dosage of Yy1, derepression of Cdkn2a may not be responsible for the deficiency of Yy1 heterozygous HSC/progenitors. To confirm we could detect gene expression changes in LSK cells from Yy1 or Suz12 mutants, we analyzed the expression of Prr6, a gene we previously identified to be highly responsive to Suz12 dosage in adult LSK cells,26 and G1ME cells (supplemental Figure 5); Prr6 was up-regulated in fetal liver LSK cells mutant for PRC1, PRC2, or PhoRC (supplemental Figure 6B).
Because we could not account for the defects in Suz12Plt8/+ or Yy1+/− HSCs via altered Cdkn2a expression, we performed global gene expression analyses to identify more genes differentially responsive to each PRC in HSC/progenitors. Fetal liver LSK cells from Bmi1−/−, Suz12Plt8/+ and Yy1+/− E14.5 embryos and their wild-type littermates (all Mpl+/+, n = 3-4 replicates for each mutant, n = 8 wild-type) were analyzed on Illumina mouse whole-genome expression arrays. The gene expression changes, identified by moderated t statistics,32 were modest, similar to those reported by us and others for global gene expression analyses in the absence of PcG proteins.10,15,26 Nevertheless, the expression signatures found previously tended to be recapitulated; genes up-regulated in our Suz12Plt8/+ fetal liver LSK cells were enriched for genes previously found up-regulated in Suz12Plt8/+ adult LSK cells (P < .001, supplemental Table 1) or Suz12 knockdown adult Lin− c-Kit+ progenitors (P < .001, supplemental Table 1).26 Genes differentially over- and underexpressed in our Suz12Plt8/+ fetal liver LSKs were enriched for genes bound by PRC2 members Suz12 and Ezh2 in ES cells (P < .001; supplemental Table 1).19 Genes differentially expressed in our Bmi1−/− fetal liver LSKs were enriched for genes bound by PRC1 member Ring1B in ES cells (P < .001; supplemental Table 1).19
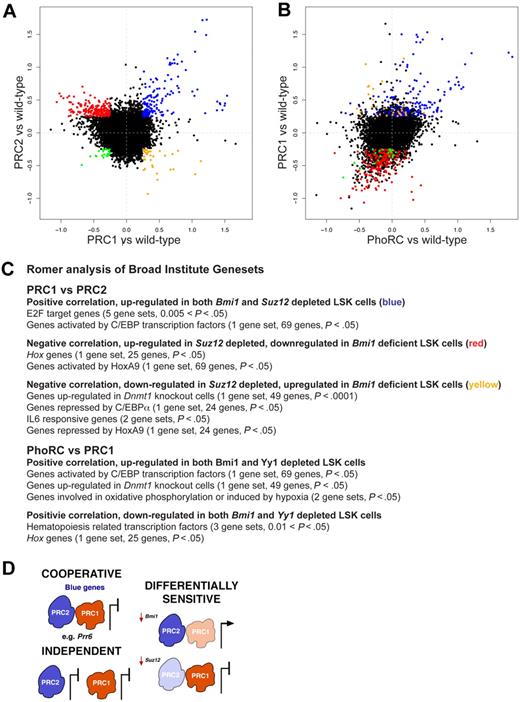
To analyze the multitude of genes regulated by PcG proteins, we devised 2 new statistical methods for data analysis. The first method, GENAS, analyses the correlation between specific comparisons (eg, Bmi1−/− vs wild-type and Suz12Plt8/+ vs wild-type), to identify classes of genes coordinately regulated by more than one PRC. We removed all probes with no evidence of differential expression in any of our 3 comparisons, and plotted fold change for the remaining 7824 probes (5907 unique genes) for each comparison against the others. The PRC1 versus PRC2 plot (Figure 4A) shows a clearly visible group of genes (blue) that are repressed by both Suz12 and Bmi1. There are also less obvious clusters of genes with more modest fold changes that exhibit opposing changes in expression (red and yellow). The GENAS analysis showed that the majority of probes follow this opposing trend (P < .001), reminiscent of the opposing phenotypes displayed by these cells, even though the individual genes with largest fold changes are in the concordant group. In contrast, there was a strong positive correlation between genes differentially regulated in Bmi1−/− and Yy1+/− LSKs (P < .001; Figure 4B) and very few genes with opposing expression changes, consistent with these factors cooperating to regulate gene expression in HSC/progenitors. To select specific genes, we applied a fold-change cutoff of log2 = 0.25. This identified 128 genes that were up-regulated in fetal liver LSK cells depleted for either Bmi1 or Suz12 (blue, Figure 4A, and supplemental Table 2), that is, repressed by both PRC1 and PRC2. A larger set of 172 genes are up-regulated in fetal liver LSK cells depleted for Suz12 but down-regulated in Bmi1-deficient LSK cells (red, Figure 4A, and supplemental Table 2). PRC2 represses expression of these genes, whereas PRC1, directly or indirectly, enhances their transcription. Reassuringly, these genes behave similarly in cells depleted for Yy1 as those depleted in Bmi1 (Figure 4B).
PRC1, PRC2, and PhoRC control different gene sets in HSC/progenitors. (A-B) Log2-fold changes for 7824 differentially expressed Illumina expression array probes in fetal liver LSK cells, comparing Bmi1−/− and Bmi1+/+ (PRC1 vs wild-type), Suz12Plt8/+ and Suz12+/+ (PRC2 vs wild-type), and Yy1+/− and Yy1+/+ (PhoRC vs wild-type). Probes (blue) in panel A are the same as probes (blue) in panel B, and similarly for those colors shown (red, yellow, or green). (C) Romer analysis of gene expression comparisons shown in panel A. Broad Institute gene sets that are enriched in particular comparisons are listed (in supplemental material for full analysis). (D) Image showing the multifaceted regulation of gene expression by PRCs. PRC1, and PRC2 may bind some targets cooperatively to repress expression; these genes are named blue genes shown in panel A. PRC2 and PRC1 may act independently at some loci, and other loci may be differentially sensitive to depletion of PRC1 (eg, Bmi1) or PRC2 (eg, Suz12).
PRC1, PRC2, and PhoRC control different gene sets in HSC/progenitors. (A-B) Log2-fold changes for 7824 differentially expressed Illumina expression array probes in fetal liver LSK cells, comparing Bmi1−/− and Bmi1+/+ (PRC1 vs wild-type), Suz12Plt8/+ and Suz12+/+ (PRC2 vs wild-type), and Yy1+/− and Yy1+/+ (PhoRC vs wild-type). Probes (blue) in panel A are the same as probes (blue) in panel B, and similarly for those colors shown (red, yellow, or green). (C) Romer analysis of gene expression comparisons shown in panel A. Broad Institute gene sets that are enriched in particular comparisons are listed (in supplemental material for full analysis). (D) Image showing the multifaceted regulation of gene expression by PRCs. PRC1, and PRC2 may bind some targets cooperatively to repress expression; these genes are named blue genes shown in panel A. PRC2 and PRC1 may act independently at some loci, and other loci may be differentially sensitive to depletion of PRC1 (eg, Bmi1) or PRC2 (eg, Suz12).
Our second novel statistical method is called Romer; a powerful tool that takes into account the large number of moderate gene expression changes, and performs competitive gene set tests using all of the gene sets from the Broad Institute's Molecular Signatures database.33 We used Romer to analyze the classes of genes enriched in those coordinately or differentially regulated by PRC1, PRC2 and PhoRC (supplemental Table 3). All comparisons had significant enrichment for genes sensitive to histone deacetylase inhibitor treatment, indicating the deregulated genes were sensitive to epigenetic control. Within the class of genes coordinately regulated by PRC1 and PRC2, genes repressed by Bmi1 and Suz12 (blue genes, Figure 4A) were significantly enriched for E2F target genes, consistent with the known roles for PRCs in this pathway. Genes up-regulated by Bmi1 and repressed by Suz12 (red genes, Figure 4A) were enriched for Hox genes, specifically genes activated by HoxA9 (Figure 4C). Genes repressed by Bmi1 and up-regulated by Suz12 (yellow genes, Figure 4A) were enriched for genes up-regulated in the absence of Dnmt1, along with genes repressed by HoxA9 and C/EBPα (Figure 4C). Genes responsive to interleukin-6, a downstream target of C/EBPα, were also enriched within this group of genes. We validated a small selection of the genes identified as coordinately or differentially regulated by PRC1 and PRC2 by qPCR (supplemental Figure 7). There were also gene sets coordinately regulated by PRC1 and PhoRC. Genes activated by C/EBP transcription factors were up-regulated in the context of PRC1 or PhoRC depletion, as they were for PRC2 depletion (Figure 4C), unlike genes repressed by C/EBPα, which appear to be differentially regulated by PRC1 and PRC2. Genes that were coordinately down-regulated in the absence of Bmi1 or reduction of Yy1 (Figure 4B) were enriched for hematopoiesis-related transcription factors and Hox genes (Figure 4C), in keeping with the defective HSC/progenitor activity of LSK cells depleted in Bmi1 or Yy1. These results confirm that depletion of PhoRC produces a similar molecular phenotype to deficiency in PRC1.
We extended our analyses to include adult LSK cells from Suz12Plt8/+ animals, either replete or deficient for c-Mpl. We found a positive correlation between the gene expression changes identified in fetal and adult LSK cells (genes shown in blue, green, red, and yellow in Figure 4A, and shown in supplemental Figure 8 and supplemental Table 4), independent of Mpl genotype (correlation of 0.39, P < .001 for Mpl−/− and 0.31, P < .001 for Mpl+/+). These studies confirm the expression changes we observe in the fetal liver LSK compartment, and suggest that similar mechanisms govern the activity of PRCs in the adult stem cell compartment.
Discussion
In recent years, the role of PcG proteins in HSCs has been a subject of interest because of the clinical relevance of understanding the molecular mechanisms underpinning HSC activity, and the relationship between stem cell biology and cancer.43 One puzzling finding has been that PRC1 components appear to enhance HSC function, whereas PRC2 components Suz12 and Eed restrict HSC and progenitor function, respectively,26,27 results that are inconsistent with the predominant hierarchical model for PRC function. The molecular basis for the apparently opposing effects of PRC1 and PRC2 has remained elusive. Furthermore, any potential contribution of PhoRC to HSC biology had not been studied.
Here we analyzed the role of PRC1, PRC2, and PhoRC in HSCs, and found that all core PRC2 components (Suz12, Ezh2, and Eed) are involved in restricting HSC/progenitor function. Overexpression of Ezh2 was previously shown to enhance HSC activity.44 One potential explanation for this discrepancy was that Suz12 has a role outside of PRC2; however, careful testing has revealed a similar phenotype in mutants of all 3 PRC2 components. We have confirmed that PRC1 and PRC2 have opposing roles in HSC/progenitors and shown that PRC1 and PRC2 do not synergize in their contribution to HSC/progenitor function. This finding is similar to previous observations in myeloid and B-lineage progenitors, using mutant alleles of Eed and the same knockout allele of Bmi1.27 Therefore, at least at some loci critical to the function of HSCs (and progenitors), PRC1 may be targeted to chromatin via PRC2-independent mechanisms, consistent with previously mentioned studies in other cell types.9,15,23 Our results suggest the loci at which PRC1 and PRC2 act independently are important for HSC/progenitor function, and both the loci themselves and the mechanism of PRC action at these loci are worthy of further consideration.
It is unclear how PRC1 may be targeted by PRC2-independent mechanisms or indeed how PRC2 is directed to specific loci, so we investigated the role of the only DNA-binding PcG protein, Yy1, in HSC function. Yy1 has a role in determining HSC/progenitor number and enhances HSC/progenitor function; PhoRC appears to play a PRC1-like role in HSC biology. Previous studies in other mammalian cell types have shown that Yy1 can associate with PRC1, PRC2, or both.45,46 Because Yy1 plays a similar role to PRC1 in HSC/progenitors, perhaps PhoRC directs PRC1 binding.
Given the differing roles of each of the PRCs in HSC/progenitors, we examined the genes controlled by each. We found that the Ink4a/Arf locus (specifically Cdkn2a expression) is differentially sensitive to the activity of PRC1, PRC2, and PhoRC in fetal liver LSK cells. Our genome-wide expression analysis of these cells has led us to propose that PRCs have multiple potential modes of action, which can vary locus by locus (Figure 4D); some genes are differentially sensitive to PRC dosage (eg, Cdkn2a), some are independently controlled by PRC1 with PhoRC or PRC2 alone (red and yellow genes), and others are coordinately controlled by PRC1, PhoRC, and PRC2 (cooperative, blue genes). Interestingly, genes that are coordinately regulated by all 3 PRCs show the largest fold changes, which may explain the focus on the hierarchical model. This observation has been misleading, however, as the opposing phenotypes we have observed suggest that the more numerous but smaller fold changes seen in the genes independently controlled by each PRC are more important for HSC/progenitor function. Our results highlight the biologic importance of small expression changes across gene sets.
Within the genes that are activated in LSK cells depleted of PRC1 or PhoRC but not PRC2, we found enrichment for genes activated in the absence of Dnmt1. A recent study in fibroblasts has identified a link between H2AK119Ub and DNA methylation in regions that are not necessarily enriched in H3K27me3.15 Therefore one intriguing possibility is that DNA methylation, along with PhoRC, plays a role in targeting PRC1 in HSC/progenitors.
The genes differentially regulated by PRC1 and PRC2 were enriched for HoxA9 targets. Furthermore, there was good correlation between the behavior of PRC2 depleted HSCs and HSCs that overexpress HoxA9,47 and PRC1- or PhoRC-depleted HSCs and those deficient in HoxA9.48 Expression changes in C/EBP targets were generally consistent between the mutant genotypes; however, genes repressed by C/EBPα, specifically, responded differentially to inhibition of PRC2 versus PRC1. One possible explanation for this result is that Bmi1 cooperates with C/EBPα to mediate silencing, and that this function is independent of PRC2. The described role for C/EBPα in HSCs fits with the differential changes we see in its targets in PRC1 compared with PRC2 mutants.49 Therefore, modest changes in the HoxA9 and C/EBPα transcriptional networks may explain some of the differences in the roles of the 3 PRCs in HSC/progenitors. One key challenge for the future will be to address the binding patterns of each complex in this rare but important cell type.
In addition to HSCs, PcG proteins have important functions in other stem cell populations, and in cancer. Whether our findings are more broadly applicable to neural or mammary stem cells, will be important to test. Our findings may also have implications for hematopoietic malignancies. A recent study reports frequent heterozygous somatic mutation of EZH2 in diffuse large B cell lymphomas,50 a class of lymphoma in which BMI1 overexpression correlates with poor prognosis.51 Both HoxA9 and C/EBPα are involved in hematopoietic malignancies, and may be differentially regulated by PRC1 and PRC2 in malignant cells, as they are in HSC/progenitors. Recent work shows that overexpression of PRC1 or PRC2 members inversely correlates with prognosis in breast cancer.52 Therefore understanding whether the PRCs have multiple potential modes of action in stem cells and cancer is critical not only to our understanding of stem cell biology, but also for optimal treatment of cancers that overexpress or possess mutated forms of these proteins.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Yang Shi, Miguel Vidal, Maarten van Lohuizen, Anton Berns, Steve Reiner, Alexander Tarakhovsky, Yoshihiro Takihara, and Kakutani Shimada for knockout mice.
This work was supported by a program grant (461219), fellowships (W.S.A., D.J.H., M.E.B), and an Independent Research Institutes Support Scheme Grant (361646) from the National Health and Medical Research Council, a Fellowship from the Cancer Council Victoria (I.J.M.), the Australian Cancer Research Fund, and a Victoria State Government Operational Infrastructure Support grant.
Authorship
Contribution: I.J.M. and M.E.B. designed and performed research, analyzed data, and wrote the paper; M.E.R., B.P., Y.H., and G.K.S. contributed new analytical tools and analyzed data; J.C. and M.P. performed research; A.E., M.B., and H.K. contributed reagents; and W.S.A. and D.J.H. supervised research and edited the manuscript
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marnie E. Blewitt, Molecular Medicine Division, The Walter and Eliza Hall Institute of Medical Research, 1G Royal Parade, Parkville, Victoria 3052, Australia; e-mail: blewitt@wehi.edu.au.