Abstract
The BCL11B transcription factor is required for normal T-cell development, and has recently been implicated in the pathogenesis of T-cell acute lymphoblastic leukemia (T-ALL) induced by TLX overexpression or Atm deficiency. To comprehensively assess the contribution of BCL11B inactivation to human T-ALL, we performed DNA copy number and sequencing analyses of T-ALL diagnostic specimens, revealing monoallelic BCL11B deletions or missense mutations in 9% (n = 10 of 117) of cases. Structural homology modeling revealed that several of the BCL11B mutations disrupted the structure of zinc finger domains required for this transcription factor to bind DNA. BCL11B haploinsufficiency occurred across each of the major molecular subtypes of T-ALL, including early T-cell precursor, HOXA-positive, LEF1-inactivated, and TAL1-positive subtypes, which have differentiation arrest at diverse stages of thymocyte development. Our findings provide compelling evidence that BCL11B is a haploinsufficient tumor suppressor that collaborates with all major T-ALL oncogenic lesions in human thymocyte transformation.
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) can be subclassified into distinct molecular subtypes based on dominant oncogenic alterations that lead to differentiation arrest at specific stages of T-cell development.1,2 These include the HOXA/MEIS1, TLX1/3, and TAL1-overexpressing subtypes,2,3 the LEF1-inactivated subtype,4 and the recently identified group of T-ALL cases with developmental arrest at very early stages of T-cell development defined by a characteristic early T-cell precursor (ETP) phenotype or by absence of biallelic TCRγ deletions (ABD).5,6 On the other hand, other T-ALL oncogenic alterations, such as deletions of CDKN2A or mutations of NOTCH1 and PTEN, occur across all T-ALL subtypes,2,7 suggesting that they can contribute to thymocyte transformation at diverse stages of T-cell development.
The BCL11B transcription factor plays key roles in normal T-cell development. In murine thymocytes, Bcl11b inactivation leads to developmental arrest at a DN2-DN3 stage,8-10 acquisition of NK-like features,8,11 and aberrant self-renewal activity.10 In human T-ALL, BCL11B is involved in recurrent cryptic t(5;14)(q35;q32) translocations with the TLX3 locus, in which BCL11B gene regulatory elements drive aberrant overexpression of the TLX3 oncogene.12-15 However, several lines of evidence suggest that BCL11B haploinsufficiency may also be an important pathogenetic consequence of this translocation. For example, we have recently identified monoallelic Bcl11b deletions in most T-ALLs arising in Atm-deficient mice,16 and Bcl11b has been shown to suppress murine T-lymphoblastic malignancies induced by p53 haploinsufficiency, radiation, or the BCR-ABL oncogene.17,18 Furthermore, recent work has also revealed monoallelic BCL11B lesions in TLX1-induced murine T-ALL, and in TLX1- or TLX3-overexpressing human T-ALL.19
Here, we show results of array CGH and sequencing analyses in T-ALL, which reveal heterozygous BCL11B mutations and deletions across each of the major molecular subtypes of T-ALL, indicating that BCL11B is a haploinsufficient tumor suppressor that can collaborate with diverse oncogenic lesions during human thymocyte transformation.
Methods
Patient samples
T-ALL diagnostic specimens were collected with informed consent in accordance with the Declaration of Helsinki and IRB approval from a cohort of children treated on Children's Oncology Group (COG) P9404 and Dana-Farber Cancer Institute (DFCI) 00-01 clinical trials (n = 47),4,6,7 as well as from a second cohort of independent samples from St Jude Children's Research Hospital (SJCRH), COG AALL0434, and Associazione Italiana Ematologia Oncologia Pediatrica (AIEOP) clinical trials (n = 70; J.R.D. and C.G.M., manuscript submitted, May 2011).
DNA copy number and expression analysis
DNA copy number was assessed by microarray-based CGH in the first cohort of cases,4,6,7 and by whole-genome sequencing or SNP array in the second cohort (J.R.D. and C.G.M., manuscript submitted, May 2011). Gene expression was assessed using Affymetrix U133 Plus 2.0 microarrays. Complete DNA copy number and expression analysis is available in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article) and are available in the NCBI GEO website under accession number GSE14618 and GSE28703.
Mutation detection
Structural homology modeling
Structural homology modeling was used to model BCL11B tandem ZF2-ZF3 zinc finger binding to a canonical GC-rich DNA oligonucleotide sequence, based on the high-resolution crystal structure of Egr1 (Zif268) in complex with DNA.20 This structural model was calculated using SWISS-MODEL,21 producing a high quality structure with a model-template C-α root mean square deviation of 2.9 Angstroms.
Statistical analysis
Differences in continuous data were assessed via the Mann-Whitney (rank-sum) test. Differences in event-free and overall survival were assessed by the log-rank test, and time-to-event distributions were plotted via the Kaplan-Meier method.
Results and discussion
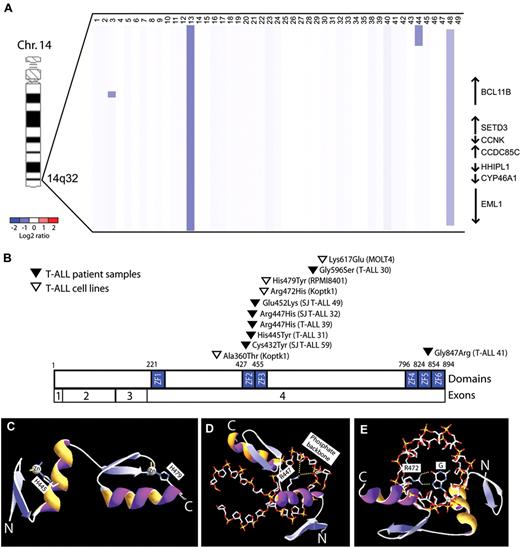
To further define the role of BCL11B inactivation in human T-ALL, we analyzed DNA copy number at the BCL11B locus in a cohort of primary T-ALL lymphoblast specimens, identifying monoallelic BCL11B deletions in 6% of cases (n = 3 of 47). These included 1 microdeletion within the BCL11B locus, 1 small deletion involving BCL11B and 6 additional genes, and 1 large 26 Mbp deletion of the distal arm of chromosome 14 (Figure 1A, supplemental Figure 1). BCL11B resequencing was performed in 43 of these cases together with an additional cohort of 70 T-ALL specimens with matched germ line DNA, revealing heterozygous missense BCL11B mutations in an additional 7 cases, as well as in 19% (n = 3 of 16) of T-ALL cell lines (Figure 1B). None of these represent known single nucleotide polymorphisms based on the NCBI (dbSNP131) or the 1000 Genomes databases (accessed November 12, 2010), and we confirmed that BCL11B mutations were somatically acquired in the 3 cases in which germ line DNA was available. Taken together, we thus identified monoallelic BCL11B lesions in 9% (n = 10 of 117) of primary T-ALL patient samples.
BCL11B inactivation in human T-ALL. (A) Array CGH was performed on genomic DNA from diagnostic lymphoblast specimens collected from 47 children with T-ALL. The CGH data are shown as a dChip plot of segmented log2 copy number ratios. Heterozygous deletions involving BCL11B were identified in 6% (n = 3 of 47) primary T-ALL specimens. (B) Sequencing of BCL11B coding sequence was performed in 43 of these cases together with an additional cohort of 70 T-ALL specimens, revealing heterozygous missense BCL11B mutations in 7 primary patient samples, as well as in 19% (n = 3 of 16) of T-ALL cell lines. Taken together, we thus identified monoallelic BCL11B lesions in 9% (n = 10 of 117) of primary T-ALL patient samples. Mutations are shown based on the full-length CCDS9950.1 BCL11B isoform. (C-E) Homology structural modeling of canonical DNA binding by the BCL11B ZF2-ZF3 zinc fingers was performed based on the high-resolution crystal structure of Zif268 in complex with DNA oligonucleotide.20 (C) His445 and His479 are required for coordination of the structural zinc ions of the ZF2 and ZF3 domains, respectively, and their mutations to Tyr are predicted to unfold of the BCL11B zinc fingers. (D-E) Arg447 and Arg472 form salt bridge and hydrogen bonds with the phosphate backbone and nucleotide bases of DNA, respectively, and their mutations to His are predicted to disrupt DNA binding. Yellow dotted lines depict salt bridge and hydrogen bonds.
BCL11B inactivation in human T-ALL. (A) Array CGH was performed on genomic DNA from diagnostic lymphoblast specimens collected from 47 children with T-ALL. The CGH data are shown as a dChip plot of segmented log2 copy number ratios. Heterozygous deletions involving BCL11B were identified in 6% (n = 3 of 47) primary T-ALL specimens. (B) Sequencing of BCL11B coding sequence was performed in 43 of these cases together with an additional cohort of 70 T-ALL specimens, revealing heterozygous missense BCL11B mutations in 7 primary patient samples, as well as in 19% (n = 3 of 16) of T-ALL cell lines. Taken together, we thus identified monoallelic BCL11B lesions in 9% (n = 10 of 117) of primary T-ALL patient samples. Mutations are shown based on the full-length CCDS9950.1 BCL11B isoform. (C-E) Homology structural modeling of canonical DNA binding by the BCL11B ZF2-ZF3 zinc fingers was performed based on the high-resolution crystal structure of Zif268 in complex with DNA oligonucleotide.20 (C) His445 and His479 are required for coordination of the structural zinc ions of the ZF2 and ZF3 domains, respectively, and their mutations to Tyr are predicted to unfold of the BCL11B zinc fingers. (D-E) Arg447 and Arg472 form salt bridge and hydrogen bonds with the phosphate backbone and nucleotide bases of DNA, respectively, and their mutations to His are predicted to disrupt DNA binding. Yellow dotted lines depict salt bridge and hydrogen bonds.
BCL11B is a zinc finger transcription factor that binds DNA via its Cys2His2 zinc finger domains.22,23 Eight of the 11 BCL11B missense mutations we identified, including 6 of the 7 in primary patient samples, affected residues within BCL11B zinc finger domains. To determine whether these mutations might disrupt zinc finger domain-mediated transcriptional activity, structural homology modeling of canonical DNA binding of the BCL11B zinc fingers was performed based on the high-resolution crystal structure of the zinc finger domains of the Egr1 (Zif268) transcription factor in complex with DNA.20 Strikingly, all but 1 of the mutations identified within ZF2-ZF3 zinc finger domains disrupted conserved amino acids modeled to be required for the structural stability of the zinc finger domain or its binding to DNA (Figure 1E-G and supplemental Figure 2; mutation E532K was of indeterminate predicted phenotype).
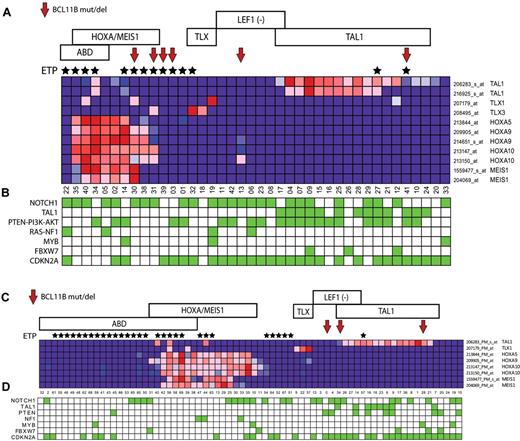
To identify characteristics associated with BCL11B inactivation, we analyzed gene expression data we had available on 110 of the 117 patient samples analyzed in our study, including 8 of the 9 cases with BCL11B genetic alterations, which revealed BCL11B haploinsufficiency across the major molecular subtypes of T-ALL, including the early T-cell precursor (ETP), HOXA/MEIS1-positive, LEF1-inactivated, and TAL1-positive cases (Figure 2A-B). These molecular subtypes of T-ALL are associated with differentiation arrest at distinct stages of T-cell development,3-6 thus suggesting that the pathogenic role of BCL11B haploinsufficiency is not solely because of induction of T-cell differentiation arrest, which occurs at the DN2-DN3 stage on complete BCL11B inactivation in normal thymocytes.8,10
BCL11B haploinsufficiency across the major molecular subtypes of T-ALL. Gene expression profiling was previously performed on 40 of the 47 T-ALL cases in the first DFCI/COG cohort, and on all 70 cases in the second cohort of cases from St Jude/COG/AIEOP analyzed in our study, both representing cohorts that are enriched for high-risk ETP/ABD T-ALL cases. Red arrows point to cases with BCL11B mutations or deletions, and black stars mark cases with the ETP phenotype. Samples are classified by T-ALL subtype as follows: ABD, absence of biallelic TCRγ deletion. ETP indicates early T-cell precursor phenotype. HOXA/MEIS, cases with over-expression of genes of the HOXA/MEIS1 cluster, defined as expression values > 100. TLX, cases with TLX1 or TLX3 overexpression. LEF1 (−), cases with LEF1 deletions or truncating mutations.4 TAL1, cases with TAL1 expression values > 100 or TAL1 activating deletions. (A-B) Heatmap depiction of gene expression and mutation data from the first cohort of 47 children with T-ALL. (C-D) Heatmap depiction of gene expression and mutation data from the second cohort of 70 children with T-ALL. The expression pattern of selected T-ALL oncogenes in each cohort is shown in panels A and C, based on the expression microarrays applied. Note that probe sets that showed no detectable expression in any sample (defined as expression values < 100) were excluded. Key T-ALL oncogenic alterations in each sample are depicted by green boxes in panels B and D, which represent the presence of NOTCH1 mutations, TAL1 activating deletions, PTEN-PI3K-AKT mutations or PTEN deletions, RAS mutation or NF1 deletion, MYB duplications, FBXW7 mutations or deletions, and CDKN2A deletions (supplemental Table and J.R.D. and C.G.M., manuscript submitted, May 2011).
BCL11B haploinsufficiency across the major molecular subtypes of T-ALL. Gene expression profiling was previously performed on 40 of the 47 T-ALL cases in the first DFCI/COG cohort, and on all 70 cases in the second cohort of cases from St Jude/COG/AIEOP analyzed in our study, both representing cohorts that are enriched for high-risk ETP/ABD T-ALL cases. Red arrows point to cases with BCL11B mutations or deletions, and black stars mark cases with the ETP phenotype. Samples are classified by T-ALL subtype as follows: ABD, absence of biallelic TCRγ deletion. ETP indicates early T-cell precursor phenotype. HOXA/MEIS, cases with over-expression of genes of the HOXA/MEIS1 cluster, defined as expression values > 100. TLX, cases with TLX1 or TLX3 overexpression. LEF1 (−), cases with LEF1 deletions or truncating mutations.4 TAL1, cases with TAL1 expression values > 100 or TAL1 activating deletions. (A-B) Heatmap depiction of gene expression and mutation data from the first cohort of 47 children with T-ALL. (C-D) Heatmap depiction of gene expression and mutation data from the second cohort of 70 children with T-ALL. The expression pattern of selected T-ALL oncogenes in each cohort is shown in panels A and C, based on the expression microarrays applied. Note that probe sets that showed no detectable expression in any sample (defined as expression values < 100) were excluded. Key T-ALL oncogenic alterations in each sample are depicted by green boxes in panels B and D, which represent the presence of NOTCH1 mutations, TAL1 activating deletions, PTEN-PI3K-AKT mutations or PTEN deletions, RAS mutation or NF1 deletion, MYB duplications, FBXW7 mutations or deletions, and CDKN2A deletions (supplemental Table and J.R.D. and C.G.M., manuscript submitted, May 2011).
To identify candidate therapeutic targets in BCL11B-mutant T-ALL, we also performed gene set enrichment analysis (GSEA), which revealed an inverse association between BCL11B-mutant T-ALL and genes down-regulated by rapamycin treatment in the BJAB Burkitt lymphoma cell line (supplemental Figure 3),24 suggesting the need for additional studies to evaluate the potential therapeutic utility of mTOR inhibitors in BCL11B-mutant T-ALL. Low BCL11B expression predicted failure of induction chemotherapy, but we were unable to identify a BCL11B expression threshold that accurately predicted long-term outcome, while BCL11B mutations/deletions similarly did not predict treatment response (supplemental Figure 4).
Recurrent cryptic t(5;14)(q35;q32) translocations juxtaposing BCL11B and TLX3 have been described in T-ALL, which result in BCL11B gene regulatory elements driving the aberrant over-expression of TLX3.12-15,25 These translocations have been thought to be pathogenic because of the resultant overexpression of the TLX3 oncogene. However, our data suggest that the inactivation of BCL11B is also an important pathogenic consequence of these translocations. Our findings provide compelling evidence that BCL11B is a haploinsufficient tumor suppressor across the major molecular subtypes of T-ALL, which are blocked at diverse stages of thymocyte development, suggesting that BCL11B loss may have pathogenic roles extending beyond differentiation arrest.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors would like to gratefully acknowledge the children with T-ALL and their families, as well as members of the COG, DFCI, SJCRH, and AIEOP member institutions, for the samples analyzed in these studies.
This work was funded by National Institute of Health grants 2P01CA109901 (A.T.L. and F.W.A.), 5P01CA68484 (S.E.S. and A.T.L.), and 1RO1CA158073 (S.Z.). COG cell banking and sample distribution were supported by the COG 9900 cell biology study and grants CA98543, CA114766, CA98413, CA30969, and CA29139 from the National Institutes of Health. Sequence and analysis of St Jude, AEIOP, and COG samples was supported by the St Jude Children's Research Hospital–Washington University Pediatric Cancer Genome Project. A.G. is supported by National Institutes of Health grant 1K08CA133103 and is a scholar of the American Society of Hematology–Amos Medical Faculty Development program. L.H. is supported by the Swedish Research Council. S.S.W. is the T. John Gribble Chair of Pediatric Hematology/Oncology at the University of New Mexico. S.P.H. is the Ergen Family Chair in Pediatric Cancer at The Children's Hospital, Aurora, CO. S.Z. is supported by the Leukemia & Lymphoma Society, the St Baldrick Foundation, and the Johns M. Driscoll Jr Children's Fund. C.G.M. is a Pew Scholar in the Biomedical Sciences. The array CGH analyses were performed and supported in part by the Belfer Institute for Applied Cancer Science.
National Institutes of Health
Authorship
Contribution: A.G. designed, performed and analyzed research, and wrote the paper; A.K. designed, performed, and analyzed research; T.S. performed and analyzed research; J.Z., A.P., and L.C. developed vital CGH analytical tools and analyzed data; S.E.D. and D.S.N. analyzed data; L.B.S., S.W., S.P.H., and S.E.S. provided samples and analyzed data; S.Z. and F.W.A. designed research and analyzed data; L.H., S.-C.C., J.R.D., and C.G.M. provided samples, designed and performed research, and analyzed data; and A.T.L. supervised research, analyzed data, and cowrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr A. Thomas Look, Department of Pediatric Oncology, Dana-Farber Cancer Institute, Mayer 630, 44 Binney St, Boston, MA 02215; e-mail: thomas_look@dfci.harvard.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal