Abstract
Fanconi anemia (FA) is a recessive syndrome characterized by progressive fatal BM failure and chromosomal instability. FA cells have inactivating mutations in a signaling pathway that is critical for maintaining genomic integrity and protecting cells from the DNA damage caused by cross-linking agents. Transgenic expression of the implicated genes corrects the phenotype of hematopoietic cells, but previous attempts at gene therapy have failed largely because of inadequate numbers of hematopoietic stem cells available for gene correction. Induced pluripotent stem cells (iPSCs) constitute an alternate source of autologous cells that are amenable to ex vivo expansion, genetic correction, and molecular characterization. In the present study, we demonstrate that reprogramming leads to activation of the FA pathway, increased DNA double-strand breaks, and senescence. We also demonstrate that defects in the FA DNA-repair pathway decrease the reprogramming efficiency of murine and human primary cells. FA pathway complementation reduces senescence and restores the reprogramming efficiency of somatic FA cells to normal levels. Disease-specific iPSCs derived in this fashion maintain a normal karyotype and are capable of hematopoietic differentiation. These data define the role of the FA pathway in reprogramming and provide a strategy for future translational applications of patient-specific FA iPSCs.
Introduction
Fanconi anemia (FA) is a recessive syndrome characterized by BM failure, congenital anomalies, and a predisposition to malignancy.1 In vitro myeloid and erythroid colony growth of BM and peripheral blood cells from FA patients is decreased, suggesting the contribution of an intrinsic cellular defect to the BM failure.2,3 FA cells have a defect in DNA repair that leads to spontaneous chromosomal breakage and increased sensitivity to DNA bifunctional cross-linking agents such as mitomycin C and diepoxybutane.4 Whereas the precise biochemical function of most FA proteins and the link between defective DNA repair and BM failure remain incompletely understood, human and murine knockout FA cells display G2 phase arrest, increased sensitivity to oxidative damage, defective p53 induction, and increased apoptosis.1,5
FA can be classified into 14 complementation groups. A loss of function in any one of these 14 genes, including FANCA, FANCB, FANCC, FANCD1 (BRCA2), FANCD2, FANCE, FANCF, FANCG, FANCL, FANCI, FANCJ, FANCM, FANCN, and FANCP (SLX4), causes the disease phenotype.6,7 Expression of these cDNAs in cells from patients with FA in vitro corrects all cell-intrinsic defects, including BM progenitor growth, in vitro.8,9 Gene-transfer studies have shown that correction of defects in mice with gene-targeted deficiency of FA proteins is feasible and corrects a significant engraftment defect—and in some mouse models provides a selective advantage in vivo.10,11 Two clinical gene-therapy trials involving a total of 6 FA patients have been reported.12,13 In both studies, the harvest of CD34+ hematopoietic stem and progenitor cells (HSCs) from the BM or mobilized peripheral blood yielded lower than expected cell numbers and compromised in vitro expansion, resulting in a reduced number of cells available for gene transfer and autologous reinfusion. Therefore, whereas gene transfer per se is no longer a limitation to the therapeutic effectiveness of this approach, there are significant deficiencies in the number of autologous FA HSCs that can be collected and used in somatic gene-therapy trials.
The unlimited proliferative capacity of iPSCs is particularly attractive with regard to regenerative therapies and disease models in FA. Successful differentiation of corrected iPSCs into transplantable HSCs would allow the generation of unlimited numbers of these cells and pretransplantation molecular characterization of gene-corrected cells.14 Several recent studies have highlighted the utility of patient-specific iPSCs for in vitro disease modeling.15,16 With regard to FA, knockdown of FANCA and FANCD2 in embryonic stem cells (ESCs) leads to reduced hemogenic potential after differentiation, suggesting that FA-deficient human pluripotent stem cells may be amenable to in vitro disease modeling.17 Raya et al recently reported a failure of 4 FA-A and 2 FA-D2 patient samples to undergo direct reprogramming, concluding that restoration of the FA pathway is a prerequisite for iPSC generation from somatic cells of FA patients.18 Because of a limited number of human samples, mechanistic studies and quantification of the reprogramming efficiency of somatic FA cells are lacking to date.
Reprogramming is a stochastic and inefficient process, with reported reprogramming efficiencies of < 1% in murine systems using viral transduction of the reprogramming factors into somatic cells.19-22 Key determinants of the reprogramming efficiency include the differentiation state of the starting cell population and the ability of the somatic cells to respond to the cellular stress of reprogramming.23-27 Reprogramming induces DNA damage, resulting in the up-regulation of p53, increased double-strand DNA (dsDNA) breaks, and senescence.24 Conversely, ablation of p53 has been shown to result in an increased reprogramming efficiency, albeit at the expense of the genomic integrity of the resulting iPSCs.24,25,27 We reasoned that the DNA-repair defect that is inherent to FA cells may directly relate to the decreased efficiency of reprogramming. In this regard, FA somatic cells may have an increased frequency of preexisting DNA damage in the starting cell population or may be unable to resolve DNA lesions that are induced during the process of reprogramming. Given the significant promise of iPSCs for regenerative medicine and the study of FA biology, we sought to identify and overcome mechanisms of resistance to reprogramming of cells defective in the FA pathway. We studied direct reprogramming of murine Fanca−/− and Fancc−/− tail-tip fibroblasts (TTFs) and primary fibroblasts from 10 unique FA patients of multiple complementation types. We demonstrate that FA-deficient somatic cells can be reprogrammed, albeit at decreased efficiency, resulting in disease-specific iPSCs that fulfill stringent pluripotency criteria. Our studies define a critical role of the FA pathway in reprogramming and demonstrate that complementation of the FA gene defect rescues reprogramming of somatic FA cells to normal levels. Murine and patient-specific iPSCs derived in this fashion maintain a normal karyotype and contribute to fetal liver hematopoietic chimerism in vivo and multilineage hematopoietic differentiation in vitro, respectively.
Methods
Mouse models
Fanca−/−and Fancc−/− mice were generated by breeding heterozygous animals (C57BL/6J background) as described previously.28,29 Wild-type (WT) littermates were used as controls. All mice were maintained in a specific pathogen–free environment and the experiments were approved by the Institutional Animal Care and Use Committee at Children's Hospital Boston.
TTF isolation and transduction
Tails from 6- to 8-week-old mice were used to establish TTF cultures, as described previously.30 After removal of the dermis, the tail pieces were cultured in 3 mL of murine embryonic fibroblast (MEF) medium (DMEM, 10% FCS, 1% glutamate, and 1% penicillin/streptomycin) under normoxic (21% O2) or hypoxic (5% O2) conditions (Forma Series II incubator; Forma Scientific). Results are presented for experiments conducted under normoxic conditions unless otherwise stated. Genetic correction or retroviral marking was performed on day 5 of TTF culture. The cells were transduced with ecotropic pseudotyped retroviral vectors encoding enhanced green fluorescent protein (MIEG3-eGFP) or the human FANCA cDNA and eGFP (S11FAIEGnls).31 Transductions were performed overnight in the presence of 8 μg/mL of polybrene (Sigma-Aldrich). The viral supernatant was replaced with MEF medium and all tail pieces were removed on day 6. On day 14, GFP+ TTFs were isolated by FACS (FACSVantage; BD Biosciences; 20 ψ sorting pressure, 100μM nozzle). On day 20 after harvest, 1 × 105 TTFs were seeded into 6-well tissue culture plates for reprogramming. 2.5 × 104 cells were concurrently seeded into sterile glass chamber slides (Lab-Tek II; Nalge Nunc International) for senescence-associated β-galactosidase (β-Gal) or γH2AX immunofluorescence staining.
Generation of murine iPSCs and assessment of reprogramming efficiency
pMXs-based retroviral vectors (Klf4, Oct3/4, Sox2, c-Myc) were obtained from Addgene (www.addgene.org). To ensure consistency across multiple experiments, a master stock of ecotropic retroviral supernatant was generated by transient transfection of GP Phoenix cells using standard methods.32 On the day before infection with the reprogramming viruses, 1 × 105 TTFs were seeded into 6-well tissue culture plates. On the day of infection, the media was aspirated and replaced with 1.5 mL of viral supernatant from each of the 4 reprogramming vectors (6 mL total volume) supplemented with 8 μg/mL of polybrene. The 6-well plate was then subjected to a 90 minutes of spin-infection at room temperature (1400g; Allegra 6R centrifuge; Beckman Coulter). The viral supernatant was replaced with fresh MEF medium on the following day. Three days after transduction, the transduced progeny of 1 × 105 TTFs was transferred to ESC conditions, as described previously.30 Then, 10 mL of fresh ESC medium (DMEM, 15% FCS, 1% glutamine, 1% nonessential amino acids, 1% penicillin/streptomycin, and 3.5 μL of β-mercaptoethanol [≥ 99%; Sigma-Aldrich]) and 1000 units/mL of leukemia-inhibitory factor (ESGRO; Millipore) were applied daily. On day 16 after reprogramming, representative 10-cm dishes were stained for alkaline phosphatase expression (Leukocyte Alkaline Phosphatase Kit; Sigma-Aldrich). The number of colonies with iPSC-like morphology appearing in the 10-cm dish was enumerated in a blinded fashion. Individual iPSC clones were subcloned for further propagation and characterization.
Generation of human iPSCs
De-identified human fibroblast samples were obtained with local institutional review board approval (NS 08-08-0378). Dermal or BM stroma-derived fibroblasts were procured locally at Children's Hospital Boston (samples 1-4) or were obtained from the Coriell Institute for Medical Research in Camden, NJ (samples 5-11). The fibroblasts were reprogrammed using individual retroviral vectors33 or a single multicistronic lentiviral vector,34 as described previously.35
Teratoma, chimaera, karyotype, and CGH analysis
Teratomas were assessed by injecting 3 × 106 iPSCs subcutaneously (murine cells) or intramuscularly (human cells) into SCID mice (NOD.CB17-Prkdcscid/J strain; The Jackson Laboratory). Teratomas were dissected after 4-8 weeks, fixed in 10% paraformaldehyde, embedded in paraffin, and 4-μm sections were stained with H&E (Rodent Histopathology Core, Dana-Farber Cancer Institute /Harvard Cancer Center). Images were obtained using a Nikon Eclipse 90i microscope. Chimaera analysis of iPSCs was conducted by injecting Fanca−/− iPSCs (derived from CD45.1+ Bl-6 mice) into blastocysts isolated from C57 (CD45.2+ Bl-6 mice) embryos collected from the uterus. The reconstituted blastocysts were implanted into 2.5-day pseudopregnant Crl:CD1(ICR) female mice (Charles River Laboratories). Embryonic day 14.5 fetal livers were isolated and homogenized to assess the percentage of CD45.1+ (iPSC-derived) hematopoietic cells or chimera were allowed to develop to adulthood to gauge skin chimerism. Cytogenetic analysis of g-banded metaphases was performed by Cell Line Genetics. Comparative genomic hybridization (CGH) was performed by the Cytogenetics Core of the Dana-Farber Harvard Cancer Center as described previously.36 The array CGH assay used the oligo-based 44-k array platform (Agilent Technologies). Briefly, a genomic imbalance between the parental fibroblasts and the resultant iPSCs was reported when 3 or more adjacent oligos show at least a single copy number change from that expected with a karyotypically normal sample.
Fluorescence immunostaining of human iPSCs
Human iPSCs were grown in 96-well plates (Thermo Scientific) coated with γ-irradiated CF-1 MEFs (GlobalStem) and fixed for 30 minutes with 4% paraformaldehyde in PBS, washed 3 times with PBS, and incubated overnight at 4°C with primary Ab and Hoechst stain (1:10 000; Invitrogen) diluted in PBS supplemented with 3% donkey serum, 3% BSA fraction V, and 0.01% Triton X-100. Primary Abs included: Alexa Fluor 488–coupled anti–Tra-1-81 (clone TRA-1-81, 1:100; BD Biosciences), Alexa Fluor 488–coupled anti-stage–specific embryonic antigen 3 (anti–SSEA-3; clone MC813-70, 1:100; eBioscience,), Alexa Fluor 647–coupled anti–SSEA-4 (clone MC813-70, 1:100; BD Biosciences), Alexa Fluor 647–coupled anti–Tra-1-60 (clone TRA-1-60, 1:75; BD Biosciences); rabbit polyclonal anti-Nanog (1:400; Abcam), and rabbit polyclonal anti-Oct4 (1:400; Abcam). After 3 washes with PBS, the samples were incubated with the secondary Ab (1:2000, Alexa Fluor 555–coupled donkey anti–rabbit; Molecular Probes) for 3 hours. After 3 washes with PBS, images were acquired using a BD Pathway 435 imager equipped with a 10× objective. Murine iPSCs were stained using the ES Cell Characterization Kit (Chemicon). Images were obtained with a Nikon Eclipse TE2000-U microscope.
γH2AX and FANCD2 immunofluorescence
The day before staining, 2.5 × 104 cells were seeded in gelatinized chamber slides (Lab-Tek II). The cells were fixed with 4% paraformaldehyde and permeabilized with 0.25% Triton X-100. The primary anti-γH2AX Ab (ab11174; Abcam) was incubated at a 1:400 dilution for 1 hour at room temperature. Alexa Fluor 488– or Alexa Fluor 459–conjugated goat anti–rabbit IgG (Invitrogen) were used as a secondary Abs. FANCD2 foci were detected in murine cells by immunofluorescence as described previously.37 FANCD2 foci in human cells were detected using an anti-FANCD2 Ab (clone FI17; Santa Cruz Biotechnology), Hoechst stain (1:10 000; Invitrogen), and an Alexa Fluor 488–conjugated goat anti–rabbit IgG secondary Ab (Invitrogen). The foci were detected using a Nikon Eclipse Ti microscope (60× magnification) and 100 cells per sample were scored in a blinded fashion.
Senescence assay
On the day before analysis, 2.5 × 104 fibroblasts were plated onto gelatinized sterile glass chamber slides (Lab-Tek II). β-Gal activity was detected at pH 6 using a senescence β-Gal staining kit (Cell Signaling Technology). One hundred cells per sample were scored in a blinded fashion using an Olympus CKX41 inverted microscope at a 20× magnification.
Western blot analysis
Primary fibroblasts or iPSCs were lysed in 2× SDS lysis buffer (Bio-Rad). The proteins were electrophoresed on SDS-polyacrylamide gels, transferred to nitrocellulose, and immunoblotted with Abs against p53 (Ab-3, clone PAb240, 1:100; Calbiochem), p21 (clone SX118, 1:500; Santa Cruz Biotechnology), p19 (clone M-20, 1:1000; Santa Cruz Biotechnology), or tubulin (1:3000; Sigma-Aldrich).
Flow cytometry
Flow cytometry was performed using a FACSCalibur or LSRII flow cytometer (BD Biosciences). Reactive oxygen species (ROS) were measured by flow cytometry using the 2′,7′-di-chlorofluorescein ROS detection agent as per the manufacturer's instructions (Molecular Probes).
Hematopoietic differentiation
Hematopoietic colony-forming activity of human iPSC lines was assayed as described previously.38 Briefly, embryoid bodies were dissociated after 15 days and 1 × 104 or 3 × 104 cells were seeded in methylcellulose-containing recombinant cytokines (MethoCult H4434; StemCell Technologies). Hematopoietic progenitor (CFU) colonies were scored in a blinded fashion 14 days after seeding and photographs were taken using a Nikon Eclipse TS100 microscope.
Statistical analysis
A Wilcoxon rank-sum test was used to compare continuous measures between 2 groups. If the direction of effect was hypothesized, a 1-sided test was used; otherwise, the test was 2-sided. P < .05 was considered statistically significant. No adjustment was made for multiple comparisons.
Results
Reprogramming of murine FA knockout fibroblasts occurs with decreased efficiency
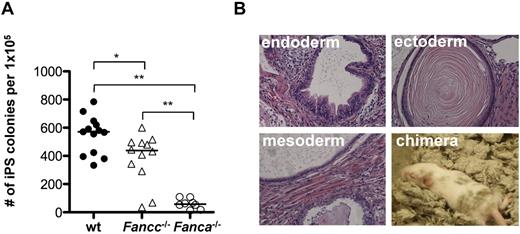
FANCA and FANCC cooperate with 6 additional FA proteins to form the FA core complex, which acts to monoubiquitinate FANCD2 and FANCI in the presence of DNA damage.39 Fanca−/−- and Fancc−/−-knockout mice closely mimic the cellular phenotype of human FA cells.28,29 Whereas neither Fanca−/− nor Fancc−/− mice develop spontaneous BM failure, competitive BM transplantation assays revealed a significant engraftment defect of BM cells from both knockout strains. This engraftment defect was more pronounced in Fanca−/− HSCs (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). To investigate the role of the FA proteins in reprogramming, Fanca−/−, Fancc−/−, and littermate WT TTFs were reprogrammed with retroviral vectors expressing Oct4, Sox2, Klf4, and c-Myc as described previously.30 The number of resulting iPSC colonies was enumerated 16 days after reprogramming (Figure 1A). In the case of the Fanca−/− TTFs, an approximately 10-fold reduction in the reprogramming efficiency was observed (mean efficiency, 0.55% vs 0.061%, WT vs Fanca−/−, P < .01, n = 8). Fancc−/− TTFs also showed a significant reduction in the reprogramming efficiency (mean efficiency, 0.38%, P < .05 compared with WT, n = 12), suggesting that a functional FA pathway is required for efficient reprogramming. Omission of c-Myc from the reprogramming factors resulted in a significant reduction of the overall reprogramming efficiency and did not increase the relative reprogramming efficiency of Fanca−/− TTFs (supplemental Figure 1B). Retroviral transduction requires actively dividing cells.40 To assess whether the observed difference in reprogramming efficiency resulted from a difference in retroviral transduction frequency, WT and Fanca−/− TTFs were transduced with a vector encoding eGFP at the time of reprogramming. There was no difference in the transduction efficiency between WT and Fanca−/− TTFs (supplemental Figure 1C).
Reprogramming efficiency of Fanca−/− and Fancc−/− fibroblasts is reduced. (A) Number of iPSC colonies derived from 1 × 105 TTFs 16 days after reprogramming. Filled circles, WT; triangles, Fancc−/−; open circles, Fanca−/−. Each symbol represents an individual animal. Horizontal line represents the median. (B) H&E stains of teratomas obtained by subcutaneous injection of Fanca−/− iPSCs (40× magnification) and fur chimerism in an animal derived by Fanca−/− iPSC blastocyst injection. **P < .01; *P < .05 by Wilcoxon rank-sum test.
Reprogramming efficiency of Fanca−/− and Fancc−/− fibroblasts is reduced. (A) Number of iPSC colonies derived from 1 × 105 TTFs 16 days after reprogramming. Filled circles, WT; triangles, Fancc−/−; open circles, Fanca−/−. Each symbol represents an individual animal. Horizontal line represents the median. (B) H&E stains of teratomas obtained by subcutaneous injection of Fanca−/− iPSCs (40× magnification) and fur chimerism in an animal derived by Fanca−/− iPSC blastocyst injection. **P < .01; *P < .05 by Wilcoxon rank-sum test.
Despite the reduced reprogramming efficiency, Fanca−/− and Fancc−/− iPSC colonies were successfully subcloned, resulting in iPSC lines that maintained an ESC-like morphology for > 20 passages (data not shown). Injection of Fanca−/− and Fancc−/− iPSC lines into immunodeficient mice resulted in the formation of teratomas containing derivatives of all 3 germ layers (Figure 1B and supplemental Figure 1D). Injection of Fanca−/− iPSC lines into blastocysts yielded animals with detectable fur chimerism (Figure 1B). Fanca−/− iPSC lines expressed SSEA-1, Oct3/4, and Nanog (supplemental Figure 1E) and maintained the FA cellular phenotype characterized by loss of monoubiquitination of FANCD2 after treatment with mitomycin C (data not shown). Therefore, the absence of a functional FA pathway in murine Fanca−/− and Fancc−/− TTFs significantly decreased the efficiency of direct reprogramming, but did not absolutely abolish the capacity to generate and maintain pluripotent iPSCs, providing the first demonstration of noncomplemented FA iPSCs.
Reprogramming activates the FA pathway and induces DNA damage
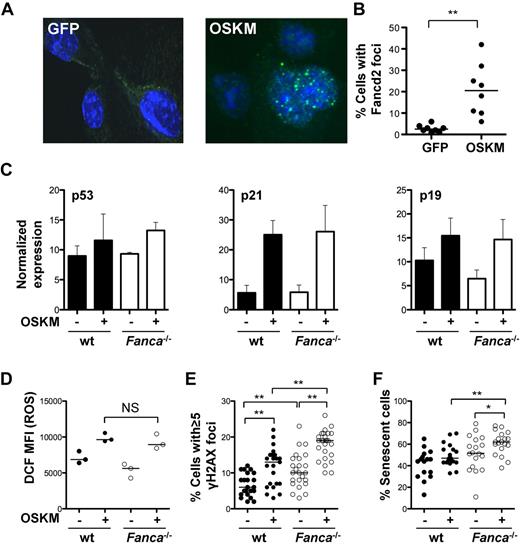
Activation of the DNA-damage response (DDR), the p53-p21 pathway, and cellular senescence have been identified as major barriers to efficient reprogramming.24-27 DNA damage results in the activation of the FA pathway via the upstream regulator ataxia telangiectasia and Rad3-related protein (ATR) and monoubiquitination of FANCD2 and FANCI.41,42 We hypothesized that reprogramming-induced cellular stress results in the activation of the FA pathway. To test this hypothesis, the frequency of FANCD2 foci were analyzed as a surrogate for FA pathway activation in WT TTFs at baseline and 7 days after reprogramming. Transduction of WT TTFs with reprogramming vectors (Klf4, Oct4, Sox2, and c-Myc) but not with an eGFP-encoding γ-retroviral control vector, led to increased FANCD2 foci formation (Figure 2A-B), confirming activation of the FA pathway during reprogramming.
Reprogramming activates the FA pathway and induces DNA damage. (A) Representative photomicrograph of FANCD2 immunofluorescence in control transduced or reprogrammed WT TTFs (60× magnification). (B) Percentage of cells containing FANCD2 foci as assessed by immunofluorescence 7 days after reprogramming. (C) Normalized expression of p53, p21, and p19 as determined by immunoblot in WT and Fanca−/− TTFs at baseline and 4 days after reprogramming (n = 4). Data are ± SEM. (D) Level of ROS measured by 2′,7′-di-chlorofluorescein (DCF) FACS analysis at baseline and 2 days after reprogramming. (E-F) Percentage of cells with ≥ 5 γH2AX foci (E) and percentage of cells undergoing senescence at baseline and 4 days after reprogramming (F). Filled circles, WT; open circles, Fanca−/−. OSKM indicates Oct3/4, Sox2, Klf4, c-Myc. *P < .05; **P < .01; NS, not significant by Wilcoxon rank-sum test. Horizontal line in panels B, D, E, and F represents the median.
Reprogramming activates the FA pathway and induces DNA damage. (A) Representative photomicrograph of FANCD2 immunofluorescence in control transduced or reprogrammed WT TTFs (60× magnification). (B) Percentage of cells containing FANCD2 foci as assessed by immunofluorescence 7 days after reprogramming. (C) Normalized expression of p53, p21, and p19 as determined by immunoblot in WT and Fanca−/− TTFs at baseline and 4 days after reprogramming (n = 4). Data are ± SEM. (D) Level of ROS measured by 2′,7′-di-chlorofluorescein (DCF) FACS analysis at baseline and 2 days after reprogramming. (E-F) Percentage of cells with ≥ 5 γH2AX foci (E) and percentage of cells undergoing senescence at baseline and 4 days after reprogramming (F). Filled circles, WT; open circles, Fanca−/−. OSKM indicates Oct3/4, Sox2, Klf4, c-Myc. *P < .05; **P < .01; NS, not significant by Wilcoxon rank-sum test. Horizontal line in panels B, D, E, and F represents the median.
To investigate the possible relationship between defective DNA repair and the reduced reprogramming efficiency of FA cells, the activation status of the DDR was evaluated at baseline and after reprogramming. p53, p21, and p19 were expressed at similar levels in WT and Fanca−/− TTFs at baseline. Reprogramming resulted in a comparable increase in p53, p21, and p19 in WT and Fanca−/− TTFs (Figure 2C), suggesting that the initial response to the integration of reprogramming vectors and expression of reprogramming proteins was equivalent in these cells. Previous data suggest that reprogramming induces ROS.43 Given the specific sensitivity of FA cells to ROS,44,45 the abundance of ROS at baseline and after reprogramming was measured. Reprogramming resulted in a similar induction of ROS in both WT and Fanca−/− cells (Figure 2D).
DNA damage can lead to several outcomes in mammalian cells, such as dsDNA breaks, cell-cycle arrest, apoptosis, and senescence. We analyzed the frequency of dsDNA breaks by γH2AX immunofluorescence and found a significantly higher incidence of dsDNA breaks in Fanca−/− TTFs before reprogramming, with a median percentage of TTFs containing ≥ 5 dsDNA breaks per nucleus of 6% versus 10% for WT versus Fanca−/−, respectively (P < .01; Figure 2E). Reprogramming led to an additional increase in dsDNA breaks in Fanca−/− TTFs (median 19% vs 13% in WT cells, P < .01). The addition of the ROS scavenger N-acetylcysteine resulted in a significant reduction of dsDNA breaks (γH2AX foci) of Fanca−/− cells, suggesting that ROS contribute to increased DNA damage in Fanca−/− cells during reprogramming (supplemental Figure 2A). Karyotype analysis of g-banded TTF metaphases revealed an increased incidence of chromosomal aberrations, including chromatid breaks, in Fanca−/− TTFs before reprogramming. One of 5 early-passage Fanca−/− iPSC clones revealed clonal autosomal aberration (supplemental Figure 2B and supplemental Table 1), highlighting the need for careful cytogenetic analysis of FA iPSCs. CGH was performed to assess imbalances arising in early-passage iPSCs in DNA derived from either WT or Fanca−/− fibroblasts. Using this sensitive technology, 1 of 3 uncorrected Fanca−/− iPSC lines was found to have a significant mosaic loss of entire chromosomes 4, 14, and 17 (supplemental Table 2). Two WT and 3 gene-corrected Fanca−/− (Fanca−/− and FANCA) samples revealed either no significant imbalances or minor subchromosomal imbalances of unclear significance. These data suggest that early complementation of the somatic cells may effectively protect FA iPSCs from chromosomal aberrations.
Because activation of the DDR and dsDNA breaks can lead to cell senescence, the percentage of cells staining positive for the senescence marker β-Gal was determined (Figure 2F). Reprogramming resulted in a modest, but not statistically significant, increase in senescence in WT TTFs. However, a significant increase in senescence of Fanca−/− TTFs was observed. Four days after reprogramming, the median percentage of senescent WT and Fanca−/− TTFs was 47% versus 62%, respectively (P < .01).
These data show that reprogramming resulted in the formation of FANCD2 foci and increased ROS. Fanca−/− TTFs displayed increased dsDNA breaks and chromosomal aberrations at baseline. Reprogramming significantly increased the frequency of dsDNA breaks and the level of senescence in Fanca−/− TTFs, suggesting that an impaired ability to repair reprogramming-induced DNA damage contributes to decreased reprogramming efficiency in FA cells.
Complementation rescues reprogramming of Fanca−/− fibroblasts
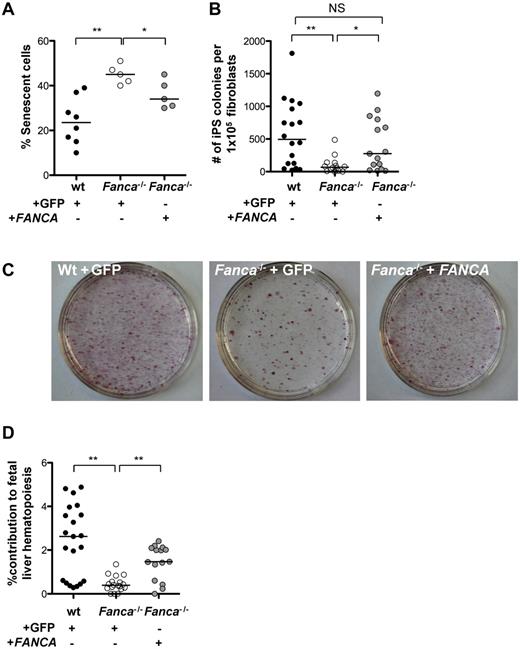
Given the increased sensitivity of FA cells to oxidative stress,5 the induction of ROS during reprogramming, and previous studies indicating enhanced reprogramming efficiency of WT cells in hypoxia,46 we hypothesized that hypoxia could enhance the reprogramming efficiency of Fanca−/− TTFs. We observed that the growth of WT fibroblasts was significantly increased at 5% O2, whereas Fanca−/− fibroblasts showed a trend toward increased growth at 5% O2 compared with 21% O2 (supplemental Figure 3A). Under hypoxic conditions, the reprogramming efficiency was 1.8- and 5.8-fold increased in WT and Fanca−/− cells, respectively (supplemental Figure 3B), suggesting a disease-associated advantage for reprogramming of FA cells under hypoxic conditions. Based on these results, subsequent experiments were carried out in 5% O2. To determine directly the role of the FA pathway in reprogramming, TTFs were transduced with retroviral vectors coexpressing FANCA and eGFP or encoding only eGFP as a control. Before reprogramming, GFP+ cells were prospectively isolated by FACS. Complementation of the Fanca−/− TTFs with FANCA resulted in a significant reduction of senescence 4 days after reprogramming (Figure 3A) and rescued the reprogramming efficiency of Fanca−/− TTFs to WT levels (Figure 3B-C). Whereas silencing of retroviral vectors occurs in ESCs and iPSCs,47 we were able to demonstrate the expression of FANCA in the complemented iPSC lines by immunoblotting, suggesting a possible selective pressure for continued correction of the FA pathway during the reprogramming process (supplemental Figure 3C). To assess whether murine FA iPSCs derived in this fashion can contribute to hematopoiesis in vivo, iPSCs derived from CD45.1+Fanca−/− animals were injected into CD45.2+ WT blastocysts. The reconstituted blastocysts were implanted into pseudopregnant females, and embryonic day 14.5 fetal livers were isolated and homogenized to assess the percentage of CD45.1+ (Fanca−/− iPSC–derived) hematopoietic cells (Figure 3D). Analysis of CD45.1 chimerism in individual fetal livers revealed a median contribution of iPSC-derived CD45.1+ cells of 2.6%, 0.4%, and 1.5% for WT, noncomplemented Fanca−/−, and gene-corrected Fanca−/− iPSCs, respectively (Figure 3D), demonstrating that gene-corrected Fanca−/− iPSCs can contribute to blood formation in vivo.
Complementation of Fanca−/− fibroblasts with FANCA rescues the reprogramming efficiency. (A) Percentage of senescent cells 4 days after reprogramming under hypoxic conditions (5% O2). (B) Frequency of iPSC colonies 16 days after reprogramming (5% O2). (C) Alkaline phosphatase–positive iPSC colonies on representative 10-cm tissue-culture dishes 16 days after reprogramming. (D) Percent contribution of CD45.1+Fanca−/− iPSC-derived cells to day 14.5 embryonic fetal liver hematopoietic cells. Shown is the percentage CD45.1+Fanca−/− cells of all CD45+ hematopoietic cells as ascertained by flow cytometry analysis of individual fetal livers. Filled circles, WT; open circles, Fanca−/− + GFP; gray circles, Fanca−/− + FANCA. Each symbol represents an individual animal. Horizontal lines represent the median. **P < .01; *P < .05; NS indicates not significant by Wilcoxon rank-sum test.
Complementation of Fanca−/− fibroblasts with FANCA rescues the reprogramming efficiency. (A) Percentage of senescent cells 4 days after reprogramming under hypoxic conditions (5% O2). (B) Frequency of iPSC colonies 16 days after reprogramming (5% O2). (C) Alkaline phosphatase–positive iPSC colonies on representative 10-cm tissue-culture dishes 16 days after reprogramming. (D) Percent contribution of CD45.1+Fanca−/− iPSC-derived cells to day 14.5 embryonic fetal liver hematopoietic cells. Shown is the percentage CD45.1+Fanca−/− cells of all CD45+ hematopoietic cells as ascertained by flow cytometry analysis of individual fetal livers. Filled circles, WT; open circles, Fanca−/− + GFP; gray circles, Fanca−/− + FANCA. Each symbol represents an individual animal. Horizontal lines represent the median. **P < .01; *P < .05; NS indicates not significant by Wilcoxon rank-sum test.
Derivation and hematopoietic differentiation of patient-specific FA iPSCs
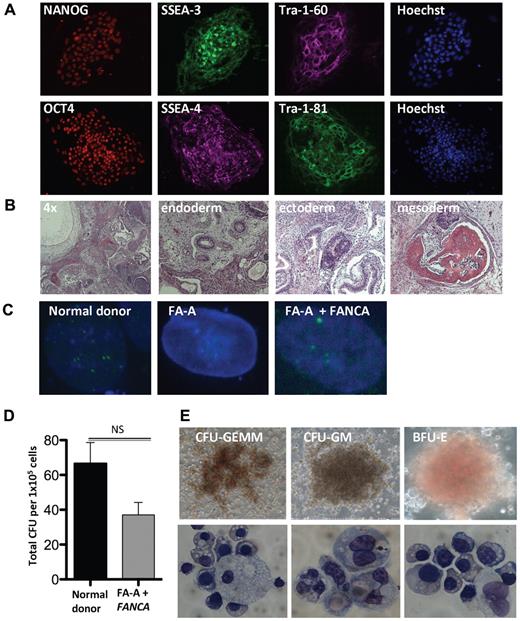
To extend our findings, we attempted to reprogram 10 unique human FA primary fibroblasts of the FA-A, FA-C, FA-G, and FA-D2 complementation groups. In contrast to the complete reprogramming failure of human FA cells reported by Raya et al,18 we derived 3 FA-A and FA-C iPSC lines from 2 individual patients without complementation under hypoxic conditions (Table 1). In agreement with our data in murine FA knockout TTFs, complementation of 3 unique FA patient samples significantly enhanced the reprogramming efficiency. Normal donor and patient-specific FA iPSC lines were examined and expressed protein markers of pluripotency, including NANOG, SSEA-3 and SSEA-4, Tra-1-60, OCT4, and Tra-1-81 (Figure 4A and supplemental Figure 4A). Intramuscular injection of these lines into SCID mice resulted in the formation of teratomas displaying derivatives of the endoderm, mesoderm, and ectoderm germ layers (Figure 4B). To determine whether the derivation of iPSCs from FA patients under these conditions yielded lines that could be therapeutically useful, karyotype analysis was performed on 3 normal donor and complemented FA-A iPSC lines. These 6 iPSC lines revealed a normal 46, XY karyotype (supplemental Figure 4B-C and supplemental Table 3). Treatment with hydroxyurea (HU) results in activation of the FA pathway in normal human cells, leading to the formation of FANCD2 foci. Whereas FA cells are unable to form FANCD2 foci, this phenotype can be rescued by complementation, indicating restoration of FA pathway function. We confirmed the presence or absence of FANCD2 foci in HU-treated normal donor and FA patient iPSCs (Figure 4C). Complemented FA-A patient iPSCs treated with HU maintained the ability to form FANCD2 foci, indicating that the FA pathway remained functional in these cells. The complete lack of D2 foci in uncorrected FA patient iPSCs confirmed that the FA pathway remained defective and ruled out mosaicism or reversion of a pathogenic allele to WT48 as a mechanism of successful reprogramming. While not showing any evidence of replicative crisis, uncorrected disease-specific lines appear to have slightly delayed growth kinetics compared with normal donor and complemented FA iPSC lines (data not shown).
Reprogramming of human Fanconi anemia cells is impaired
| Sample no. . | Complementation group* . | Vector . | Efficiency % (no. iPSC clones) . | Oxygen tension† . |
|---|---|---|---|---|
| 1 | Normal donor | Lenti | 0.01 (12) | 5 |
| 2 | FA-A | Lenti | 0.001 (2) | 5 |
| 2 | FA-A + FANCA | Lenti | 0.003 (6) | 5 |
| 3 | FA-A | Lenti | 0 | 5 |
| 3 | FA-A + FANCA | Lenti | 0.003 (3) | 5 |
| 4 | FA-C | Lenti | 0.001(1) | 5 |
| 4 | FA-C + FANCC | Lenti | 0.002 (2) | 5 |
| 5-7 | FA-A | Retro | 0 | 21 |
| 8,9 | FA-C | Retro | 0 | 21 |
| 10 | FA-G | Retro | 0 | 21 |
| 11 | FA-D2 | Retro | 0 | 21 |
| Sample no. . | Complementation group* . | Vector . | Efficiency % (no. iPSC clones) . | Oxygen tension† . |
|---|---|---|---|---|
| 1 | Normal donor | Lenti | 0.01 (12) | 5 |
| 2 | FA-A | Lenti | 0.001 (2) | 5 |
| 2 | FA-A + FANCA | Lenti | 0.003 (6) | 5 |
| 3 | FA-A | Lenti | 0 | 5 |
| 3 | FA-A + FANCA | Lenti | 0.003 (3) | 5 |
| 4 | FA-C | Lenti | 0.001(1) | 5 |
| 4 | FA-C + FANCC | Lenti | 0.002 (2) | 5 |
| 5-7 | FA-A | Retro | 0 | 21 |
| 8,9 | FA-C | Retro | 0 | 21 |
| 10 | FA-G | Retro | 0 | 21 |
| 11 | FA-D2 | Retro | 0 | 21 |
Fibroblasts (1 × 105) were reprogrammed. The number of independent reprogramming experiments, type of reprogramming vector used, and oxygen tension are shown. Efficiency is expressed as the number of iPSC clones derived per number of input fibroblasts.
Plus or minus genetic correction.
Precent O2.
Derivation and hematopoietic differentiation of patient-specific FA iPSCs. (A) Immunofluorescence of complemented patient-derived FA-A iPSCs. (B) Representative H&E stains of teratomas obtained by intramuscular injection of a complemented FA-A iPSC clone (4× and 20× magnification). (C) FANCD2 foci formation of normal donor (ND) or patient-specific complemented FA iPSC lines after exposure to vehicle or 2mM HU for 16 hours (60× magnification). (D) Number of CFUs per 1 × 105 normal donor and complemented patient-derived FA-A input cells (n = 3 independent clones, mean ± SD). (E) Photomicrographs depicting CFUs derived from patient-specific FA-A iPSCs and cytology of erythroid precursor cells, macrophages, and granulocytes (20× and 60× magnification). Images were obtained from FA-A iPSCs derived from patient sample number 2. GEMM indicates granulocyte/erythroid/macrophage/megakaryocyte; GM, granulocyte/macrophage; and BFU-E, burst-forming unit-erythroid.
Derivation and hematopoietic differentiation of patient-specific FA iPSCs. (A) Immunofluorescence of complemented patient-derived FA-A iPSCs. (B) Representative H&E stains of teratomas obtained by intramuscular injection of a complemented FA-A iPSC clone (4× and 20× magnification). (C) FANCD2 foci formation of normal donor (ND) or patient-specific complemented FA iPSC lines after exposure to vehicle or 2mM HU for 16 hours (60× magnification). (D) Number of CFUs per 1 × 105 normal donor and complemented patient-derived FA-A input cells (n = 3 independent clones, mean ± SD). (E) Photomicrographs depicting CFUs derived from patient-specific FA-A iPSCs and cytology of erythroid precursor cells, macrophages, and granulocytes (20× and 60× magnification). Images were obtained from FA-A iPSCs derived from patient sample number 2. GEMM indicates granulocyte/erythroid/macrophage/megakaryocyte; GM, granulocyte/macrophage; and BFU-E, burst-forming unit-erythroid.
To effect hematopoietic differentiation, fully characterized normal donor and patient-specific FA iPSC lines were allowed to aggregate to embryoid bodies as described previously.38 After 15 days of culture, the embryoid bodies were disaggregated and seeded in hematopoietic growth factor–containing semisolid methylcellulose. The complemented FA disease–specific iPSC lines showed a robust multilineage hematopoietic differentiation potential, resulting in erythroid and myeloid hematopoietic CFUs to a similar degree compared with normal donor controls (Figure 4D-E).
Discussion
Translation of FA somatic gene therapy into the clinical setting has thus far failed to result in measurable efficacy, primarily because of the markedly reduced numbers of CD34+ cells available for gene correction and autologous reinfusion.12,13 Disease-specific FA iPSCs provide an expandable source of autologous stem cells and therefore hold significant promise for the development of cellular therapies and disease models for genetic BM failure diseases. Raya et al recently reported a failure of 4 FA-A and 2 FA-D2 patient samples to undergo direct reprogramming. FA iPSCs could only be derived from 3 patient samples if the fibroblasts were first complemented by retroviral transduction before reprogramming, suggesting that restoration of the FA pathway is a prerequisite for iPSC generation from the somatic cells of FA patients.18
We sought to further elucidate the putative function of the FA pathway during reprogramming and pluripotent stem cell maintenance to identify strategies for overcoming the decreased reprogramming efficiency of somatic FA cells. Given the defective DNA repair and genomic instability of FA cells, we reasoned that FA somatic cells may undergo reprogramming less efficiently as a result of increased cellular and genomic damage accrued before or during reprogramming. We took advantage of 2 knockout mouse strains to evaluate the putative reprogramming block of FA cells in a reproducible and quantitative fashion. We observed that the reprogramming efficiency of Fanca−/− and Fancc−/− TTFs was significantly reduced compared with isogenic, age-matched WT controls, thus providing a model system for studying mechanisms of reprogramming resistance. Whereas both knockout mouse strains failed to express a functional FA core complex protein (FANCA and FANCC, respectively), the reprogramming efficiency of Fanca−/− TTFs was reduced more severely than that of Fancc−/− fibroblasts, and was correlated with the severity of the engraftment defect in these mice. Nonetheless, FA-knockout iPSCs were fully pluripotent and could be maintained in culture for more than 20 passages without any apparent replicative crisis.
Activation of the FA pathway results in FA core complex–mediated monoubiquitination of FANCD2, causing FANCD2 to localize to chromatin foci, which contain repair factors including Rad51, BRACA1, BRACA2, NBS1, PCNA, and γH2AX. Cells deficient in any 1 of the 8 known FA core complex proteins are unable to form FANCD2 chromatin foci.39 We observed that reprogramming, but not transduction with a control eGFP-encoding retroviral vector, resulted in the formation of FANCD2 foci, demonstrating that the FA pathway is activated during reprogramming of normal somatic cells.
Reprogramming has been shown to result in the up-regulation of p53, causing dsDNA breaks and inducing senescence.24,27 MEFs harboring increased DNA damage as a consequence of telomerase (Terc) or ataxia telangiectasia mutated (ATM) deficiency, undergo reprogramming with a significantly decreased efficiency.25 Reprogramming induced similar levels of p53-p21 and p19 in both WT and Fanca−/− fibroblasts at an early time point 4 days after reprogramming. This suggests that activation of the p53 program is not a key mediator of the FA reprogramming resistance, even though we cannot rule out that p53 may become hyperactivated in FA cells at a later time point. Fanca−/− fibroblasts contained increased dsDNA and chromosome breaks, raising the possibility that Fanca−/− cells that exceed a critical threshold of DNA and genomic damage are eliminated during the reprogramming process, possibly through senescence. Fanca−/− iPSCs did not contain chromosome breaks and fulfilled stringent pluripotency criteria. We did, however, observe significant chromosomal aberrations that occurred in uncorrected Fanca−/− iPSC lines, suggesting that FA pathway complementation of somatic cells promotes genome stability during reprogramming.
To determine the significance of the FA pathway in reprogramming directly, we undertook complementation of Fanca−/− early in the culture of primary fibroblasts. Complementation reduced senescence and rescued the reprogramming efficiency of Fanca−/− fibroblasts to WT levels. CGH analysis also revealed the lack of significant chromosomal imbalances in 3 complemented Fanca−/− iPSCs obtained in this fashion. Additional analysis of FA iPSCs with regard to chromosomal damage will be needed to confirm that complementation significantly reduces genomic alterations associated with reprogramming of FA somatic cells. Hypoxia has been demonstrated to increase the reprogramming efficiency of murine and human somatic cells.46 We noted a disease-associated advantage of hypoxic tissue-culture conditions, resulting in a 1.8- and 5.8-fold increase in the reprogramming efficiency of WT and Fanca−/− TTFs, respectively. Whereas the implications of hypoxia in reprogramming of human FA patient cells warrants further study, it is likely that the combination of hypoxic tissue culture conditions and early complementation prevents oxidative cellular damage in FA cells,49 thus facilitating direct reprogramming.
Our results demonstrate that somatic cells harboring mutations that render the FA pathway defective are resistant but not refractory to reprogramming. Complementation increases the reprogramming efficiency of murine and human FA-deficient somatic cells. The complemented disease-specific iPSC lines obtained in this fashion showed a normal, 46 XY karyotype; retained the ability to form FANCD2 foci, indicating the persistence of FA pathway correction; and were capable of yielding multilineage myeloid and erythroid hematopoietic progenitors in vitro (human iPSCs) and contributing to embryonic day 14.5 fetal liver hematopoietic chimerism in vivo (murine iPSCs). The inability of human iPSCs to yield transplantable hematopoietic progenitors, in combination with concerns about viral integration sites in iPSCs and teratoma formation in vivo, currently precludes the clinical translation of this technology.50 However, disease-specific iPSCs are of significant value for in vitro disease modeling.15-17 As the field progresses to overcome these barriers, iPSCs may in the future provide an important source of autologous stem cells for regenerative medicine approaches to acquired and inherited BM failure.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the Dana-Farber/Harvard Cancer Center Rodent Histopathology Core for assisting with teratoma histology and the Cytogenetics Core for carrying out the CGH analysis, and the Children's Hospital Boston Intellectual and Developmental Disabilities Research Center Imaging Core (P30HD 18655) for assisting with photomicrographs of the FAND2 foci. They would also like to thank Dr Yuko Fujiwara (Mouse Embryonic Stem Cells and Gene Targeting Core, Children's Hospital Boston; NKDDK Hematology center grant P30 DK049216-17) for assistance with the generation of chimeric mice.
This work was supported by a fellowship grant from the St Baldrick's Foundation (to L.M.), by the Harvard Stem Cell Institute (to L.M.), by the Hood Foundation (to L.M.), and by the National Institutes of Health (NIH R01 HL081499 and DK074310 to D.A.W. and NIH 1RC4DK090913-01 to D.A.W., A.D., and G.Q.D.).
National Institutes of Health
Authorship
Contribution: L.U.W.M. and M.D.M. designed and executed the experiments and wrote the manuscript; C.E.H., R.V., K.M.B., and L.A.M. provided technical assistance; K.P. provided advice and assistance pertaining to DNA damage response assays; A.S. provided the polycistronic lentiviral reprogramming vector; I.-H.P., T.S., and A.L.D. performed the human reprogramming experiments; E.G. provided human FA samples; W.B.L. and K.S. performed the statistical analyses; and A.D., G.Q.D., and D.A.W. designed the experiments and participated in writing the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for M.D.M. is Division of Stem Cells and Cancer, HI-STEM (Heidelberg Institute for Stem Cell Technology and Experimental Medicine), German Cancer Research Center, Heidelberg, Germany. The current affiliation for I.-H.P. is Yale Stem Cell Center, Department of Genetics, Yale School of Medicine, New Haven, CT.
Correspondence: David A. Williams, MD, Children's Hospital Boston, 300 Longwood Ave, Karp 08125.3, Boston, MA 02115; e-mail: dawilliams@childrens.harvard.edu.
References
Author notes
L.U.W.M. and M.D.M. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal