Abstract
Patients with a t(9;11) translocation (MLL-AF9) develop acute myeloid leukemia (AML), and while in mice the expression of this fusion oncogene also results in the development of myeloid leukemia, it is with long latency. To identify mutations that cooperate with Mll-AF9, we infected neonatal wild-type (WT) or Mll-AF9 mice with a murine leukemia virus (MuLV). MuLV-infected Mll-AF9 mice succumbed to disease significantly faster than controls presenting predominantly with myeloid leukemia while infected WT animals developed predominantly lymphoid leukemia. We identified 88 candidate cancer genes near common sites of proviral insertion. Analysis of transcript levels revealed significantly elevated expression of Mn1, and a trend toward increased expression of Bcl11a and Fosb in Mll-AF9 murine leukemia samples with proviral insertions proximal to these genes. Accordingly, FOSB and BCL11A were also overexpressed in human AML harboring MLL gene translocations. FOSB was revealed to be essential for growth in mouse and human myeloid leukemia cells using shRNA lentiviral vectors in vitro. Importantly, MN1 cooperated with Mll-AF9 in leukemogenesis in an in vivo BM viral transduction and transplantation assay. Together, our data identified genes that define transcription factor networks and important genetic pathways acting during progression of leukemia induced by MLL fusion oncogenes.
Introduction
The mixed lineage leukemia or myeloid/lymphoid leukemia (MLL) gene found on chromosome 11q23 is involved in oncogenic translocations in adult and infant leukemia.1 Translocations involving MLL are also frequently found in therapy-related leukemia when patients have received topoisomerase II inhibitors as part of their treatment.2 MLL is reported to be involved in translocations with > 60 genes, all of which are thought to result in fusion proteins.3
The MLL-AF9 translocation, t(9;11)(p22;q23), is the most common MLL translocation observed in patients with de novo and therapy-related acute myeloid leukemia (AML) and indicates an intermediate to poor prognosis with a high risk of relapse.4,5 A knock-in mouse model for the MLL-AF9 translocation was generated and mice heterozygous for the Mll-AF9 knock-in allele were reported to develop AML, with 50% of the mice developing disease by 5 months of age.6-8 However, mice presented with leukemia only after a relatively long latency, indicating that cooperating mutations are needed to contribute to leukemia progression.
Murine leukemia viruses (MuLV) have been used to identify important leukemia-associated genes in various leukemia-predisposed mutant strain backgrounds, such as Eμ-Myc, Cdkn2a(−/−), inversion 16, and Nf1−/+ mouse models.9-11 A recombinant MuLV that induces myeloid leukemia with a broad tropism in inbred mouse strains has been engineered by combining sequences from the amphotropic virus strain 4070A and Moloney murine leukemia virus (Mo-MuLV), designated MOL4070LTR.12 MOL4070LTR (abbreviated here as M4070) functions as an insertional mutagen to accelerate leukemia in at least 2 mouse models of AML.13,14 Here we use M4070 to identify mutations that collaborate with Mll-AF9 to drive leukemogenesis in mice. To identify genes relevant to human MLL leukemia, a comparative oncogenomics approach with clinical patient samples was used. Functional validation of 2 genes (FOSB and MN1) support their role in leukemogenesis in concert with Mll-AF9.
Methods
Mice and retroviral infection
Heterozygous Mll-AF9 C57BL/6J mice (provided by Dr Terence Rabbitts, Section of Experimental Therapeutics, Leeds Institute of Molecular Medicine) were bred to wild-type (WT) 129/SvJ mice (The Jackson Laboratory) to produce F1 offspring.13 Two- to 4-day-old F1 offspring were inoculated intraperitoneally with 1 to 2 × 105 infectious particles in 0.1 mL of media. Control mice were injected with 0.1 mL of a nonviral supernatant. Four experimental cohorts were established: infected Mll-AF9, infected WT, noninfected Mll-AF9, and noninfected WT. Mice were observed daily for signs of morbidity such as labored breathing, immobility, and organomegaly, at which time they were killed. Mice were housed, bred, and manipulated according to specific pathogen-free conditions set out by the University of Minnesota's Institutional Animal Care and Use Committee.
Immunophenotyping
Immunophenotyping included morphology, histopathology, flow cytometry, Southern blot, cytology, and IHC (supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Proviral insertion site sequencing
PCR amplification of M4070 proviral insertions was performed essentially as described previously14 (see supplemental Methods). Splinkerette PCR products were shotgun cloned into pCR4-Topo vector (Invitrogen), transformed into electrocompetent DH10B Escherichia coli (ElectroMax; Invitrogen), and plated onto selective medium with ampicillin (120 μg/mL). Plasmid DNA was prepped from bacteria after 24-hour growth using alkaline lysis. Plasmid DNA was sequenced using an M13R primer and BigDye 3.1 (Invitrogen), on 3730 DNA analyzer machines (Applied Biosystems Inc).
Sequence processing and annotation
A total of 26 160 initial ABI sequence reads were processed and analyzed using a custom, semiautomated processing pipeline first described in Starr et al.15 Nonredundant (NR) insertion positions were annotated using the EnsEMBL API16 with the name of the gene whose start site was closest to the proviral insertion position. Common insertion site (CIS) positions were annotated similarly using the median insertion position within the CIS as a reference point (see supplemental Methods).
Quantitative real-time PCR
For quantitative real-time PCR, see supplemental Methods.
Lentiviral production and shRNA knockdown analysis
293T cells were transfected with TRIPZ lentiviral shRNAmir plasmids from Open Biosystems (Thermo Fisher Scientific) encoding shRNA to the human FOSB gene or a scrambled control. Lentiviral supernatant was collected after 24 and 48 hours, filtered with a 45-μm filter, and concentrated using the LentiX Concentrator (Clontech). U937 leukemia cells17,18 were transduced with the lentivirus for 6 hours, followed by puromycin selection to produce cell lines that stably express the shRNA plasmid. To induce knockdown with the shRNA, cells were plated at 0.5 million cells per well in 6-well plates with 2 mL of media containing puromycin for 24 hours. The cells were then treated with 4 μg/μL doxycycline to induce the shRNAs.
Transduction of Tet-On MLL-AF9;NrasG12D cells and shRNA induction
Experiments were performed as previously described.19
Western blot analysis
Cells were incubated with lysis buffer for 20 minutes on a rotator and centrifuged at 20 817g for 10 minutes, both at 4°. Samples were analyzed with a Bradford reaction and 40 μg of total protein were run with sample buffer on a prepared 10% Bis-Tris gel using an Invitrogen gel box at 200 V and transferred to a nitrocellulose membrane using the iBlot system (Invitrogen). Membranes were blocked with 5% nonfat milk, incubated with anti-Fosb rabbit polyclonal Ab (Cell Signaling Technology), and anti–rabbit secondary Ab (DAKO) before developing using ECL (Thermo Scientific).
BM transduction/transplantation assay
BM cells were harvested from 8- to 13-week-old Mll-AF9/+ and WT littermate control mice treated with 150 mg/kg 5-fluorouracil (5-FU; InvivoGen) 6 days earlier. E86 cells (stably expressing retroviruses encoding either pSF91-MN1 or a GFP-only construct)20 were sublethally irradiated and plated in nutrient-rich medium with a cytokine cocktail of 10 ng/mL mouse recombinant IL-6 (mIL-6), 50 ng/mL murine recombinant stem cell factor (mSCF; Invitrogen), and 5 ng/mL murine recombinant IL-3 (mIL-3; Thermo Fisher Scientific). Twenty-four hours later, the BM cells were added with 5 μg/mL polybrene (Millipore). After 48 hours, BM cells were collected and replated for expansion for 24 hours in nutrient-rich media. Recipient 8-week-old C57BL/6J.BoyJ mice were lethally irradiated at 900 rad the day before being injected intravenously with 106 transduced BM cells each and followed for disease progression.
Results
Infection with M4070 retrovirus accelerates leukemia development in Mll-AF9/+ mice
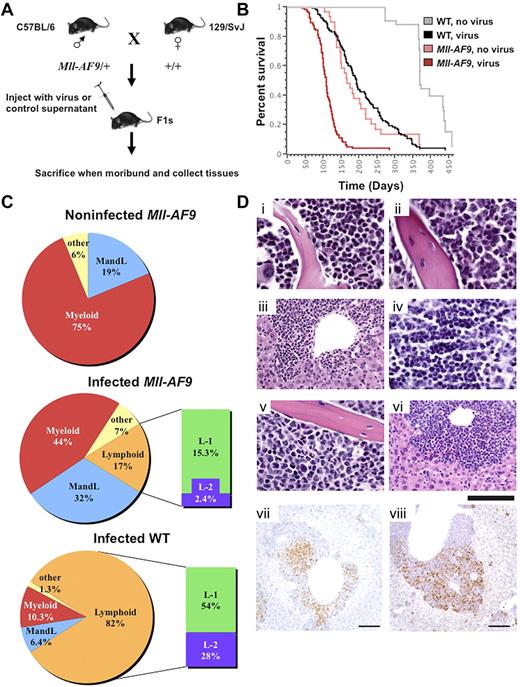
F1 animals, heterozygous for Mll-AF9 or WT, were generated as shown (Figure 1A). Neonates were injected IP with the M4070 virus or mock infected.21 Ninety-eight percent of the animals in both M4070-infected groups and the mock-infected Mll-AF9 group succumbed to overt leukemia within 1 year. Infected Mll-AF9 mice had a significantly decreased latency of disease with a median survival of 108 days compared with 165 days in noninfected Mll-AF9 mice (P < .0001). Infected WT mice also developed leukemia but with a much longer latency (191 days median survival) compared with the infected Mll-AF9 group (P < .0001; Figure 1B). Interestingly, Mll-AF9 female mice died significantly faster than their male littermates, regardless of whether they were injected with the M4070 virus or not (supplemental Figure 1A-B). This finding may be related to the higher frequency of MLL rearrangements observed in human female infant leukemia.22
Insertion mutagenesis screen accelerates leukemia in experimental mice. (A) Mll-AF9/+ mice on a C57BL/6 genetic background were crossed to WT mice of the 129/SvJ genetic background to generate experimental cohorts. The resulting pups were infected by IP injection and followed for disease progression. Mice were killed when moribund, and tissues were collected for cloning insertions, FACS analysis, and histology. (B) Log-rank (Mantel-Cox) tests performed on a Kaplan-Meier survival plot indicate that infected Mll-AF9 mice develop disease with a reduced latency compared with infected WT mice (P < .0001) and noninfected Mll-AF9 mice (P < .0001). A total of 279 mice were used in the study: infected WT (n = 114), infected Mll-AF9 (n = 97), noninfected WT (n = 40), and noninfected Mll-AF9 (n = 28). (C) Infected WT animals develop mostly lymphoid leukemia and Mll-AF9/+ animals develop mostly myeloid leukemia. Pie charts depict the percentage of each leukemia type in the experimental cohorts. Myeloid disease is displayed in red, both myeloid and lymphoid disease is blue, only lymphoid disease is orange, and other diseases are yellow. Lymphoid disease is further divided into L-1 (green) and L-2 (purple). A high CD4 or CD8 population characterizes both L-1 and L-2 but L-2 also has Mac1 positively on its surface. (D) Disease phenotype in MLL-AF9 mice infected with retrovirus. Mouse 410 showed myeloid predominance with differentiated forms in marrow (i) and spleen (not shown): myeloid neoplasm common in Mll-AF9 mice. Mouse 529 showed immature myeloid forms in marrow (ii), infiltrate in the liver (iii), lymphoma in the thymus (iv), and a mixture of myeloid leukemia and lymphoma in the spleen (flow immunophenotype shown in supplemental Figure 3B): mixed AML and T-cell lymphoma/leukemia. Mouse 522 showed moderate myeloid differentiation in the marrow (v), infiltrate in the liver (vi), and myeloid leukemia in the spleen (flow immunophenotype shown in supplemental Figure 3C): AML. H&E. IHC of mouse 539 shows CD3 (vii) and myeloperoxidase (viii) positivity in infiltrating cells in the liver; chromogen is DAB. Scale bar represents 20 μm in subpanels i, ii, iv, and v; 50 μm in iii and vi; and 100 μm in vii and viii. Image acquisition details can be found in supplemental Methods.
Insertion mutagenesis screen accelerates leukemia in experimental mice. (A) Mll-AF9/+ mice on a C57BL/6 genetic background were crossed to WT mice of the 129/SvJ genetic background to generate experimental cohorts. The resulting pups were infected by IP injection and followed for disease progression. Mice were killed when moribund, and tissues were collected for cloning insertions, FACS analysis, and histology. (B) Log-rank (Mantel-Cox) tests performed on a Kaplan-Meier survival plot indicate that infected Mll-AF9 mice develop disease with a reduced latency compared with infected WT mice (P < .0001) and noninfected Mll-AF9 mice (P < .0001). A total of 279 mice were used in the study: infected WT (n = 114), infected Mll-AF9 (n = 97), noninfected WT (n = 40), and noninfected Mll-AF9 (n = 28). (C) Infected WT animals develop mostly lymphoid leukemia and Mll-AF9/+ animals develop mostly myeloid leukemia. Pie charts depict the percentage of each leukemia type in the experimental cohorts. Myeloid disease is displayed in red, both myeloid and lymphoid disease is blue, only lymphoid disease is orange, and other diseases are yellow. Lymphoid disease is further divided into L-1 (green) and L-2 (purple). A high CD4 or CD8 population characterizes both L-1 and L-2 but L-2 also has Mac1 positively on its surface. (D) Disease phenotype in MLL-AF9 mice infected with retrovirus. Mouse 410 showed myeloid predominance with differentiated forms in marrow (i) and spleen (not shown): myeloid neoplasm common in Mll-AF9 mice. Mouse 529 showed immature myeloid forms in marrow (ii), infiltrate in the liver (iii), lymphoma in the thymus (iv), and a mixture of myeloid leukemia and lymphoma in the spleen (flow immunophenotype shown in supplemental Figure 3B): mixed AML and T-cell lymphoma/leukemia. Mouse 522 showed moderate myeloid differentiation in the marrow (v), infiltrate in the liver (vi), and myeloid leukemia in the spleen (flow immunophenotype shown in supplemental Figure 3C): AML. H&E. IHC of mouse 539 shows CD3 (vii) and myeloperoxidase (viii) positivity in infiltrating cells in the liver; chromogen is DAB. Scale bar represents 20 μm in subpanels i, ii, iv, and v; 50 μm in iii and vi; and 100 μm in vii and viii. Image acquisition details can be found in supplemental Methods.
Virally infected Mll-AF9/+ mice develop predominantly myeloid leukemia
Moribund mice that died within 1 year presented with hematopoietic malignancies, often exhibiting splenomegaly, enlarged lymph nodes, thymus, and occasionally liver. Noninfected Mll-AF9 mice showed 75% myeloid disease, characterized by a high Gr-1 and/or Mac-1 population, and 19% with distinct myeloid and lymphoid populations (MandL). The mixed myeloid and lymphoid disease in this Mll-AF9 model is a novel discovery not described in previous reports. Phenotypic analysis revealed different lineage distribution on infection with M4070. Forty-four percent of infected Mll-AF9 mice exhibited myeloid disease, 32% MandL, and only 17% had lymphoid disease. Conversely, the majority of the infected WT mice presented with lymphoid disease (82%), 10.3% showed myeloid disease, and only 6.4% of mice had MandL mixed disease (Figure 1C). The lymphoid populations were characterized by an extrathymic high CD4+ or CD8+ single-positive population, or dominant CD4+ and CD8+ double-positive population in the spleen or lymph nodes (supplemental Figure 3B and data not shown). This defines the first of 2 distinct classes of lymphoid disease in both virally infected groups, herein called L-1 (Figure 1C). The second class of mice were found to have T-cell lineage lymphoid disease by IHC but also expressed the Mac1 myeloid marker on the blast cell surface, herein called L-2, a phenotype not found in noninfected Mll-AF9 animals (Figure 1C). The total number and percentage of mice with each phenotype classification from each experimental cohort is shown in supplemental Figure 2A.
A common method to determine lineage is to assess TCR or BCR rearrangements. However, when we examined leukemias in all 3 cohorts (infected Mll-AF9, noninfected Mll-AF9, infected WT) for clonal Jβ1 or Jβ2 (T-cell) or JH (B-cell) receptor gene rearrangements, we frequently saw evidence of TCR and/or BCR rearrangements even if they were phenotypically determined to be myeloid disease. In fact, most Mll-AF9 mice had myeloid disease but almost 50% of myeloid tumors were positive for clonal TCR or BCR rearrangements. Moreover, 20% of animals with lymphoid disease were negative for clonal T- or B-cell rearrangements (supplemental Figure 2B). Thus, the presence or absence of a TCR and/or BCR rearrangement is neither completely sensitive nor specific to a lymphoid phenotype, and is on its own insufficient to assess lineage. To determine which phenotype qualifiers were statistically significantly associated, Fisher exact tests were performed on each pairwise comparison of the total phenotype condition and each phenotype variable (supplemental Table 1A). A myeloid or MandL phenotype was positively correlated with a larger spleen weight, the Mll-AF9/+ genotype, and only in the case of myeloid disease, an extremely elevated white blood cell (WBC) count whereas there were multiple reciprocal positive correlations between lymphoid phenotypes, BCR and/or TCR rearrangements, a normal spleen weight, and WT leukemias.
Pathologic analyses illustrated the disruption of normal tissue architecture by tissue infiltration of leukemic blast cells in virally infected Mll-AF9 mice (Figure 1D). Mll-AF9 mice, both infected and noninfected, commonly showed a differentiated myeloid neoplasm in the marrow (Figure 1Di). A subset of both infected Mll-AF9 and infected WT mice presented with mixed myeloid leukemia and T-cell lymphoma/leukemia in the liver, marrow, thymus, and spleen of the animal, with CD3 and myeloperoxidase-positive cells detectable in infiltrating cells by IHC (Figure 1Dii-iv,vii-viii). There were also Mll-AF9 mice that exhibited classic AML in the marrow and liver (Figure 1Dv-vi). Cytology was also performed to confirm the maturity and cell type of the disease in infected MLL-AF9 mice. Examples are shown in supplemental Figure 3A which includes cytology of a well-differentiated myeloid leukemia, a blastic myeloid leukemia, a lymphoid leukemia/lymphoma, and a moderately differentiated myeloid leukemia. Supplemental Figure 3B depicts flow cytometry data from a representative mouse with 2 different diseases (mixed AML and T-cell lymphoma/leukemia) as was observed in both infected WT and infected Mll-AF9 cohorts. Its corresponding pathology is shown in Figure 1Dii-iv. The myeloid immunophenotype characteristic of the Mll-AF9 mice is shown in supplemental Figure 3C, whose corresponding pathology is in Figure 1Di.
Survival curves were generated to determine whether the reduced latency in M4070-infected Mll-AF9 mice was associated with a particular phenotype. The survival of Mll-AF9 mice was significantly decreased compared with WT mice regardless of phenotypic class (supplemental Figure 1C-E). Importantly, the disease latency did not show dependence on phenotype within the WT or Mll-AF9 groups of mice (supplemental Figure 1F-H). Thus, the genetic mutations induced by the M4070 virus influenced leukemia progression as governed by the initiating mutation. Therefore, infection accelerated mostly myeloid leukemia Mll-AF9/+ mice, while causing mostly lymphoid leukemia in WT mice.
Cloning retroviral integrations leads to identification of CISs
Ligation-mediated PCR was used to amplify the M4070-genomic DNA junctions in 165 independent M4070-infected leukemia samples (both Mll-AF9 and WT). A total of 1745 nonredundant insertions or unique reference sequences, representing an average of 10.8 insertions per tumor, were recovered. Sequences are available through the public mouse Retroviral Tagged Cancer Gene Database (RTCGD).23 In total, 88 unique CISs were identified by established statistical analysis and the additional criterion that proviral insertions in leukemia samples must come from at least 3 mice (see supplemental Methods and Table 1). Sixty-nine CISs were recovered by combining all insertions from both infected Mll-AF9 leukemia and infected WT leukemia. Additional CISs came from analysis of only insertions in Mll-AF9 leukemia (15) and infected WT leukemia (4). The rest of the CISs from the WT leukemia group (supplemental Table 2) and Mll-AF9 leukemia group (supplemental Table 3) overlapped with the combined dataset. Southern blotting using an M4070-specific probe showed that the proviral insertions in infected animals were clonal to oligoclonal with one internal proviral fragment represented by a band common to all tumors and additional unique M4070 insertions (supplemental Figure 4). No proviral insertions were detected in noninfected animals, and M4070 insertions were consistent throughout each diseased animal (data not shown).
CIS list
| Chromosome . | CIS position . | CIS range, kb . | Mll-AF9 mice with insert . | WT mice with insert . | RTCGD . | Cancer Gene Census . | COSMIC . | All genes in/near CIS . |
|---|---|---|---|---|---|---|---|---|
| 1 | 4485458-4486321 | 1 | 2 | 1 | YES | Sox17* | ||
| 1 | 36823950-37002868 | 179 | 3 | 1 | YES | Tmem131,*Zap70, AC123854.2 | ||
| 1 | 135756752-135974231 | 217 | 2 | 2 | AC157924.6,*Prelp, Fmod, Btg2, Optc | |||
| 1 | 173849175-174012079 | 163 | 4 | 1 | YES | YES | Slamf6,*Copa, Vangl2, Nhlh1, AC158930.1, Ncstn | |
| 2 | 11547443-11577841 | 30 | 3 | 0 | YES | YES | Il2ra* | |
| 2 | 26315310-26525572 | 210 | 0 | 11 | YES | YES | YES | Notch1,*mmu-mir-126, Egfl7, Agpat2, Fam69b, AL732311 |
| 2 | 90344091-90491625 | 148 | 3 | 1 | YES | Ptprj* | ||
| 2 | 90919486-90925879 | 6 | 3 | 1 | YES | Slc39a13,*Sfpi1 | ||
| 2 | 101463219-101464091 | 1 | 4 | 0 | B230118H07Rik,*Rag2 | |||
| 2 | 103601727-103784302 | 183 | 1 | 3 | YES | YES | YES | Lmo2,*Nat10, Caprin1, BX537331.1, AL928544.7 (miRNA), AL928544.5 (miRNA) |
| 2 | 103601727-103784302 | 183 | 0 | 3 | YES | Nat10,*†Lmo2, Caprin1, BX537331.1, AL928544.7 (miRNA), AL928544.5 (miRNA) | ||
| 2 | 117168903-117367506 | 199 | 3 | 10 | YES | YES | Rasgrp1* | |
| 2 | 152601718-152828604 | 227 | 2 | 2 | YES | YES | Bcl2l1,*Tpx2, Mylk2, Ttll9, Foxs1, Dusp15 | |
| 2 | 165712889-165837434 | 125 | 4 | 0 | Prkcbp1,*Zmynd8, Ncoa3 | |||
| 2 | 165781678-165837434 | 56 | 3 | 0 | RP23-108D12.5*‡ | |||
| 2 | 167625651-167785437 | 160 | 4 | 1 | A530013C23Rik,*Ptpn1 | |||
| 2 | 167750534-167785437 | 35 | 3 | 0 | YES | Ptpn1*‡ | ||
| 2 | 169958430-170046828 | 88 | 2 | 2 | YES | YES | Zfp217* | |
| 3 | 94945270-95035953 | 91 | 3 | 0 | YES | Tnfaip8l2,*‡Cdc42se1, Sema6c, Gabpb2, Mlt11, Gm128, Bnipl, Lysmd1, Scnm1 | ||
| 4 | 32341351-32513306 | 172 | 3 | 3 | YES | YES | Bach2* | |
| 4 | 133220770-133371434 | 151 | 2 | 2 | YES | Pigv,*Aridla | ||
| 4 | 133652224-133867470 | 215 | 1 | 2 | YES | YES | Cd52,*Ubxn11, Aim1, Sh3bgrl3, Ccdc21, Gm7534, Catsper4, Cnksr1, Zfp593, Grrp1, Pdik1 | |
| 4 | 149076009-149079348 | 3 | 2 | 1 | YES | Pik3cd* | ||
| 5 | 34038901-34279652 | 241 | 1 | 3 | YES | YES | YES | Fgfr3,*Tacc3 |
| 5 | 108078270-108186428 | 108 | 5 | 19 | YES | YES | Gfi1,*Evi5, Rpap2 | |
| 5 | 111845647-112004142 | 158 | 10 | 3 | YES | YES | YES | Mn1,*C130026L21Rik |
| 6 | 48598556-48720382 | 122 | 6 | 0 | YES | Gimap8,*Gimap cluster | ||
| 6 | 48630644-48720382 | 90 | 5 | 0 | YES | Gimap4,*‡Gimap cluster | ||
| 6 | 127104034-127281434 | 177 | 2 | 5 | YES | YES | YES | Ccnd2,*AC161597.1 |
| 7 | 19858212-20077994 | 220 | 6 | 0 | YES | Fosb,*mmu-mir-343, Ercc1, Ercc2, Ppplrl3, Rtn2, Ckm, Exoc3l2, C79127, Vasp, Cd3eap, Klc3, Mark4 | ||
| 7 | 29165895-29228553 | 63 | 4 | 0 | YES | Gmfg,*Paf1, Samd4b, Med29 | ||
| 7 | 121285480-121354575 | 69 | 5 | 11 | YES | YES | Rras2,*Copb1 | |
| 7 | 136903269-136957718 | 54 | 2 | 2 | YES | Brwd2* | ||
| 7 | 152126941-152233702 | 107 | 3 | 2 | YES | YES | YES | Ccnd1,*Oraov1 |
| 8 | 10856578-11099413 | 243 | 3 | 3 | AC116499.9,*3930402G23Rik, Irs2 | |||
| 8 | 10910498-11099413 | 189 | 0 | 3 | YES | 3930402G23Rik,*†Irs2 | ||
| 8 | 86266601-86296089 | 29 | 2 | 1 | YES | Cd97* | ||
| 8 | 131049443-131116479 | 67 | 3 | 0 | YES | Nrp1*‡ | ||
| 9 | 32416427-32624588 | 208 | 3 | 5 | YES | YES | Ets1* | |
| 9 | 44135577-44376194 | 241 | 2 | 1 | YES | Dpagt1,*Bcl9l, H2afx, Hyou1, Rps25, Slc37a4, AC122428.3, C030014l23Rik, C2cd2l, Hmbs, Vps11, Trappc4, Ccdc84, Foxr1, Upk2, AC122428.2, Cxcr5 | ||
| 9 | 110800877-110812241 | 11 | 2 | 1 | YES | Als2cl* | ||
| 9 | 123758644-123985636 | 227 | 3 | 1 | YES | Fyco1,*Xcrl, CAAA01140679.1.6228.1, Ccr1, Ccr1l1, Ccr3 | ||
| 10 | 20763054-20972634 | 210 | 8 | 9 | YES | YES | YES | Myb,*Ahi1, AC153556.5 (miRNA) |
| 10 | 20811983-20972634 | 161 | 0 | 9 | AC153556.5,*†Myb | |||
| 10 | 41658107-41790233 | 132 | 1 | 3 | YES | Armc2* | ||
| 10 | 59615644-59634420 | 19 | 2 | 1 | YES | Chst3,*Spock2 | ||
| 10 | 76990210-77085753 | 96 | 3 | 0 | YES | Itgb2,*‡181008A18Rik, Pttglip, Sumo3, Ube2g2 | ||
| 10 | 79452089-79621648 | 170 | 4 | 0 | YES | Cnn2,*Abca7, Hmha1, Gpx4, Stk11, Atp5d, Midn, ORF61, Polr2e, Sbno2, Dos | ||
| 10 | 79895567-80140544 | 245 | 2 | 2 | YES | Mknk2,*Tcf3, Onecut3, Atp8b3, Rexo1, Klf16, Fam108a, Scamp4, Adat3, Csnk1g2, Btbd2 | ||
| 10 | 92532859-92627678 | 95 | 3 | 0 | 4930485B16Rik,*‡Cdk17 | |||
| 11 | 11465042-11679166 | 214 | 2 | 8 | YES | YES | YES | Ikzf1,*4930512M02Rik, AL596450.1, RP23-373H2 |
| 11 | 23642090-23679371 | 37 | 2 | 1 | YES | YES | YES | Rel* |
| 11 | 24098976-24156602 | 58 | 6 | 2 | YES | YES | YES | Bcl11a* |
| 11 | 51713117-51817285 | 104 | 4 | 0 | YES | Phf15,*Cdkn2aipn1, Ube2b, Cdkl3 | ||
| 11 | 52118189-52155248 | 37 | 2 | 2 | YES | Tcf7,*Vdac1 | ||
| 11 | 68174513-68271352 | 97 | 1 | 4 | YES | YES | Pik3r5,*Ntn1, AL606831.2 miRNA | |
| 11 | 77601654-77615704 | 14 | 2 | 1 | YES | Myo18a,*AL591065.2 | ||
| 11 | 79467420-79566569 | 99 | 3 | 0 | YES | Rab11fip4,*‡mmu-mir-193 (miRNA), mmu-mir-365-2 (miRNA), AL731726.1 | ||
| 11 | 86407328-86438598 | 31 | 3 | 0 | Tmem49* | |||
| 11 | 87565872-87567954 | 2 | 2 | 1 | YES | mmu-mir-142* | ||
| 11 | 100752419-100753004 | 1 | 2 | 1 | YES | Stat3* | ||
| 11 | 106548821-106782459 | 234 | 2 | 3 | Gm885,*Pecam1, Polg2, AL593847.1, Smurf2, Ddx5, Ccdc45 | |||
| 11 | 115872987-116120321 | 247 | 2 | 3 | YES | Galk1,*Trim65, Srp68, Itgb4, H3f3b, Unk, Uncl3d, Trim47, Mrpl38, Wbp2, Fbf1, Acox1, Evpl, Cdk3, 2310004N24Rik | ||
| 11 | 117205625-117212544 | 7 | 2 | 1 | YES | YES | Sept9* | |
| 11 | 120491729-120493009 | 1 | 3 | 0 | YES | Mafg* | ||
| 12 | 86976896-87168328 | 191 | 2 | 2 | YES | Jdp2,*Ttll5, 0610007P14Rik, Batf, Mfsd7c | ||
| 12 | 108362175-108363890 | 2 | 1 | 2 | AC163345.1* | |||
| 13 | 28644507-28873369 | 229 | 3 | 2 | RP23-45H23.1,*Sox4, RP23-371K8 | |||
| 14 | 70133928-70229580 | 96 | 3 | 0 | YES | Chmp7,*‡Tnfrsf10b, Rhobtb2, Pebp4 | ||
| 15 | 61815622-62044652 | 229 | 6 | 9 | YES | YES | YES | Myc,*Pvt1 |
| 15 | 62000405-62022073 | 22 | 3 | 0 | Pvt1*‡ | |||
| 15 | 66633426-66693812 | 60 | 3 | 1 | YES | Sla,*Tg | ||
| 15 | 66646986-66693812 | 47 | 3 | 0 | YES | Tg,*‡Sla | ||
| 15 | 80346987-80525653 | 179 | 4 | 2 | YES | Enthd1,*Grap2, Fam83f | ||
| 15 | 96373651-96486074 | 112 | 3 | 0 | AC123606.11,*‡Slc38a1, Slc38a2 | |||
| 15 | 96373651-96540760 | 167 | 3 | 2 | YES | YES | Slc38a1,*Slc38a2, AC123606.1 | |
| 15 | 97443447-97669490 | 226 | 4 | 0 | YES | Rpap3,*Pp11r, Hdac7, Rapgef3, AC104225.2, Slc48a1, Vdr | ||
| 16 | 32433625-32549358 | 116 | 3 | 1 | Pcyt1a,*Zdhhc19, Osta, Tctex1d2 | |||
| 16 | 32517604-32549358 | 32 | 3 | 0 | YES | Zdhhc19*‡ | ||
| 16 | 49771352-49938369 | 167 | 4 | 0 | YES | Ift57,*Cd47G, m5486, AC107830.1, AC107830.2 | ||
| 16 | 49839166-49938369 | 99 | 3 | 0 | YES | Cd47*‡ | ||
| 17 | 29534871-29639166 | 104 | 6 | 7 | YES | YES | YES | Pim1,*Fgd2 |
| 17 | 47649930-47875555 | 226 | 1 | 4 | YES | Taf8,*Tcfeb, Med20, Frs3, Pgc, Usp49, Ccnd3, Bysl, Tomm6, Prickle4 | ||
| 17 | 52420984-52490384 | 69 | 3 | 0 | AC121600.2,*‡AC121600.1 (miRNA) | |||
| 18 | 35900301-36089864 | 190 | 2 | 3 | YES | Tmem173,*Cxxc5 | ||
| 18 | 60962858-60989942 | 27 | 2 | 1 | YES | YES | Cd74,*Tcof1 | |
| 19 | 37514331-37569767 | 55 | 4 | 6 | YES | Exoc6,*Hhex | ||
| 19 | 37514331-37569373 | 55 | 0 | 6 | YES | YES | Hhex,*†Exoc6 |
| Chromosome . | CIS position . | CIS range, kb . | Mll-AF9 mice with insert . | WT mice with insert . | RTCGD . | Cancer Gene Census . | COSMIC . | All genes in/near CIS . |
|---|---|---|---|---|---|---|---|---|
| 1 | 4485458-4486321 | 1 | 2 | 1 | YES | Sox17* | ||
| 1 | 36823950-37002868 | 179 | 3 | 1 | YES | Tmem131,*Zap70, AC123854.2 | ||
| 1 | 135756752-135974231 | 217 | 2 | 2 | AC157924.6,*Prelp, Fmod, Btg2, Optc | |||
| 1 | 173849175-174012079 | 163 | 4 | 1 | YES | YES | Slamf6,*Copa, Vangl2, Nhlh1, AC158930.1, Ncstn | |
| 2 | 11547443-11577841 | 30 | 3 | 0 | YES | YES | Il2ra* | |
| 2 | 26315310-26525572 | 210 | 0 | 11 | YES | YES | YES | Notch1,*mmu-mir-126, Egfl7, Agpat2, Fam69b, AL732311 |
| 2 | 90344091-90491625 | 148 | 3 | 1 | YES | Ptprj* | ||
| 2 | 90919486-90925879 | 6 | 3 | 1 | YES | Slc39a13,*Sfpi1 | ||
| 2 | 101463219-101464091 | 1 | 4 | 0 | B230118H07Rik,*Rag2 | |||
| 2 | 103601727-103784302 | 183 | 1 | 3 | YES | YES | YES | Lmo2,*Nat10, Caprin1, BX537331.1, AL928544.7 (miRNA), AL928544.5 (miRNA) |
| 2 | 103601727-103784302 | 183 | 0 | 3 | YES | Nat10,*†Lmo2, Caprin1, BX537331.1, AL928544.7 (miRNA), AL928544.5 (miRNA) | ||
| 2 | 117168903-117367506 | 199 | 3 | 10 | YES | YES | Rasgrp1* | |
| 2 | 152601718-152828604 | 227 | 2 | 2 | YES | YES | Bcl2l1,*Tpx2, Mylk2, Ttll9, Foxs1, Dusp15 | |
| 2 | 165712889-165837434 | 125 | 4 | 0 | Prkcbp1,*Zmynd8, Ncoa3 | |||
| 2 | 165781678-165837434 | 56 | 3 | 0 | RP23-108D12.5*‡ | |||
| 2 | 167625651-167785437 | 160 | 4 | 1 | A530013C23Rik,*Ptpn1 | |||
| 2 | 167750534-167785437 | 35 | 3 | 0 | YES | Ptpn1*‡ | ||
| 2 | 169958430-170046828 | 88 | 2 | 2 | YES | YES | Zfp217* | |
| 3 | 94945270-95035953 | 91 | 3 | 0 | YES | Tnfaip8l2,*‡Cdc42se1, Sema6c, Gabpb2, Mlt11, Gm128, Bnipl, Lysmd1, Scnm1 | ||
| 4 | 32341351-32513306 | 172 | 3 | 3 | YES | YES | Bach2* | |
| 4 | 133220770-133371434 | 151 | 2 | 2 | YES | Pigv,*Aridla | ||
| 4 | 133652224-133867470 | 215 | 1 | 2 | YES | YES | Cd52,*Ubxn11, Aim1, Sh3bgrl3, Ccdc21, Gm7534, Catsper4, Cnksr1, Zfp593, Grrp1, Pdik1 | |
| 4 | 149076009-149079348 | 3 | 2 | 1 | YES | Pik3cd* | ||
| 5 | 34038901-34279652 | 241 | 1 | 3 | YES | YES | YES | Fgfr3,*Tacc3 |
| 5 | 108078270-108186428 | 108 | 5 | 19 | YES | YES | Gfi1,*Evi5, Rpap2 | |
| 5 | 111845647-112004142 | 158 | 10 | 3 | YES | YES | YES | Mn1,*C130026L21Rik |
| 6 | 48598556-48720382 | 122 | 6 | 0 | YES | Gimap8,*Gimap cluster | ||
| 6 | 48630644-48720382 | 90 | 5 | 0 | YES | Gimap4,*‡Gimap cluster | ||
| 6 | 127104034-127281434 | 177 | 2 | 5 | YES | YES | YES | Ccnd2,*AC161597.1 |
| 7 | 19858212-20077994 | 220 | 6 | 0 | YES | Fosb,*mmu-mir-343, Ercc1, Ercc2, Ppplrl3, Rtn2, Ckm, Exoc3l2, C79127, Vasp, Cd3eap, Klc3, Mark4 | ||
| 7 | 29165895-29228553 | 63 | 4 | 0 | YES | Gmfg,*Paf1, Samd4b, Med29 | ||
| 7 | 121285480-121354575 | 69 | 5 | 11 | YES | YES | Rras2,*Copb1 | |
| 7 | 136903269-136957718 | 54 | 2 | 2 | YES | Brwd2* | ||
| 7 | 152126941-152233702 | 107 | 3 | 2 | YES | YES | YES | Ccnd1,*Oraov1 |
| 8 | 10856578-11099413 | 243 | 3 | 3 | AC116499.9,*3930402G23Rik, Irs2 | |||
| 8 | 10910498-11099413 | 189 | 0 | 3 | YES | 3930402G23Rik,*†Irs2 | ||
| 8 | 86266601-86296089 | 29 | 2 | 1 | YES | Cd97* | ||
| 8 | 131049443-131116479 | 67 | 3 | 0 | YES | Nrp1*‡ | ||
| 9 | 32416427-32624588 | 208 | 3 | 5 | YES | YES | Ets1* | |
| 9 | 44135577-44376194 | 241 | 2 | 1 | YES | Dpagt1,*Bcl9l, H2afx, Hyou1, Rps25, Slc37a4, AC122428.3, C030014l23Rik, C2cd2l, Hmbs, Vps11, Trappc4, Ccdc84, Foxr1, Upk2, AC122428.2, Cxcr5 | ||
| 9 | 110800877-110812241 | 11 | 2 | 1 | YES | Als2cl* | ||
| 9 | 123758644-123985636 | 227 | 3 | 1 | YES | Fyco1,*Xcrl, CAAA01140679.1.6228.1, Ccr1, Ccr1l1, Ccr3 | ||
| 10 | 20763054-20972634 | 210 | 8 | 9 | YES | YES | YES | Myb,*Ahi1, AC153556.5 (miRNA) |
| 10 | 20811983-20972634 | 161 | 0 | 9 | AC153556.5,*†Myb | |||
| 10 | 41658107-41790233 | 132 | 1 | 3 | YES | Armc2* | ||
| 10 | 59615644-59634420 | 19 | 2 | 1 | YES | Chst3,*Spock2 | ||
| 10 | 76990210-77085753 | 96 | 3 | 0 | YES | Itgb2,*‡181008A18Rik, Pttglip, Sumo3, Ube2g2 | ||
| 10 | 79452089-79621648 | 170 | 4 | 0 | YES | Cnn2,*Abca7, Hmha1, Gpx4, Stk11, Atp5d, Midn, ORF61, Polr2e, Sbno2, Dos | ||
| 10 | 79895567-80140544 | 245 | 2 | 2 | YES | Mknk2,*Tcf3, Onecut3, Atp8b3, Rexo1, Klf16, Fam108a, Scamp4, Adat3, Csnk1g2, Btbd2 | ||
| 10 | 92532859-92627678 | 95 | 3 | 0 | 4930485B16Rik,*‡Cdk17 | |||
| 11 | 11465042-11679166 | 214 | 2 | 8 | YES | YES | YES | Ikzf1,*4930512M02Rik, AL596450.1, RP23-373H2 |
| 11 | 23642090-23679371 | 37 | 2 | 1 | YES | YES | YES | Rel* |
| 11 | 24098976-24156602 | 58 | 6 | 2 | YES | YES | YES | Bcl11a* |
| 11 | 51713117-51817285 | 104 | 4 | 0 | YES | Phf15,*Cdkn2aipn1, Ube2b, Cdkl3 | ||
| 11 | 52118189-52155248 | 37 | 2 | 2 | YES | Tcf7,*Vdac1 | ||
| 11 | 68174513-68271352 | 97 | 1 | 4 | YES | YES | Pik3r5,*Ntn1, AL606831.2 miRNA | |
| 11 | 77601654-77615704 | 14 | 2 | 1 | YES | Myo18a,*AL591065.2 | ||
| 11 | 79467420-79566569 | 99 | 3 | 0 | YES | Rab11fip4,*‡mmu-mir-193 (miRNA), mmu-mir-365-2 (miRNA), AL731726.1 | ||
| 11 | 86407328-86438598 | 31 | 3 | 0 | Tmem49* | |||
| 11 | 87565872-87567954 | 2 | 2 | 1 | YES | mmu-mir-142* | ||
| 11 | 100752419-100753004 | 1 | 2 | 1 | YES | Stat3* | ||
| 11 | 106548821-106782459 | 234 | 2 | 3 | Gm885,*Pecam1, Polg2, AL593847.1, Smurf2, Ddx5, Ccdc45 | |||
| 11 | 115872987-116120321 | 247 | 2 | 3 | YES | Galk1,*Trim65, Srp68, Itgb4, H3f3b, Unk, Uncl3d, Trim47, Mrpl38, Wbp2, Fbf1, Acox1, Evpl, Cdk3, 2310004N24Rik | ||
| 11 | 117205625-117212544 | 7 | 2 | 1 | YES | YES | Sept9* | |
| 11 | 120491729-120493009 | 1 | 3 | 0 | YES | Mafg* | ||
| 12 | 86976896-87168328 | 191 | 2 | 2 | YES | Jdp2,*Ttll5, 0610007P14Rik, Batf, Mfsd7c | ||
| 12 | 108362175-108363890 | 2 | 1 | 2 | AC163345.1* | |||
| 13 | 28644507-28873369 | 229 | 3 | 2 | RP23-45H23.1,*Sox4, RP23-371K8 | |||
| 14 | 70133928-70229580 | 96 | 3 | 0 | YES | Chmp7,*‡Tnfrsf10b, Rhobtb2, Pebp4 | ||
| 15 | 61815622-62044652 | 229 | 6 | 9 | YES | YES | YES | Myc,*Pvt1 |
| 15 | 62000405-62022073 | 22 | 3 | 0 | Pvt1*‡ | |||
| 15 | 66633426-66693812 | 60 | 3 | 1 | YES | Sla,*Tg | ||
| 15 | 66646986-66693812 | 47 | 3 | 0 | YES | Tg,*‡Sla | ||
| 15 | 80346987-80525653 | 179 | 4 | 2 | YES | Enthd1,*Grap2, Fam83f | ||
| 15 | 96373651-96486074 | 112 | 3 | 0 | AC123606.11,*‡Slc38a1, Slc38a2 | |||
| 15 | 96373651-96540760 | 167 | 3 | 2 | YES | YES | Slc38a1,*Slc38a2, AC123606.1 | |
| 15 | 97443447-97669490 | 226 | 4 | 0 | YES | Rpap3,*Pp11r, Hdac7, Rapgef3, AC104225.2, Slc48a1, Vdr | ||
| 16 | 32433625-32549358 | 116 | 3 | 1 | Pcyt1a,*Zdhhc19, Osta, Tctex1d2 | |||
| 16 | 32517604-32549358 | 32 | 3 | 0 | YES | Zdhhc19*‡ | ||
| 16 | 49771352-49938369 | 167 | 4 | 0 | YES | Ift57,*Cd47G, m5486, AC107830.1, AC107830.2 | ||
| 16 | 49839166-49938369 | 99 | 3 | 0 | YES | Cd47*‡ | ||
| 17 | 29534871-29639166 | 104 | 6 | 7 | YES | YES | YES | Pim1,*Fgd2 |
| 17 | 47649930-47875555 | 226 | 1 | 4 | YES | Taf8,*Tcfeb, Med20, Frs3, Pgc, Usp49, Ccnd3, Bysl, Tomm6, Prickle4 | ||
| 17 | 52420984-52490384 | 69 | 3 | 0 | AC121600.2,*‡AC121600.1 (miRNA) | |||
| 18 | 35900301-36089864 | 190 | 2 | 3 | YES | Tmem173,*Cxxc5 | ||
| 18 | 60962858-60989942 | 27 | 2 | 1 | YES | YES | Cd74,*Tcof1 | |
| 19 | 37514331-37569767 | 55 | 4 | 6 | YES | Exoc6,*Hhex | ||
| 19 | 37514331-37569373 | 55 | 0 | 6 | YES | YES | Hhex,*†Exoc6 |
List of all CISs from insertion site analysis. Columns 1 and 2 are the chromosome and position of each CIS according to NCBI build 37. Column 3 shows the range of the CIS in kilobases. Columns 4 and 5 are the number of each genotype with contributing insertions to each CIS. Columns 6 to 8 refer to the gene shown with an asterisk in column 9. Column 6 indicates whether the gene has previously been identified in the RTCGD,23 column 7 indicates whether the human homolog of the gene has been identified as a cancer gene in the Cancer Gene Census,25 and column 8 indicates whether a mutation in the human homolog of the gene has been identified as a recurring somatic mutation in cancer (COSMIC).24 Column 9 shows all the genes in or near the CIS region that may be affected by proviral insertions.
CIS indicates common insertion site; RTCGD, Retroviral Tagged Cancer Gene Database; COSMIC, Catalog of Somatic Mutations In Cancer; and WT, wild type.
The gene whose transcriptional start site is closest to the median of the CIS region, which was called the CIS-associated candidate gene, annotated using Ensembl release 55.
Genes are only found in the infected WT CIS analysis.
Genes are found only when analyzing CIS from infected Mll-AF9 mice.
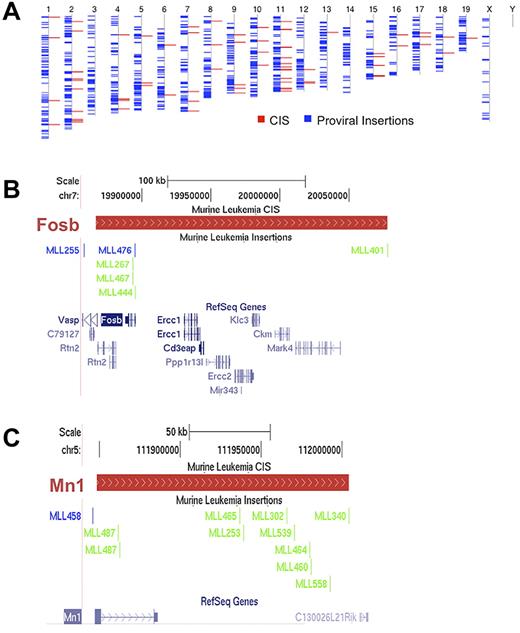
The CISs were distributed throughout the mouse genome (Figure 2A). Examples of 2 CIS regions and the proviral insertions that define them are displayed in Figure 2B and C. We defined a CIS-associated gene as the gene with the transcription start site closest to the median of the CIS region, which became a candidate gene for a role in leukemia progression.
Proviral insertions are randomly distributed throughout the genome and define CISs. (A) Blue lines represent all the insertions recovered from the screen overlaid on the mouse chromosomes, labeled above. Red lines represent all the resulting CISs. (B-C) Two representative CISs with the distribution of insertions. (B) Insertions defining Fosb; (C) insertions defining Mn1. The red bars define a CIS region. The scale and chromosome region are above the CIS region while the insertions from the different mice in the screen are below. With respect to the primary CIS-associated genes, blue lines on the insertion track indicate positive orientation and green lines indicate a negative orientation. RefSeq genes within the CIS are in blue at the bottom of each figure.
Proviral insertions are randomly distributed throughout the genome and define CISs. (A) Blue lines represent all the insertions recovered from the screen overlaid on the mouse chromosomes, labeled above. Red lines represent all the resulting CISs. (B-C) Two representative CISs with the distribution of insertions. (B) Insertions defining Fosb; (C) insertions defining Mn1. The red bars define a CIS region. The scale and chromosome region are above the CIS region while the insertions from the different mice in the screen are below. With respect to the primary CIS-associated genes, blue lines on the insertion track indicate positive orientation and green lines indicate a negative orientation. RefSeq genes within the CIS are in blue at the bottom of each figure.
Twenty-nine candidate genes, of the 88 CIS-associated genes identified, have been previously identified in other genetic screens according to the RTCGD.23 Sixty-nine of the human homologs of candidate genes are mutated in human cancer according to the Catalog of Somatic Mutations In Cancer (COSMIC),24 and 13 are known cancer genes according to the Cancer Gene Census,25 including genes associated with leukemia such as Cyclin D1, Fgfr3, Myc, Ikaros, Notch1, Myb, Bcl11a, and Mn1 (Table 1). Ingenuity pathway analysis shows that our candidate gene list falls into 3 main functional categories: cell transformation, blood cell development, and cell differentiation (supplemental Table 4A). In addition, the top 5 overrepresented canonical pathways from this list of candidate genes are linked to acute and chronic myeloid leukemia genes (supplemental Table 4B).
Analysis of candidate leukemia genes leads to identification of Mn1, Fosb, and Bcl11a as candidate MLL-AF9 cooperating genes
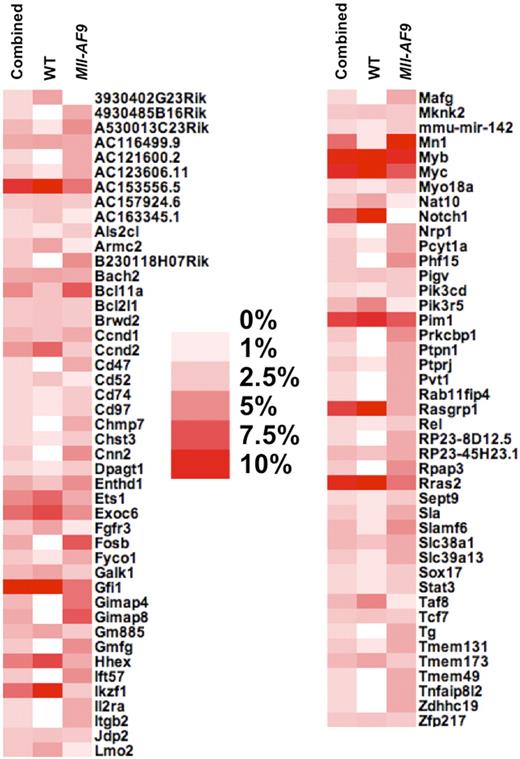
To prioritize genes that may cooperate with Mll-AF9 to cause leukemia progression from the 88 candidate genes as listed in Table 1, we determined the percentage of leukemias containing an insertion within each independent gene for each of the 3 analysis groups (WT, Mll-AF9, and combined). The candidate genes were sorted in alphabetical order and visualized in a heat map (Figure 3). We labeled genes as low priority if they had frequent insertions in all 3 groups, such as 3 of the 4 most frequently mutated genes (Gfi1, Myc, and Myb), or those only found in a high percentage of leukemia in the WT CIS list but not the Mll-AF9 CIS list, such as Hhex, Ikaros (Ikzf1), and Notch1, a CIS without a single insertion in Mll-AF9 mice. We then selected genes as a top priority those found in a high percentage of mice in the Mll-AF9 CIS list but not the WT CIS list include Bcl11a, Fosb, Mn1, and a Gimap gene cluster.
Heat map shows the percentage of mice containing an insertion in each CIS of the total number of mice with at least one insertion. Combined, WT, and Mll-AF9 refer to the 3 different CIS analyses that were performed, where combined means all insertions from both genotypes were used. The darkest red color indicates that more than 10% of the mice in that CIS list have insertions in a given CIS-associated gene, while white indicates no mice had insertions near a given gene in that CIS list, as indicated in the legend.
Heat map shows the percentage of mice containing an insertion in each CIS of the total number of mice with at least one insertion. Combined, WT, and Mll-AF9 refer to the 3 different CIS analyses that were performed, where combined means all insertions from both genotypes were used. The darkest red color indicates that more than 10% of the mice in that CIS list have insertions in a given CIS-associated gene, while white indicates no mice had insertions near a given gene in that CIS list, as indicated in the legend.
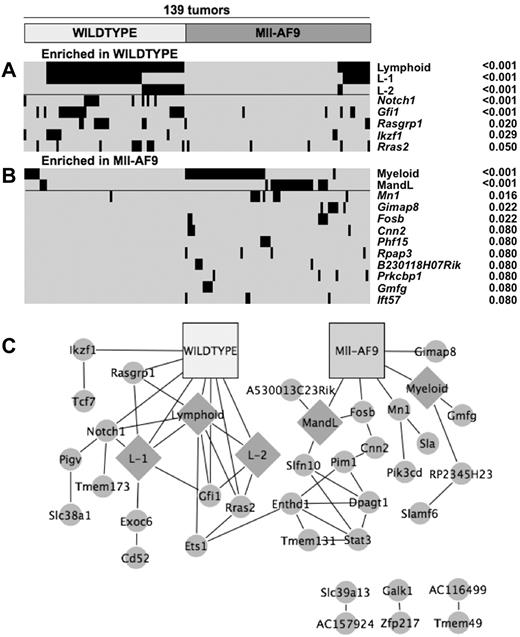
Fisher exact tests were performed on each pairwise comparison of the genotype, phenotype, and CIS-associated gene. A modified heat map shows the statistically significant pairs in each genotype. The lymphoid phenotypes (lymphoid, L1 and L2) were enriched in the WT leukemia as well as 5 CIS-associated candidate genes, including Notch1 and Ikaros (Figure 4A). Conversely, the Mll-AF9 mice showed enrichment in the myeloid and MandL phenotype groups as well as 3 candidate genes, including Mn1 and Fosb genes (Figure 4B). The significant associations between phenotypes, genotypes, and genes are displayed in a network where each line represents a significant pairwise connection that allows visualization of all the associations simultaneously (Figure 4C). The significant associations between phenotype variables and specific candidate genes are listed in supplemental Table 1B. Several genes associated with myeloid disease or Mll-AF9 mice (Gmfg, Bcl11a, Mn1) were positively associated with a high spleen weight or high WBC count. As expected, several genes already shown to be associated with the WT genotype and lymphoid disease such as Rasgrp1, Rras2, and Notch1 were also associated with Jβ1 or Jβ2 rearrangement. Fisher exact tests were also performed between each candidate gene pair to determine whether 2 genes occurred more frequently together than would be expected by chance. These connections are also included in the enrichment network (Figure 4C). Taken together, these data led us to pursue Mn1, Fosb, and Bcl11a for further study as potential MLL-AF9 cooperating genes.
There are significant associations between CIS-associated candidate genes and disease phenotypes, experimental cohorts, and other CIS-associated candidate genes. Modified heat map of significant candidate gene and phenotype enrichment in (A) WT and (B) Mll-AF9 mice. Each horizontal tick mark represents a proviral insertion identifying a CIS or a positive marker for a given phenotype in a given mouse leukemia. Mice are divided into the 2 infected genotypes across the x-axis; significant phenotypes and candidate genes are along the y-axis. Positive significant associations of phenotypes and candidate genes are shown (P < .05). In addition, candidate genes are shown that were found in 4 separate animals only in Mll-AF9 leukemias (P = .08). (C) Network of significant associations between phenotypes, genotypes, and candidate genes. All associations with P values < .05 were used. Genotypes are shown in squares, phenotypes are shown in diamonds, and CIS-associated candidate genes are shown in circles.
There are significant associations between CIS-associated candidate genes and disease phenotypes, experimental cohorts, and other CIS-associated candidate genes. Modified heat map of significant candidate gene and phenotype enrichment in (A) WT and (B) Mll-AF9 mice. Each horizontal tick mark represents a proviral insertion identifying a CIS or a positive marker for a given phenotype in a given mouse leukemia. Mice are divided into the 2 infected genotypes across the x-axis; significant phenotypes and candidate genes are along the y-axis. Positive significant associations of phenotypes and candidate genes are shown (P < .05). In addition, candidate genes are shown that were found in 4 separate animals only in Mll-AF9 leukemias (P = .08). (C) Network of significant associations between phenotypes, genotypes, and candidate genes. All associations with P values < .05 were used. Genotypes are shown in squares, phenotypes are shown in diamonds, and CIS-associated candidate genes are shown in circles.
Candidate leukemia genes have aberrant expression in human leukemia
To determine the relevance of the genes detected in this screen in human leukemia, we compared our CIS-associated candidate genes with published data on gene expression and copy number in human leukemia samples. First, we used gene expression profile (GEP) analysis performed on data from a cohort of 461 patients with AML26,27 to look for expression of the human homologs of our top 3 candidate MLL-AF9 cooperating genes: FOSB, MN1, and BCL11A. We found expression of all 3 genes in AML patients at varying levels, illustrating the heterogeneity of gene expression in human AML (supplemental Figure 5A). MN1 expression was low overall in human AML but higher on average in 2 AML subsets: patients with 3q rearrangements and inversion 16 patients. The inversion 16 observation is consistent with a previous publication.28 Expression of FOSB was detected in ∼ 50% of AML patients analyzed with either MLL-AF9 translocations or with other 11q23 translocations. BCL11A was also expressed in all MLL rearranged AMLs at varying levels (supplemental Figure 5B).
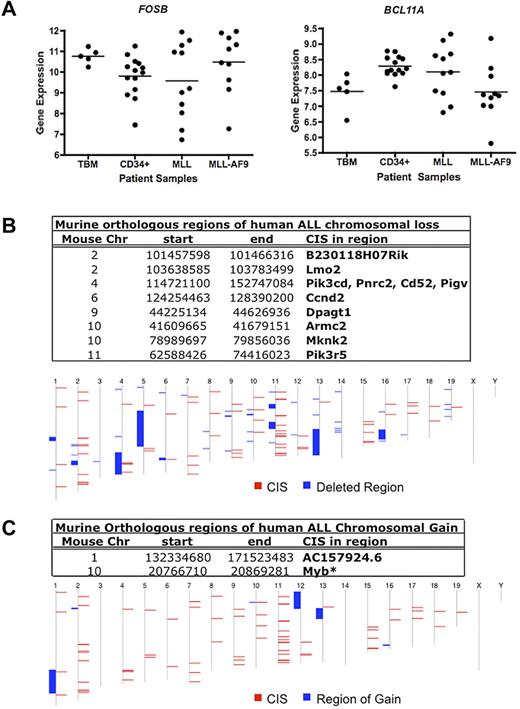
We then analyzed expression in a subset of the primary AMLs with MLL rearrangements.27 No significant differences in mean expression for FOSB and BCL11A were observed in patients with MLL-AF9 AMLs or other MLL rearrangements compared with normal total BM (TBM) and CD34+ cells (Figure 5A). However, in the case of each gene, some leukemia samples had higher expression than the highest expressing TBM and/or CD34+ samples. These data suggest a possible role for FOSB and BCL11A as MLL-AF9–cooperating genes.
Human GEP and gene copy number data for candidate Mll-AF9 cooperating genes. (A) Dot plots of expression of 2 CIS-associated candidate gene homologs in AML patients: FOSB and BCL11A. A representative probe for BCL11A is shown. Total BM (TBM) and CD34+ cells from healthy patients (n = 5 and 14, respectively) were used as controls compared with AMLs with any MLL rearrangement (n = 11), and patients with MLL-AF9 translocations (n = 10). The horizontal line represents the mean for each patient sample group. Data were log2 transformed. (B) Murine genomic regions orthologous to regions of deletion in human patients with ALL. In the bottom panel, the human chromosome map is shown. Blue blocks represent regions of chromosome deletion in ALL patients, and red blocks represent the combined CISs from our murine screen. Genes annotated using Ensembl release 55. (C) Murine genomic regions orthologous to regions of duplication or gain in human patients with ALL. In the bottom panel, the human chromosome map is shown. Blue blocks represent regions of chromosome gain in ALL patients, and red blocks represent the combined CISs from our murine screen. Genes annotated using Ensembl release 55. *Gene in region of gain in human AML that overlapped with a CIS from our murine screen.32
Human GEP and gene copy number data for candidate Mll-AF9 cooperating genes. (A) Dot plots of expression of 2 CIS-associated candidate gene homologs in AML patients: FOSB and BCL11A. A representative probe for BCL11A is shown. Total BM (TBM) and CD34+ cells from healthy patients (n = 5 and 14, respectively) were used as controls compared with AMLs with any MLL rearrangement (n = 11), and patients with MLL-AF9 translocations (n = 10). The horizontal line represents the mean for each patient sample group. Data were log2 transformed. (B) Murine genomic regions orthologous to regions of deletion in human patients with ALL. In the bottom panel, the human chromosome map is shown. Blue blocks represent regions of chromosome deletion in ALL patients, and red blocks represent the combined CISs from our murine screen. Genes annotated using Ensembl release 55. (C) Murine genomic regions orthologous to regions of duplication or gain in human patients with ALL. In the bottom panel, the human chromosome map is shown. Blue blocks represent regions of chromosome gain in ALL patients, and red blocks represent the combined CISs from our murine screen. Genes annotated using Ensembl release 55. *Gene in region of gain in human AML that overlapped with a CIS from our murine screen.32
In addition, we looked for recurrent changes in gene copy number in human AML and acute lymphoblastic leukemia (ALL) of other genes from this screen. Regions of chromosome gain and loss in pediatric ALL samples from St Jude's Children's Hospital29 were compared with the map positions of the human homologs of all CIS-associated candidate genes. Surprisingly, we found that eleven of our candidate genes overlap with 8 different areas of chromosomal deletion (Figure 5B), suggesting some pathways may be deregulated in lymphoid leukemia at least in part by deletion. There were also 2 regions of chromosomal gain that overlapped with 2 of our candidate leukemia genes (Figure 5C). One is a novel gene (AC157924.6) and the other is MYB, the amplification of which has been implicated in lymphoid and myeloid leukemia development.30,31 Similarly, one CIS overlapped with a region of chromosomal gain containing MYB when using data from human AML samples collected at Washington University in St Louis.32 However, minimal recurring copy number alterations were found in human AML overall, suggesting other mechanisms may lead to aberrant AML gene expression, such as epigenetic modifications.
Candidate MLL-AF9 cooperating genes have aberrant expression in mouse myeloid leukemia samples
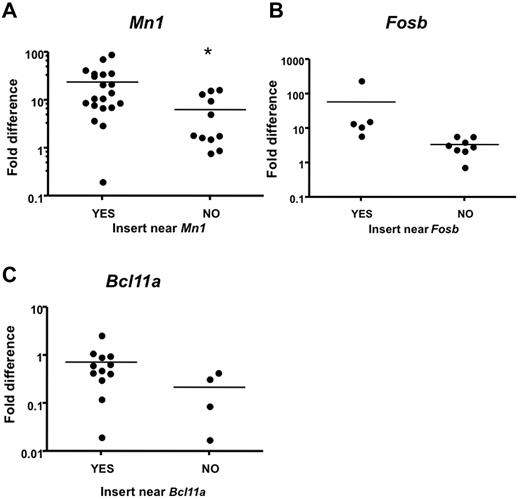
To confirm that proviral insertions affect expression of CIS-associated candidate genes in leukemia samples with insertions near those genes, we compared expression to leukemia samples that do not contain proviral insertions near these genes. Intron-spanning primers were designed for 15 candidate genes, and quantitative real-time PCR (qRT-PCR) was performed on RNA isolated from leukemias and from the corresponding tissue (lymph nodes, thymus, or spleen) from normal WT animals as baseline. Notably, Mn1 expression was significantly increased in Mll-AF9 myeloid leukemia tissues with insertions near Mn1 compared with Mll-AF9 myeloid leukemia tissues without insertions near Mn1 (P < .05; Figure 6A). In this case, we found that distal insertions 100 kb downstream of Mn1, but near a novel gene called C130026L21Rik, also affected Mn1 expression. A trend toward increased expression in myeloid leukemia from mice with insertions near Fosb and Bcl11a was also observed (Figure 6B-C). We found that Rras2 (P < .05) and Notch1 (P < .05) expression was significantly increased in infected WT leukemic tissues with insertions near those specific genes compared with infected WT leukemic tissues without insertions near those genes (data not shown).
Quantitative real-time PCR analysis indicates higher expression of CIS-associated candidate genes in mice with insertions near those genes. Candidate gene transcript expression in spleens or lymph nodes of mice with insertion events near or away from those genes. Intron-spanning primers were designed. Expression was calculated using the ΔΔCT method41 and is shown in log scale. Expression level was normalized to the respective WT tissue. Each dot represents one PCR. Reactions were performed on cDNA isolated from leukemia with insertions near other CIS genes labeled with “NO.” (A) Reactions performed on cDNA isolated from leukemia with insertions near Mn1 and the novel gene C130026L21Rik labeled with “YES” (*significant P value < .05). (B) Reactions performed on cDNA isolated from leukemia with insertions near Fosb labeled with YES (P = .153). (C) Reactions performed on cDNA isolated from leukemia with insertions near Bcl11a labeled with YES (P = .172).
Quantitative real-time PCR analysis indicates higher expression of CIS-associated candidate genes in mice with insertions near those genes. Candidate gene transcript expression in spleens or lymph nodes of mice with insertion events near or away from those genes. Intron-spanning primers were designed. Expression was calculated using the ΔΔCT method41 and is shown in log scale. Expression level was normalized to the respective WT tissue. Each dot represents one PCR. Reactions were performed on cDNA isolated from leukemia with insertions near other CIS genes labeled with “NO.” (A) Reactions performed on cDNA isolated from leukemia with insertions near Mn1 and the novel gene C130026L21Rik labeled with “YES” (*significant P value < .05). (B) Reactions performed on cDNA isolated from leukemia with insertions near Fosb labeled with YES (P = .153). (C) Reactions performed on cDNA isolated from leukemia with insertions near Bcl11a labeled with YES (P = .172).
Functional validation of candidate genes cooperating with MLL-AF9 in AML leukemogenesis
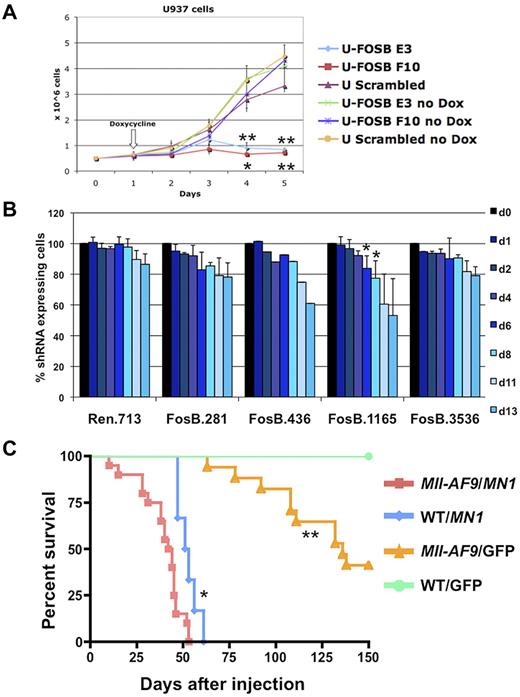
To validate the importance of FOSB in MLL-AF9 leukemia, we down-regulated expression using shRNA in leukemic cell lines. Human myeloid cell lines stably expressing doxycycline inducible shRNA versus FOSB were created. The U937 cell line, which has a CALM-AF10 translocation and up-regulation of HOXA9 and MEIS1,33 similar to MLL-AF9 fusion leukemias,34 was used as a surrogate line. U937 cells expressing shRNA constructs against FOSB or a scrambled shRNA were induced with doxycycline or treated with vehicle for 4 days. The cell lines without induction of shRNA or with the scrambled construct showed normal proliferation in exponential growth phase while the 2 cell lines expressing shRNA against FOSB had significantly impaired growth over 5 days (Figure 7A). In addition, we transduced Tet-On-competent MLL-AF9;NrasG12D murine cells19 with TRMPV-Neo-shRNA constructs targeting Fosb or Renilla luciferase as a control. After doxycycline treatment, 2 Fosb-specific shRNAs (FosB.1165 and FosB.436) depleted shRNA-expressing AML cells (Figure 7B). Western blot analysis was used to confirm knockdown for both experiments (supplemental Figure 7). This shows that continued expression of FOSB is required for leukemia maintenance of 2 AML cell lines, one expressing the MLL-AF9 fusion oncogene.
Fosb and Mn1 can contribute to AML maintenance or development of AML with MLL-AF9. (A) Growth curve of U937 cells with and without induction of shRNA against FOSB with doxycycline. The x-axis represents days after plating cells at 0.5 million cells/well; the y-axis is the number of cells per well in millions of cells. Doxycycline was added 24 hours after plating. Error bars reflect the SD of 3 wells/condition/cell line. Cell lines and conditions were labeled according to the legend (*significant P value < .05; **significant P value < .01). P values reflect paired t test comparing E3 or F10 shRNA U937 cells to U937 cells with a scrambled control, all treated with doxycycline. (B) shRNA competitive proliferation assay in Tet-On MLL-AF9;NrasG12D AMLs (expressing rtTA3) transduced with indicated TRMPV-Neo-shRNA targeting Fosb or Renilla luciferase. Cells were drug selected and mixed with 20%-40% untransduced cells, followed by shRNA induction with doxycycline. The percentage of shRNA-expressing cells is monitored over time. Error bars represent 2 or 3 independent experiments except in the case of 436 (*significant P value < .05). (C) Kaplan-Meier survival curve of mice injected with BM transduced with a retrovirus containing MN1 or GFP. The x-axis is the number of days after injection of cells into irradiated recipient mice; the y-axis is the percentage of survival. The study was ended at 150 days. Survival of mice receiving MN1-transduced Mll-AF9 BM was significantly shorter than that of mice with MN1-transduced WT BM (P = .0046) and mice with GFP-transduced Mll-AF9 BM (P < .0001); *significant P value < .001, **significant P value < .0001. A total of 52 mice were used in the study: Mll-AF9 BM/MN1 (n = 20), WT BM/MN1 (n = 6), Mll-AF9 BM/GFP (n = 21), and WT BM/GFP (n = 5).
Fosb and Mn1 can contribute to AML maintenance or development of AML with MLL-AF9. (A) Growth curve of U937 cells with and without induction of shRNA against FOSB with doxycycline. The x-axis represents days after plating cells at 0.5 million cells/well; the y-axis is the number of cells per well in millions of cells. Doxycycline was added 24 hours after plating. Error bars reflect the SD of 3 wells/condition/cell line. Cell lines and conditions were labeled according to the legend (*significant P value < .05; **significant P value < .01). P values reflect paired t test comparing E3 or F10 shRNA U937 cells to U937 cells with a scrambled control, all treated with doxycycline. (B) shRNA competitive proliferation assay in Tet-On MLL-AF9;NrasG12D AMLs (expressing rtTA3) transduced with indicated TRMPV-Neo-shRNA targeting Fosb or Renilla luciferase. Cells were drug selected and mixed with 20%-40% untransduced cells, followed by shRNA induction with doxycycline. The percentage of shRNA-expressing cells is monitored over time. Error bars represent 2 or 3 independent experiments except in the case of 436 (*significant P value < .05). (C) Kaplan-Meier survival curve of mice injected with BM transduced with a retrovirus containing MN1 or GFP. The x-axis is the number of days after injection of cells into irradiated recipient mice; the y-axis is the percentage of survival. The study was ended at 150 days. Survival of mice receiving MN1-transduced Mll-AF9 BM was significantly shorter than that of mice with MN1-transduced WT BM (P = .0046) and mice with GFP-transduced Mll-AF9 BM (P < .0001); *significant P value < .001, **significant P value < .0001. A total of 52 mice were used in the study: Mll-AF9 BM/MN1 (n = 20), WT BM/MN1 (n = 6), Mll-AF9 BM/GFP (n = 21), and WT BM/GFP (n = 5).
It has been previously reported that knockdown of MN1 in 2 human AML cell lines with MLL-AF9 impairs growth.35 To determine whether MN1 can cooperate with MLL-AF9 in vivo, retroviral BM transduction/transplantation assays were performed. BM from 5-FU–treated Mll-AF9 or WT mice was harvested, transduced with a retrovirus encoding a candidate gene (pSF91-MN120) or a GFP-only construct, and transplanted back into 129/BL6 F1 syngeneic recipient mice. Mice that received Mll-AF9 BM transduced with the MN1 gene succumbed to disease significantly faster with a median survival of 43 days compared with 52 days in mice transplanted with WT BM expressing MN1 (P = .0046) or 136 days in mice transplanted with GFP-transduced Mll-AF9 BM (P < .0001; Figure 7C). The GFP vector DNA was detected in all animals, verifying the viral transduction was successful (data not shown). The majority (25 of 27) of the animals from the 3 experimental groups with MN1 and/or Mll-AF9 had histopathology consistent with myeloid leukemia (data not shown). Thus, these data show MN1 can cooperate with MLL-AF9 in the induction of myeloid leukemia in vivo, strongly supporting a role for MN1 in myeloid transformation in MLL leukemia.
Discussion
Here, we report the findings of a large-scale MuLV insertional mutagenesis screen in an MLL translocation mouse model. We show that the M4070 chimeric retrovirus can accelerate myeloid and lymphoid disease in Mll-AF9 knock-in mice and can cause lymphoid disease in WT mice. MuLV retroviral insertions in leukemias identified 88 CISs, and the enrichment of insertion mutations in some CIS-associated candidate genes in Mll-AF9 leukemias suggested that they might cooperate with Mll-AF9 in the development of leukemia. Human microarray expression data were examined for the altered expression of candidate genes and showed a subset of MLL translocation-positive and MLL-AF9 AMLs had higher levels of these candidate genes than controls. There was aberrant expression of candidate Mll-AF9 cooperating genes Mn1, Bcl11a, and Fosb in murine leukemia samples by qRT-PCR. Finally, FOSB was shown to be essential to leukemia maintenance by in vitro shRNA knockdown and cooperation of MN1 with Mll-AF9 was confirmed with in vivo transduction/transplantation studies.
The M4070-infected WT mice developed mostly T-cell ALL whereas the M4070-infected Mll-AF9 transgenic mice mostly developed myeloid leukemia. Moreover, the latency of disease was significantly reduced for infected Mll-AF9 mice compared with infected WT mice (Figure 1B), suggesting that the mutations induced by M4070 cooperated with the Mll-AF9 allele in these mice. The differences in phenotype may be caused by differences in target cell availability in the 2 groups as Mll-AF9 mice have myeloproliferation before development of leukemia.7 Interestingly, approximately one-third of the animals in the infected Mll-AF9 group and 6% percent of the infected WT mice exhibited disease consisting of myeloid and lymphoid lineages. Thus, M4070 seems to accelerate ALL as well as AML, even in the same animal. It also appears fewer mutations are needed for transformation to overt leukemia in the presence of Mll-AF9 because of its potent oncogenic quality. WT leukemias contain on average more highly penetrant CISs (CISs with insertions from at least 5 leukemias; 2.13) than Mll-AF9 leukemias (1.43; supplemental Figure 6).
One of the goals of this article was to accurately define the phenotype of each mouse by collecting extensive complementary data using several established methods. For example, TCR and BCR gene rearrangement status has often been used to diagnose lymphoid leukemia without other data. Here, we show that this approach may be inadequate and that other methods must also be taken into account. Furthermore, approximately one-half of the mice with leukemia had a normal WBC count, thus an analysis of circulating cells also cannot be used alone to determine phenotype. However, enlargement of tissues was a reliable way to grossly define myeloid leukemia. Almost all mice with myeloid disease had a significantly higher-than-normal spleen weight, often presenting with over 10-fold enlargement, while mice with lymphoid leukemia tended to have an enlarged thymus. Flow cytometry was the main method used to define the phenotype of the mice, but was complemented with histology and IHC analyses to avoid misclassification. There were several cases in which the disease appeared to have a clear phenotype by flow but only advanced methods revealed the entire pathology of the animal, such as the L-2 animals, which had a lymphoid pathology with a myeloid surface marker. Thus, M4070 infection can induce multiple and complex phenotypes, which must be carefully scrutinized to make meaningful conclusions about the corresponding CIS-associated genes.
The use of M4070 to accelerate myeloid leukemia has been established.14,36 Similarly, we also detected leukemia acceleration in a myeloid leukemia-predisposed background and identified a longer list of CISs, all of which have at least 3 insertions in 3 mice/leukemias. Several candidate genes observed in other MuLV-induced AML models such as Bcl11a, Mn1, and Myb were found in our list. As expected, CIS-associated candidate genes identified almost exclusively in the infected WT cohort presenting with T-cell ALL were known T cell leukemia genes, such as Ikaros, Pim1, Notch1, and Lmo2.25 We also identified candidate genes in this cohort that have not been implicated in lymphoid disease before, such as Armc2 and Taf8. To determine the genes that play an important role in Mll-AF9 leukemogenesis, the extensive candidate gene list was prioritized by the Mll-AF9 genotype and myeloid phenotype using Fisher exact tests. This analysis allowed us to narrow the scope of candidate genes we chose for further study. Several candidate genes were found in our screen in Mll-AF9–positive myeloid leukemia that had not been reported before and may specifically interact with Mll-AF9 in leukemogenesis, including Gimap genes,37 Fosb, and the novel gene B230118H07Rik in the RAG locus.38 However, relatively few CISs were found exclusively in myeloid leukemia from Mll-AF9 mice. Therefore, we suspect that many of the CIS-associated candidate genes can contribute to myeloid leukemia initiated by Mll-AF9, and may also contribute to leukemia formation in other genetic contexts.
Human leukemia databases are important resources to determine whether candidate Mll-AF9 cooperating mouse genes could be clinically relevant. There was highly variable expression for the 3 top candidate genes (FOSB, MN1, and BCL11A) among all AML samples, even in the MLL rearranged AMLs. There were also many candidate genes that overlapped with the COSMIC24 or Cancer Gene Census25 databases (Table 1). These results suggest that MLL-AF9 likely cooperates with multiple genes, rather than one particularly potent gene, explaining the heterogeneity observed in patients.
Functional tests both in vitro and in vivo were used to provide evidence for a role of FOSB or MN1 in MLL-AF9 leukemia. FOSB is one of 4 genes in the FOS gene family thought to be involved in cell processes including proliferation and transformation but has not previously been implicated in AML disease progression. Remarkably, knockdown of FOSB expression impaired growth of both human and murine myeloid leukemia cells. These results strongly suggest that FOSB is an important novel leukemia gene that cooperates with MLL-AF9 in our mouse model, becoming a potential therapeutic target for MLL-AF9–driven leukemias. In addition, we show that MN1 could cooperate in vivo with Mll-AF9 in myeloid leukemia, postulating that MN1 may also be an important AML cancer gene depending on the genetic context. These data are consistent with findings that overexpressing MN1 is sufficient to cause AML in mice.20 Mn1 has also been identified as a CIS and cooperator with NUP98-HOXD13 and CALM-AF10 in 2 other mouse AML models using this same retrovirus.14,39 MN1 has been identified in the Cancer Gene Census database as a known cancer gene and overexpression of MN1 has been found in both human and mouse AML.14,20,40 Similarly, Mn1 insertions were found in leukemic mice induced by a retrovirus encoding MLL-ENL. Finally, knockdown of MN1 can impair growth of myeloid cell lines with MLL gene rearrangements and coexpression of MN1 and MLL-ENL with retroviruses can cooperate to induce a rapid AML-like disease.35
An important goal of this research was to establish novel MLL-AF9 cooperating genes that will ultimately lead to new therapeutic strategies for the treatment of AML in patients with this translocation. Some of these are known leukemia genes but their role in MLL-AF9 leukemia has not been previously reported, such as MN1, and some are novel genes that need to be explored, such as FOSB and B230118H07Rik. Therapeutic targets may also be identified by defining candidate genes from CISs that co-occur, such as CyclinD2 with Fosb and Sla or Pik3cd in the case of Mn1 (Figure 4C). Future studies are intended to test drugs on AML cell lines and in vivo to serve as preclinical testing for eventual application in human patients with MLL-AF9 leukemia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank members of the Largaespada laboratory for insightful discussions. They thank especially Dr Timothy Starr, Dr Vincent Keng, Sue Rathe, Dr Ernesto Diaz-Flores, Jonathan Linehan, Dr Sonja Nodland, Dr Keiko Akagi, Wendy Hudson, Dr Michael Heuser, Dr Bin Yin, Dr Anthony Uren, Dr Mei Lin Maunakea, and Dr Tony Cox for their technical assistance and advice as well as for providing reagents in some cases. They thank the Minnesota Supercomputing Institute for providing computational resources along with systems and database administrative support.
This work was supported by National Cancer Institute grant U01 CA84221 (D.A.L.), the Leukemia & Lymphoma Society of America LLS 7019-04 (D.A.L.), the University of Minnesota Cancer Biology training grant CA009138, and grants from Cancer Research UK and The Wellcome Trust (D.J.A.). S.C.K. is a scholar of the Leukemia & Lymphoma Society. L.S.C. is supported by fellowships from the National Cancer Institute (F32 CA106192 and K01 CA122183) and a postdoctoral fellowship from the American Cancer Society PF-05-153-01.
National Institutes of Health
Wellcome Trust
Authorship
Contribution: R.J.B. designed and performed research, analyzed and interpreted data, prepared figures and legends, and wrote the manuscript; L.S.C. designed and performed research; A.L.S., S.L., M.G.O., and S.C.K. analyzed results and prepared figures and legends; R.A.B., M.D.D., and M.J.N. performed research; J.Z. and A.R.R. performed research, analyzed results, and prepared figures; K.A.T.S., D.F., and A.-F.J.L. analyzed results; L.W. provided reagents; J.H.K. provided reagents and supported research; R.D. and S.W.L. designed research; D.J.A. designed and performed research and contributed to the editing of the manuscript; and D.A.L. designed research, interpreted data, and contributed to writing and editing the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for A.-F.J.L. is Office of the Vice President for Research, University of Minnesota Interdisciplinary Informatics, Minneapolis, MN.
Correspondence: David A. Largaespada, Department of Genetics, Cell Biology and Development, Masonic Cancer Center, University of Minnesota Twin Cities, 6-160 Jackson Hall, 321 Church St, Minneapolis, MN 55455; e-mail: larga002@umn.edu.