Key Points
The ERG stem cell enhancer is active in acute myeloid leukemia and is regulated by a heptad of transcription factors.
Expression signatures derived from ERG promoter–enhancer activity and heptad expression are associated with clinical outcome.
Abstract
Aberrant transcriptional programs in combination with abnormal proliferative signaling drive leukemic transformation. These programs operate in normal hematopoiesis where they are involved in hematopoietic stem cell (HSC) proliferation and maintenance. Ets Related Gene (ERG) is a component of normal and leukemic stem cell signatures and high ERG expression is a risk factor for poor prognosis in acute myeloid leukemia (AML). However, mechanisms that underlie ERG expression in AML and how its expression relates to leukemic stemness are unknown. We report that ERG expression in AML is associated with activity of the ERG promoters and +85 stem cell enhancer and a heptad of transcription factors that combinatorially regulate genes in HSCs. Gene expression signatures derived from ERG promoter–stem cell enhancer and heptad activity are associated with clinical outcome when ERG expression alone fails. We also show that the heptad signature is associated with AMLs that lack somatic mutations in NPM1 and confers an adverse prognosis when associated with FLT3 mutations. Taken together, these results suggest that transcriptional regulators cooperate to establish or maintain primitive stem cell–like signatures in leukemic cells and that the underlying pattern of somatic mutations contributes to the development of these signatures and modulate their influence on clinical outcome.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous disease with respect to cellular morphology, immunophenotype, cytogenetics, and clinical outcome.1 Patients with normal cytogenetics (CN-AML), a subgroup that accounts for approximately 40% of this neoplasm, have a 5-year survival rate. This varies from 24% to 42%, suggesting a genetic diversity even within this subgroup of patients.2 Gene expression signatures derived from normal cord blood and AML cell fractions that have the capacity to engraft nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice have recently been shown to correlate with poor overall survival (OS).3 The implication is that AML cells that possess a leukemic “stem cell” signature are prone to renew, persist, and evade chemotherapy. What is not clear is what drives the generation and maintenance of these stem cell signatures and the degree of similarity to transcriptional mechanisms that regulate these signatures in normal hematopoietic stem and progenitor cells (HSPCs).
We have shown that the expression of transcriptional regulators in T-cell acute lymphoblastic leukemia (T-ALL) cells is maintained by self-sustaining transcriptional circuits made up of cell-type–specific cis-regulatory elements within their loci and the gene products themselves.4,5 However, the role of stem cell enhancers in regulating gene expression in AML has not, to our knowledge, been explored in detail. Given that stem cell gene expression signatures influence the clinical outcome of AML,3 it is important to understand how these signatures are established and what relationship they have to somatic mutations in individual AMLs. High expression of the E-twenty six (ETS) transcription factor (TF) Ets Related Gene (ERG) has been widely reported as an independent predictor of poor outcome in cytogenetically normal AML (CN-AML)6 and T-ALL.7 ERG is also a member of a leukemia stem cell (LSC)/hematopoietic stem cell (HSC) signature this has been recently reported to influence clinical outcome in CN-AML3 and constitutes a powerful oncogene both in solid organ and hematological malignancies. Although the ERG locus can be rearranged and fused with EWS in Ewing sarcoma,8 TMPRSS2 in prostate cancer,9 and FUS/TLS in AML,10 high ERG expression in T-ALL and AML is most commonly found in the absence of a mutation involving its locus. Therefore, identifying aberrant mechanisms of ERG overexpression will further inform leukemia biology and highlight targets for therapeutic intervention. We recently showed that high ERG expression in T-ALL occurs in the context of SCL, LMO2, LYL1, FLI1, ERG, and GATA3 binding to a specific enhancer within the ERG locus, +85kb downstream of the translation start site.5 During normal hematopoiesis, this enhancer selectively targets myeloid stem/progenitors and immature thymocytes.5 Many of the factors that regulate this enhancer in T-ALL are components of a heptad of TFs (SCL, LYL1, LMO2, GATA2, RUNX1, FLI1 and ERG) that act combinatorially to regulate genes in HSPCs.11
Taking into account the role of ERG as a transcriptional regulator in HSPCs and leukemia and the activity of the ERG +85 stem enhancer in both normal and lymphoid leukemic stem cells, we hypothesized that it would serve as a useful candidate gene and enhancer to probe the relationships between stem cell enhancers, stem cell TFs, and the stem cell signature in AML cells. Our findings demonstrate that gene expression signatures derived from stem cell enhancer activity and heptad expression in AML are independently associated with OS and that heptad expression predicts NPM1 mutation status in CN-AML and confers an adverse prognosis when associated with FLT3 mutations.
Materials and methods
Patient and control samples
Peripheral blood (PB) or bone marrow (BM) samples were collected from patients with AML with ≥80% blasts. CD34+ control cells were obtained from patients undergoing chemotherapy and granulocyte colony-stimulating factor-stimulated apheresis for stem cell harvesting for myeloma or lymphoma. T cells and neutrophils were obtained from PB of healthy donors. Informed consent was obtained in accordance with the Declaration of Helsinki and local ethical guidelines. See supplemental Experimental Procedures for more details.
Chromatin immunoprecipitation
Quantification of ERG transcripts
Specific primers for quantitative reverse-transcription polymerase chain reaction (qRT-PCR) that amplified transcripts arising from only the distal promoter (DP) or proximal promoter (PP) were designed along with a primer set that amplified total ERG transcripts.5 ,12 The amplicons of each primer set were then subcloned into pcDNA3 (Invitrogen, Carlsbad, CA) to create a plasmid standard control containing a single copy of each ERG amplicon, thus allowing direct comparison between ERG primer sets. See supplemental Experimental Procedures for more details.
Cell culture and transfection assays
K562, KG-1, and ME-1 leukemic cell lines were maintained as specified by the distributors.
Transfection of AML cells was performed in quadruplicate with electroporation using standard conditions and repeated at least once; see supplemental Experimental Procedures for more details.
Transgenic and transplantation assays
Lin–/Sca1+/c-Kit+ (LSK) cells were sorted from L83735 CD45.2 BM using a fluorescence-activated cell sorter and split into either Venus-positive or -negative fractions. Sublethal irradiation at 2 × 450 rads was performed on recipient CD45.1/45.2 F1 mice. The LSK cells for transplantation were mixed with 2 × 105 spleen cells and transplanted through tail vein injections. Two rounds of transplantations were performed; see supplemental Experimental Procedures for details.
Cluster analysis of patients based on histone marks
The probe intensities for each region and patient were summed and normalized against the intensities across the whole ERG locus. The values were further scaled, between zero and 1, to allow for quantitative comparison across patients and loci. Patients were clustered using hierarchical and k-means unsupervised clustering algorithms into 4 groups; see supplemental Experimental Procedures for more details.
Principal component analysis
Genome-wide gene expression was acquired for 25 of the 26 AML patients (array data were missing for AML patient 26) and 5 CD34+ HSPCs using microarrays. Principal component analysis (PCA) was performed on whole array data. The 4 clusters derived earlier were applied, and the mean of the first 2 principle components (PCs) plotted; the point sizes were chosen proportional to the within-cluster variance. The most influential probes (100 probe sets, representing 84 unique genes) from the first PC were used as the promoter–stem cell enhancer signature (P-SCE-sig). Gene expression datasets from the 25 AML and 5 CD34+ HSPCs were merged with publicly available data; see supplemental Experimental Procedures for more details.
Survival analysis
In the first survival analysis, patients were split into 2 groups using the k-means optimization criteria on the expression of P-SCE-genes. OS and event-free survival (EFS) were analyzed in both groups using Kaplan-Meier statistics. In order to extrapolate survival onto the 4 H3K9 and H3K27 clusters, we applied a pattern matching strategy that associates each of the 161 and 156 patients with 1 or other of our 4 clusters. See supplemental Experimental Procedures for more details.
Pathway analysis
The core analysis tool within the Ingenuity Pathway Analysis suite (IPA, version 12402621; Ingenuity Systems, Redwood City, CA) was used to identify canonical pathways that are significantly overrepresented. To rank significance of pathways, the member genes of each pathway were extracted from IPA, gene expression levels for patients in G1 and G2 were retrieved, and the expression of all genes was compared between the 2 groups using multivariate analysis of variance; see supplemental Experimental Procedures for more details.
Results
Erg +85 hematopoietic enhancer targets HSCs
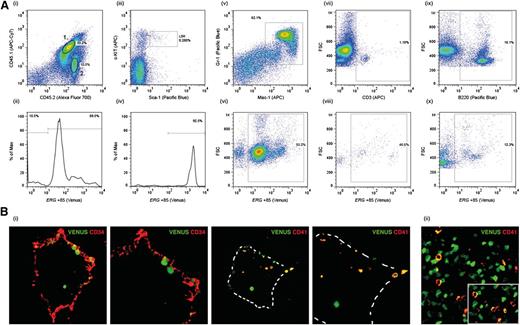
Unlike transgenic mice carrying the well-characterized Scl +19 HSC enhancer, which is expressed throughout embryogenesis but not in the adult (SV/lac/19, L58813 ), SV/Venus/Erg +85 mice (L8373) express the transgene both during embryonic development and in the adult.5 However, even in this line there is some position effect variegation, with a proportion of L8373 mice silencing the transgene in the adult (data not shown). Using L8373 mice, we previously showed that the Erg +85 enhancer is active in fetal liver progenitors and in LSK cells in adult BM.5 To establish whether Venushigh LSK cells in the BM of adult transgenic mice include HSCs, we transplanted Venushigh LSK (CD45.2) cells into sublethally irradiated CD45.1/45.2 recipients. At 8 weeks and 14 weeks post-transplant, donor-derived Venus-positive cells were detectable in 10/12 recipients (Figure 1A). The majority (>90%) of donor-derived BM LSK cells in the recipient as well as donor-derived myeloid and B- and T-lymphocytes expressed the Venus reporter at intensities proportionate to that reported previously5 (Figure 1A). Taken together, these data show that the Erg +85 enhancer targets HSCs that contribute to sustained hematopoiesis in recipient mice. The Erg +85 enhancer is also active at sites of HSC emergence, including CD41+ cells in the dorsal aorta at embryonic day (E) 10.5, which include the earliest definitive HSCs in the developing embryo14 (Figure 1B).
ERG +85 enhancer targets HSCs. (A) CD45.1/45.2 mice were transplanted with Venushigh LSK cells from CD45.2 SV40/Erg +85/Venus transgenic mice. BM cells stained for CD45.1 and CD45.2 at 14 weeks post-transplant show a mix of donor-derived (2) and recipient (1) cells. Donor-derived CD45.2 BM cells continue to express the Venus reporter 14 weeks post-transplant (i-ii). The Erg +85 enhancer remains highly active (ie, high Venus expression) in donor-derived LSK cells 14 weeks post-transplant (iii-iv). Donor-derived Mac1+/Gr1+ myeloid cells in the recipient continue to express the Venus reporter 14 weeks post-transplant (v-vi). A proportion of donor-derived CD3+ T cells in the recipient continue to express the Venus reporter 14 weeks post-transplant (vii-viii). Donor derived B220+ B cells in the recipient continue to express the Venus reporter 14 weeks post-transplant (ix-x). (B) Transverse cryosections through the aorta-gonad-mesonephros of E10.5 SV40/Erg +85/Venus transgenic embryos stained with CD34 (endothelial, HSC) and CD41 (an early marker of definitive hematopoiesis). Venus-expressing cells in the dorsal aorta express CD41 (i). A cryosection through the fetal liver of an E11.5 SV40/Erg +85/Venus transgenic embryo. CD41-expressing fetal liver cells also express the Venus reporter (ii).
ERG +85 enhancer targets HSCs. (A) CD45.1/45.2 mice were transplanted with Venushigh LSK cells from CD45.2 SV40/Erg +85/Venus transgenic mice. BM cells stained for CD45.1 and CD45.2 at 14 weeks post-transplant show a mix of donor-derived (2) and recipient (1) cells. Donor-derived CD45.2 BM cells continue to express the Venus reporter 14 weeks post-transplant (i-ii). The Erg +85 enhancer remains highly active (ie, high Venus expression) in donor-derived LSK cells 14 weeks post-transplant (iii-iv). Donor-derived Mac1+/Gr1+ myeloid cells in the recipient continue to express the Venus reporter 14 weeks post-transplant (v-vi). A proportion of donor-derived CD3+ T cells in the recipient continue to express the Venus reporter 14 weeks post-transplant (vii-viii). Donor derived B220+ B cells in the recipient continue to express the Venus reporter 14 weeks post-transplant (ix-x). (B) Transverse cryosections through the aorta-gonad-mesonephros of E10.5 SV40/Erg +85/Venus transgenic embryos stained with CD34 (endothelial, HSC) and CD41 (an early marker of definitive hematopoiesis). Venus-expressing cells in the dorsal aorta express CD41 (i). A cryosection through the fetal liver of an E11.5 SV40/Erg +85/Venus transgenic embryo. CD41-expressing fetal liver cells also express the Venus reporter (ii).
ERG expression in AML patients is proportional to the degree of H3K9/K14Ac enrichment at the ERG promoters and +85 stem cell enhancer
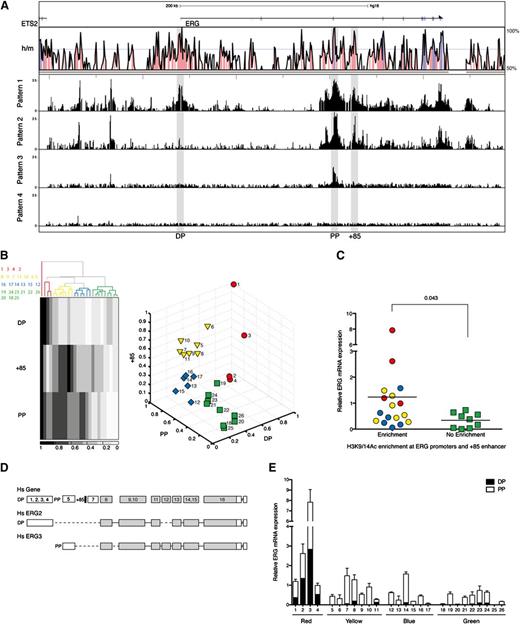
The transcriptional control of ERG expression in AML is poorly understood. To survey the chromatin accessibility profiles across the ERG locus in primary AML cells, we performed ChIP-chip using H3K9/K14Ac ChIP material from a panel of 26 AML samples hybridized to high-density Agilent custom arrays with oligomers tiled across the ERG locus to its flanking genes (Figure 2A; see supplemental Table 1 for patient characteristics and supplemental Figure 1 for individual H3K9/K14Ac ChIP-chip profiles). The human ERG gene has 2 recognized promoters separated by ∼165 kb.5 Unlike in T-ALL cells, H3K9/K14Ac enrichments at the promoters and +85 stem cell enhancer were not uniform in AML cells and broadly fell into the following 4 visually recognizable patterns: pattern 1, enrichment at both PP and DP and the +85 enhancer; pattern 2, enrichment only at PP and the +85 enhancer; pattern 3, enrichment only at PP; and pattern 4, little or no enrichment at any of these regulatory elements.
ERG expression in AML patients is proportional to the degree of H3K9/14Ac enrichment at the ERG promoters and +85 stem cell enhancer. (A) H3K9/14Ac enrichment across the ERG locus was measured in 26 AML samples by ChIP-chip. The enrichment profiles broadly fell into 4 visually recognizable patterns. Representative profiles of each pattern are shown below the Vista human/mouse sequence conservation (> 50%) plot. See supplemental Figure 1 for enrichment profiles in individual patients. (B) To group AML patients based on this epigenetic mark, 2 unsupervised clustering algorithms were used in conjunction with normalized H3K9/14Ac enrichments at the promoters and +85 stem cell enhancer. The panel to the left shows the hierarchical clustering pattern of patients together with a heat map representation of H3K9/14Ac enrichments. Sample IDs are listed to the left of the heat map and correspond to columns from left to right. The panel to the right shows the output of k-means clustering. The identity of patient samples in each group was the same irrespective of the clustering algorithm. (C) ERG transcripts in AML samples were measured by qRT-PCR and correlated with H3K9/K14Ac enrichment at the ERG promoter and +85 enhancer (right) using Mann-Whitney U test for statistical analysis. (D) Schematic diagram of the ERG locus (not to scale) showing the composite exon-intron structure of human ERG (upper). The translated exons are colored gray, and the positions of the promoter/enhancer elements relative to exons are indicated (the promoters as DP and PP and the +85 enhancer as a black bar). Exon usage of 2 transcripts originating from the distal promoter (Hs ERG2) and proximal promoter (Hs ERG3) are shown. (E) ERG transcripts originating from either the distal or proximal promoter were quantified. Their abundance is shown relative to total ERG in each AML sample.
ERG expression in AML patients is proportional to the degree of H3K9/14Ac enrichment at the ERG promoters and +85 stem cell enhancer. (A) H3K9/14Ac enrichment across the ERG locus was measured in 26 AML samples by ChIP-chip. The enrichment profiles broadly fell into 4 visually recognizable patterns. Representative profiles of each pattern are shown below the Vista human/mouse sequence conservation (> 50%) plot. See supplemental Figure 1 for enrichment profiles in individual patients. (B) To group AML patients based on this epigenetic mark, 2 unsupervised clustering algorithms were used in conjunction with normalized H3K9/14Ac enrichments at the promoters and +85 stem cell enhancer. The panel to the left shows the hierarchical clustering pattern of patients together with a heat map representation of H3K9/14Ac enrichments. Sample IDs are listed to the left of the heat map and correspond to columns from left to right. The panel to the right shows the output of k-means clustering. The identity of patient samples in each group was the same irrespective of the clustering algorithm. (C) ERG transcripts in AML samples were measured by qRT-PCR and correlated with H3K9/K14Ac enrichment at the ERG promoter and +85 enhancer (right) using Mann-Whitney U test for statistical analysis. (D) Schematic diagram of the ERG locus (not to scale) showing the composite exon-intron structure of human ERG (upper). The translated exons are colored gray, and the positions of the promoter/enhancer elements relative to exons are indicated (the promoters as DP and PP and the +85 enhancer as a black bar). Exon usage of 2 transcripts originating from the distal promoter (Hs ERG2) and proximal promoter (Hs ERG3) are shown. (E) ERG transcripts originating from either the distal or proximal promoter were quantified. Their abundance is shown relative to total ERG in each AML sample.
To evaluate whether H3K9/K14Ac enrichment at these elements is associated with ERG expression in AML, we applied 2 unsupervised clustering algorithms to group patients using normalized relative enrichment at DP, PP, and +85 elements. Hierarchical clustering of the 26 samples yielded 4 groups (Figure 2B). When these samples were divided into 4 groups by k-means clustering, the identities of individual samples that clustered into red, yellow, blue, and green groups were the same across both methods. Total ERG levels were measured by qRT-PCR and normalized to 14-3-3 protein ζ/δ in each AML sample (Figure 2C). Samples with relative H3K9/K14Ac enrichment at the ERG promoters and +85 enhancer had higher ERG levels than samples with no enhancer activity. ERG has multiple isoforms that broadly originate from either the PP or DP (Figure 2D). To assess whether active histone marks at the promoter correlated with types of transcripts originating from either the PP or DP, we quantified abundance of each using a plasmid standard control containing a single copy of both DP and PP amplicons. The distribution of transcripts originating from either the DP or PP in each AML corresponded well with the degree of H3K9/K14Ac enrichment at each promoter (Figure 2E). Taken together, these data underscore the association between access at ERG regulatory elements and the type and abundance of transcripts in AML.
ERG +85 stem enhancer is active in AML cells and its activity is dependent on conserved ETS/GATA/E-BOX TF binding sites
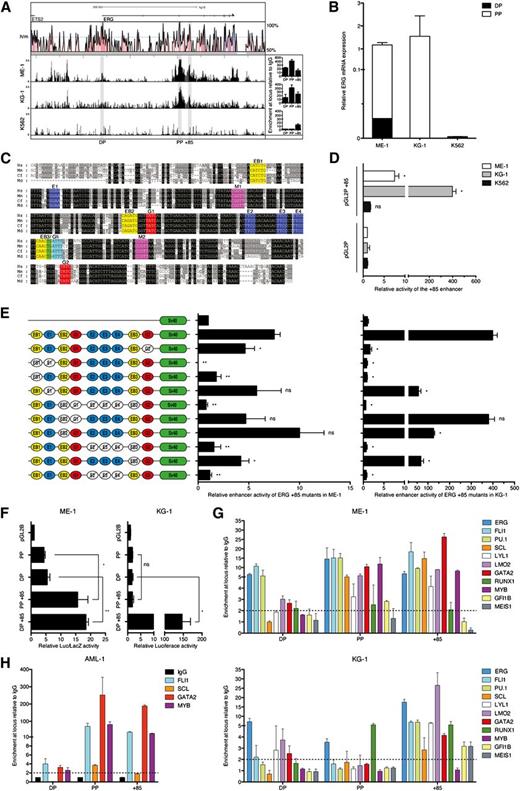
To test activity of the enhancer in AML cells, we first screened a panel of AML cell lines for ERG expression and H3K9/K14Ac ChIP-chip profiles. We selected ME-1 and KG-1 as appropriate ERG expressors and K562 as an appropriate negative control. ME-1 and KG-1 cells have active histone marks at both PP and the +85 stem cell enhancer, with ME-1 also showing modest H3K9/K14Ac enrichment at DP (Figure 3A). Therefore, they are representative of patterns 1 and 2, respectively. In line with these ChIP-chip profiles, most ERG transcripts in ME-1 and KG-1 originate from PP, with some transcripts in ME-1 also originating from DP (Figure 3B). The ERG+85 region has blocks of sequences that are highly conserved from human to opossum (Figure 3C). In accordance with endogenous ERG expression, the ERG +85 stem cell enhancer is active in ERG-expressing ME-1 and KG-1 cells but not in nonexpressing K562 cells (Figure 3D).
ERG +85 stem cell enhancer is regulated by HSC TFs in AML cells. (A) H3K9/14Ac ChIP-chip profiles across the ERG locus are shown for 2 high (ME-1 and KG-1) and 1 low (K562) ERG-expressing AML cell lines. qRT-PCR confirmation of enrichments (from an independent ChIP experiment) is shown to the right of each profile. (B) Abundance of ERG transcripts originating from either the distal or proximal promoter is shown relative to total ERG expression in each AML cell line. (C) The +85 enhancer has a number of highly conserved TF binding sites including E-box (EB, CANNTG, yellow), Ets (E, GGAW, blue), Myb (M, YAACNG, purple), Gata (G, GATA, red), and Gfi (AAATCA, cyan). (D) Stable (KG-1 and K562) and transient (ME-1) transfection assays show activity of the +85 enhancer in conjunction with a heterologous SV40 promoter. (E) Mutation of conserved TF binding sites in conjunction with transient (ME-1) and stable (KG-1) transfection assays in ERG-expressing AML cells shows the dependence of the +85 enhancer on specific binding sites for its activity in leukemic cells. (F) Activity of the endogenous ERG promoters in AML cells either alone or in conjunction with the +85 enhancer measured by transient (ME-1) and stable (KG-1) transfection assays. (G) ChIP-qPCR enrichment of HSC TFs at the +85 enhancer and promoters of ERG in ME-1 and KG-1 AML cells. (H) ChIP-qPCR enrichment of TFs at the ERG +85 stem cell enhancer and promoters in AML patient 1, a patient with H3K9/14Ac enrichment at both promoters and enhancer. *P < .05; **P < .01. NS, not significant.
ERG +85 stem cell enhancer is regulated by HSC TFs in AML cells. (A) H3K9/14Ac ChIP-chip profiles across the ERG locus are shown for 2 high (ME-1 and KG-1) and 1 low (K562) ERG-expressing AML cell lines. qRT-PCR confirmation of enrichments (from an independent ChIP experiment) is shown to the right of each profile. (B) Abundance of ERG transcripts originating from either the distal or proximal promoter is shown relative to total ERG expression in each AML cell line. (C) The +85 enhancer has a number of highly conserved TF binding sites including E-box (EB, CANNTG, yellow), Ets (E, GGAW, blue), Myb (M, YAACNG, purple), Gata (G, GATA, red), and Gfi (AAATCA, cyan). (D) Stable (KG-1 and K562) and transient (ME-1) transfection assays show activity of the +85 enhancer in conjunction with a heterologous SV40 promoter. (E) Mutation of conserved TF binding sites in conjunction with transient (ME-1) and stable (KG-1) transfection assays in ERG-expressing AML cells shows the dependence of the +85 enhancer on specific binding sites for its activity in leukemic cells. (F) Activity of the endogenous ERG promoters in AML cells either alone or in conjunction with the +85 enhancer measured by transient (ME-1) and stable (KG-1) transfection assays. (G) ChIP-qPCR enrichment of HSC TFs at the +85 enhancer and promoters of ERG in ME-1 and KG-1 AML cells. (H) ChIP-qPCR enrichment of TFs at the ERG +85 stem cell enhancer and promoters in AML patient 1, a patient with H3K9/14Ac enrichment at both promoters and enhancer. *P < .05; **P < .01. NS, not significant.
Previously we showed that activity of the ERG +85 stem cell enhancer in T-ALL cells is dependent on conserved ETS/GATA/E-BOX TF binding sites.5 The mutation series, which was previously evaluated in T-ALL cells, was tested in ME-1 and KG-1 AML cells by transient and stable transfection assays, respectively. Interestingly, many of the mutations that ablated enhancer activity in T-ALL cells showed a similar result in AML cells (Figure 3E). These included the 5′ conserved E-Box and ETS (EB1-E1) motif pair and the trio of ETS binding sites (E2/E3/E4). Taken together, these data show that the activity of the ERG +85 stem enhancer in AML cells is dependent on specific, highly conserved ETS, GATA, and E-BOX motifs. The endogenous promoters show only modest activity in the AML cell lines, but their activity is significantly boosted by the +85 enhancer (Figure 3F).
ERG +85 stem cell enhancer in AML cells is bound by HSC transcriptional regulators
Given the complementary activity of the ERG +85 stem cell enhancer in AML and T-ALL and its dependence on near identical TF binding motifs for their activity, we measured expression levels of the heptad genes and other regulators such as PU.1, MEIS1, GFI1B, and MYB, all of which have known roles in HSC and AML development (supplemental Figure 2). We then performed ChIP in ME-1 and KG-1 cells for enrichment at the ERG promoters and +85 stem cell enhancer (Figure 3G). The heptad TFs and PU.1 are enriched at the +85 enhancer in both cell lines. MYB, GFIB, and MEIS1 are enriched at the enhancer in one or the other cell line. There is variable enrichment of these TFs at the ERG promoters. Enrichment of a limited number of TFs was also assessed in AML patient 1 (Figure 3H). Taken together, transcriptional regulators that bind the ERG +85 stem enhancer in AML cells are those that bind and combinatorially regulate target genes in HSCs.
Epigenetic marks at the ERG locus are shared between normal CD34+ HSPCs and high ERG-expressing AMLs
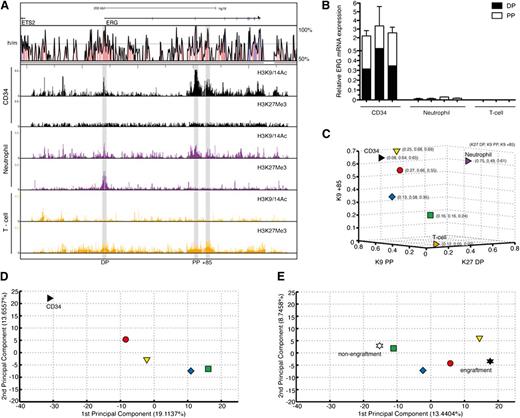
High ERG and CD34 mRNA expressions have both been reported to independently predict poor prognosis in AML.15 However, both in this patient group and in ours, ERG and CD34 expressions were correlated with Spearman correlation coefficients of 0.54 and 0.6, respectively (n = 26, P = .001; data not shown). To determine whether this correlation is due to high ERG-expressing AMLs sharing epigenetic marks at the ERG locus with normal CD34+ cells, either as a reflection of the cell of origin or a similarly perturbed epigenetic environment, we performed ChIP-chip for active (H3K9/K14Ac) and inactive (H3K27Me3) histone marks on highly purified CD34+ cells from 5 healthy donors. These marks were also assessed in neutrophils and CD3+ T cells (Figure 4A). CD34+ cells show active marks at both promoters and the +85 enhancer and no inactive marks. Neutrophils show bivalency at the DP and active marks at the PP and enhancer. T cells show no active marks and modest enrichment of inactive marks at these sites. These epigenetic marks functionally correlate with ERG transcript abundance and type (Figure 4B). To assess the epigenetic relationship between high and low ERG-expressing AML samples (in Figure 2) and these primary cells, normalized enrichment signals of selected histone marks at the ERG promoters and +85 stem enhancer in these cells were merged. Whereas the signal strengths of the high ERG-expressing red and yellow groups in particular were close to those of CD34+ cells, the low ERG-expressing green group was positioned closer to the mature cell types (Figure 4C and supplemental Figure 3).
Degree of stemness of the AML transcriptome is related to activity of the ERG +85 stem cell enhancer. (A) ChIP-chip profiles for active (H3K9/14Ac) and inactive (H3K27Me3) histone marks across the ERG locus are shown for normal CD34+ stem/progenitors, mature neutrophils, and CD3+ T cells. CD34+ cells show active marks at both promoters and the +85 enhancer and no inactive marks. Neutrophils show bivalency at the distal promoter and active marks at the proximal promoter and enhancer. T cells show no active marks and modest enrichment of inactive marks at these sites. (B) Abundance and types of ERG transcripts in each cell type shown in the previous panel. (C) A 3-dimensional display of normal CD34+ stem/progenitors and 26 AML samples plotted according to normalized levels of enrichment of specific histone marks at their ERG promoter/enhancer elements. Individual AML samples were clustered as before into 4 groups, with the arithmetic mean of each group plotted in the figure. Samples in the red and yellow groups in particular display more epigenetic similarity with normal CD34+ stem/progenitors compared with samples in the green group. (D) A PCA of global gene expression profiles of normal CD34+ stem/progenitors and 25 AML samples is shown. As before, the arithmetic mean of each group is plotted. The transcriptomes of AML samples in the red group that, along with the yellow group, shares the closest epigenetic identity (at the ERG locus) to normal CD34+ stem/progenitors also display the closest transcriptional identity. There is progressive loss of transcriptional similarity to CD34+ cells as the AMLs lose their epigenetic identity (at the ERG locus) with CD34+ cells. (E) A PCA of transcriptomes of NOD/SCID engrafting and nonengrafting AML fractions3 computed with the transcriptomes of our 25 AML samples is shown. As before, the arithmetic mean of each group is plotted. The AML samples that lack an active enhancer mark (green group) cluster closer to the nonengrafting AML fraction, whereas those samples with an active enhancer and transcriptome that is closer to normal CD34+ cells (red and yellow) cluster closer to engrafting AML fractions.
Degree of stemness of the AML transcriptome is related to activity of the ERG +85 stem cell enhancer. (A) ChIP-chip profiles for active (H3K9/14Ac) and inactive (H3K27Me3) histone marks across the ERG locus are shown for normal CD34+ stem/progenitors, mature neutrophils, and CD3+ T cells. CD34+ cells show active marks at both promoters and the +85 enhancer and no inactive marks. Neutrophils show bivalency at the distal promoter and active marks at the proximal promoter and enhancer. T cells show no active marks and modest enrichment of inactive marks at these sites. (B) Abundance and types of ERG transcripts in each cell type shown in the previous panel. (C) A 3-dimensional display of normal CD34+ stem/progenitors and 26 AML samples plotted according to normalized levels of enrichment of specific histone marks at their ERG promoter/enhancer elements. Individual AML samples were clustered as before into 4 groups, with the arithmetic mean of each group plotted in the figure. Samples in the red and yellow groups in particular display more epigenetic similarity with normal CD34+ stem/progenitors compared with samples in the green group. (D) A PCA of global gene expression profiles of normal CD34+ stem/progenitors and 25 AML samples is shown. As before, the arithmetic mean of each group is plotted. The transcriptomes of AML samples in the red group that, along with the yellow group, shares the closest epigenetic identity (at the ERG locus) to normal CD34+ stem/progenitors also display the closest transcriptional identity. There is progressive loss of transcriptional similarity to CD34+ cells as the AMLs lose their epigenetic identity (at the ERG locus) with CD34+ cells. (E) A PCA of transcriptomes of NOD/SCID engrafting and nonengrafting AML fractions3 computed with the transcriptomes of our 25 AML samples is shown. As before, the arithmetic mean of each group is plotted. The AML samples that lack an active enhancer mark (green group) cluster closer to the nonengrafting AML fraction, whereas those samples with an active enhancer and transcriptome that is closer to normal CD34+ cells (red and yellow) cluster closer to engrafting AML fractions.
AML cells with activity at the ERG promoters and +85 stem cell enhancer have a global gene expression signature that more closely resembles that of normal hematopoietic and leukemic stem cells
To explore whether ERG promoter and +85 stem cell enhancer accessibility in AML samples was informative for more than ERG expression, we performed a PCA of the combined transcriptomes of 25 AML samples and CD34+ HSPCs from 5 controls (Figure 4D). Significantly, the resemblance of the global AML transcriptome to that of CD34+ HSPCs progressively diminished as their epigenetic identity at the ERG locus diverged. In particular, samples in the green group, which lack activity at the ERG promoters and +85 stem cell enhancer, had the most divergent global expression profile to normal CD34+ HSPCs. This association was further clarified using an independent dataset of highly purified normal BM blood stem/progenitor cell fractions obtained from Gentles et al16 (supplemental Figure 4; supplemental Experimental Procedures).
The ability of sorted CD34+/–, CD38+/– AML cell fractions to initiate leukemia in NOD/SCID mice goes beyond surface expression of these stem/progenitor markers and relates more to a shared LSC signature.3 We performed a PCA overlapping the global gene expression profiles of our AML samples with those of NOD/SCID engrafting and nonengrafting AML cell fractions (CD34+/–, CD38+/–; Figure 4E). Significantly, the transcriptomes of the green group, which were most divergent from normal CD34+ HSPCs, were also farthest from the NOD/SCID–engrafting LSC transcriptomes and more closely related to those of the nonengrafting set. To validate the location of the data points in the PCA and to assess the significance of distances between groups, we performed a bootstrap analysis where we randomly assigned samples to different groups and quantified the distance between red/yellow/blue/green AML groups and NOD/SCID engraftment/nonengraftment groups. Observed distances between the red group and NOD/SCID engrafters and that of the green group and NOD/SCID nonengrafters were significantly smaller than the mean distances by bootstrap analysis (P < .01; see supplemental Experimental Procedures). This association was further clarified using an independent dataset of highly purified AML cells obtained from Gentles et al16 (supplemental Figure 5; supplemental Experimental Procedures).
The list of genes, which contributed to the first principal component in Figure 4D, was arranged in descending order of significance and a P-SCE-sig was constructed using the top 100 probes/84 genes (see supplemental Table 2, supplemental Figure 6, and supplemental Experimental Procedures). The P-SCE-sig was applied to the NOD/SCID–engrafting and –nonengrafting AML cell fractions and a second PCA was performed (supplemental Figure 7). This showed that the PCA derived from whole genome expression could be replicated using expression levels of the much smaller set of genes that comprise the P-SCE-Sig. The proximity of the data points in the PCA was again validated by bootstrap analysis against randomly selected gene sets (P < .01; see supplemental Experimental Procedures).
A gene signature derived from AMLs with ERG promoter and +85 stem cell enhancer activity is associated with clinical outcome
To test the clinical relevance of ERG promoter and +85 stem cell enhancer activity in AML, we first applied k-means clustering to separate a cohort of 161 CN-AML patients17 into 2 groups based on similarity to the P-SCE-Sig. We then evaluated OS and EFS of patients in each group. Indeed, the P-SCE-Sig was able to dichotomize this patient cohort into groups with significantly different survival characteristics (OS, P < .05 and EFS, P < .05; Figure 5A). Next we used a pattern-matching strategy (see supplemental Experimental Procedures) to match patients from this AML cohort with the cohort described in Figure 2, based on expression levels of genes in the P-SCE-Sig. Patients who shared a pattern with the red and yellow groups had a marginally worse OS (P = .0533) and significantly worse EFS (P < .001) than those with similarity to the green group (Figure 5B). The difference in EFS (P < .001) was maintained in patient groups even when the blue group was included in the clustering algorithm, but significance for OS was lost (P = .067; data not shown).
Gene signatures derived from AMLs with promoter–stem cell enhancer activity and stem cell factor expression are associated with clinical outcome. (A) Unsorted cytogenetically normal AML samples (161) were divided into 2 populations by the P-SCE-sig derived from the PCA in Figure 4D (see supplemental Figure 6 and supplemental Experimental Procedures for details) using the k-means clustering algorithm. The panel to the left shows Kaplan-Meier plots for OS and the panel to the right for EFS. Subjects represented by the blue line (G2) show worse OS and OFS than those represented by the gray line (G1). (B) The cytogenetically normal AML samples were divided based on similarity of their transcriptomes to the green group or the yellow and red groups. The panel to the left shows Kaplan-Meier plots for OS and the panel to the right for event free survival. The green line represents subjects whose AML cells show higher concordance with patients with no promoter-enhancer activity (green group) and the red line represents subjects whose AML cells show higher concordance with patients with promoter-enhancer activity (red/yellow). (C) The cytogenetically normal AML samples were divided into 2 populations based on expression levels of the Heptad genes (Heptad-sig) using the k-means clustering algorithm. The panel to the left shows Kaplan-Meier plots for OS and the panel to the right for EFS.
Gene signatures derived from AMLs with promoter–stem cell enhancer activity and stem cell factor expression are associated with clinical outcome. (A) Unsorted cytogenetically normal AML samples (161) were divided into 2 populations by the P-SCE-sig derived from the PCA in Figure 4D (see supplemental Figure 6 and supplemental Experimental Procedures for details) using the k-means clustering algorithm. The panel to the left shows Kaplan-Meier plots for OS and the panel to the right for EFS. Subjects represented by the blue line (G2) show worse OS and OFS than those represented by the gray line (G1). (B) The cytogenetically normal AML samples were divided based on similarity of their transcriptomes to the green group or the yellow and red groups. The panel to the left shows Kaplan-Meier plots for OS and the panel to the right for event free survival. The green line represents subjects whose AML cells show higher concordance with patients with no promoter-enhancer activity (green group) and the red line represents subjects whose AML cells show higher concordance with patients with promoter-enhancer activity (red/yellow). (C) The cytogenetically normal AML samples were divided into 2 populations based on expression levels of the Heptad genes (Heptad-sig) using the k-means clustering algorithm. The panel to the left shows Kaplan-Meier plots for OS and the panel to the right for EFS.
A gene signature derived from expression levels of the HSC transcriptional heptad is also associated with clinical outcome
Given the relationship between the ERG promoter and +85 stem cell enhancer activity in AML and the HSC transcriptome, we next evaluated patient survival based on expression levels of the heptad TFs that bind and regulate the +85 enhancer. Extrapolating heptad gene expression (Heptad-signature [Heptad-sig]) to the CN-AML, samples were dichotomized by k-means clustering based on similarity to the Heptad-sig into 2 patient groups (G1 and G2). These 2 groups demonstrated significantly different OS (P < .001) and EFS (P < .01; Figure 5C). ERG expression alone failed to discriminate groups with survival difference using k-means clustering (data not shown). Retrospective analysis of heptad expression in these groups showed that RUNX1, FLI1, LMO2, GATA2, ERG, and LYL1 levels were significantly higher in the poor survival group (G2), whereas SCL expression was significantly lower; see supplemental Table 3 for differential expression of heptad and supplemental Table 4 for differential expression of all genes between G1 and G2.
Heptad signature is associated with distinct molecular subtypes of AML and is an independent risk factor for poor prognosis
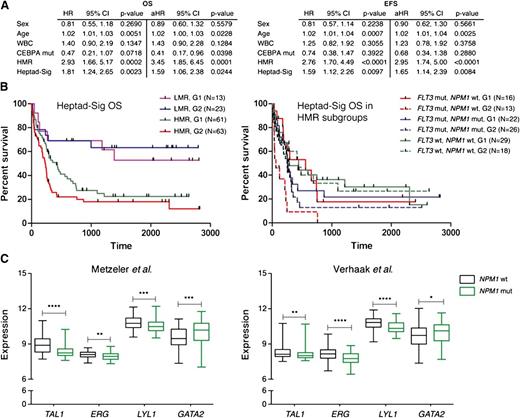
CN-AML patients who lack gross genomic alterations can be categorized into clinically relevant prognostic groups based on the mutational status of FLT3, NPM1, and CEBPα.18,19 Individuals with low molecular risk (LMR; NPM1 mut, FLT3 wt) are considered to have a favorable prognosis and receive standard therapy, whereas those with high molecular risk (HMR; FLT3-ITD or NPM1wt, FLT3 wt) may benefit from more intensive therapy, including allogeneic stem cell transplantation.2 CEBPα mutations confer a favorable outcome. To determine the effect of the Heptad-sig independent of other variables, hazard rates were assessed individually to obtain hazard ratios (HRs) and assessed jointly in a multivariable analysis to determine adjusted hazard ratios (aHRs). Statistical significance was assessed through 95% confidence intervals.20 HMR and the Heptad-sig remained statistically significant for poor OS and EFS when adjusted for the other variables (Figure 6A). We also examined whether the heptad signature affected the survival of cytogenetically normal patients within HMR and LMR groups (Figure 6B). Although there was no significant difference in the LMR group, the heptad signature conferred a worse OS on patients belonging to the HMR group (P < .05). When the HMR group was further evaluated based on its constituent FLT3 and NPM1 mutation profiles, we noted that the negative impact of the heptad signature on OS in the HMR group was limited to patients with FLT3-ITD mutations (P < .01). In fact there were no long-term survivors in the AML cohort with FLT3-ITD mutations, wild-type NPM1, and the heptad signature (Figure 6B).
The heptad signature is associated with distinct molecular subtypes of AML. (A) Multivariable analysis for OS and EFS for different variables. (B) The cytogenetically normal AML samples were further subdivided into LMR and HMR groups and survival recalculated based on the Heptad-sig. The panel to the left shows Kaplan-Meier plots for OS with the HMR group as a collective. The panel to the right shows survival for individual combinations that comprise the HMR group. (C) Distribution of heptad gene expression in NPM1wt and NPM1mut AML in 2 independent CN-AML cohorts. The box plots visualize the distribution of gene expression levels within these groups. Significance between the NPM1wt and NPM1mut groups was assessed using the Mann-Whitney U test. *P < .05; **P < .01; ***P < .001; and ****P < .0001. Black boxes, NPM1 wt cohort; green boxes, NPM1 mutant.
The heptad signature is associated with distinct molecular subtypes of AML. (A) Multivariable analysis for OS and EFS for different variables. (B) The cytogenetically normal AML samples were further subdivided into LMR and HMR groups and survival recalculated based on the Heptad-sig. The panel to the left shows Kaplan-Meier plots for OS with the HMR group as a collective. The panel to the right shows survival for individual combinations that comprise the HMR group. (C) Distribution of heptad gene expression in NPM1wt and NPM1mut AML in 2 independent CN-AML cohorts. The box plots visualize the distribution of gene expression levels within these groups. Significance between the NPM1wt and NPM1mut groups was assessed using the Mann-Whitney U test. *P < .05; **P < .01; ***P < .001; and ****P < .0001. Black boxes, NPM1 wt cohort; green boxes, NPM1 mutant.
NPM1 mutations are highly associated with a homeobox gene-specific expression signature and low CD34 expression.21,22 In contrast, NPM1 wild-type AMLs show elevated CD34 and ERG expression.15 To investigate the relationships between the Heptad signature, HOX-signature, and NPM1 mutation status, we used expression levels of the Heptad and HOX genes (HOXA5, HOXA7, HOXA9, HOXA10, and HOXB5) in a training set of samples from the German–Austrian CN-AML cohort17 with known NPM1 mutation status in order to build a regression model for each signature. The predictive value of the Heptad signature for NPM1 mutation status was comparable to that of the HOX signature (NPM1mut; HOX, 0.9 and Heptad 0.86: NPM1wt; HOX, 0.71 and Heptad 0.72; see supplemental text). To explore this association further, we grouped CN-AML samples based on mutation status and evaluated expression levels of genes that comprise the Heptad signature. As previously noted, HOX gene expression was elevated in NPM1mut AML (data not shown) and ERG expression in NPM1wt AML (Figure 6C). TAL1 and LYL1 levels were also elevated in NPM1wt, but GATA2 levels were significantly lower. Indeed, expression levels of these 4 genes could predict NPM1 mutational status as effectively as the Heptad or HOX signature (NPM1mut; 0.85: NPM1wt; 0.72; see supplemental text). To confirm this association, we evaluated expression levels of the Heptad in a second independent cohort of CN-AML patients23 and again observed that ERG, TAL1, and LYL1 levels were higher and GATA2 levels lower in NPM1wt AML (Figure 6C). The regression model developed using a training set from the German–Austrian cohort17 was applied to the Dutch cohort,23 and again expression levels of the 4 genes predicted NPM1 mutation status with comparable efficacy (NPM1mut; HOX, 0.98 and Tetrad 0.91; NPM1wt; HOX, 0.78 and Tetrad 0.65). Taken together, these data suggest that relatively high ERG, TAL1, and LYL1 and low GATA2 constitute a pattern of regulatory TF expression that is shared in NPM1wt AML.
Different stem cell signatures converge on a core set of signaling pathways
There is only modest overlap among the heptad target gene set, genes in the P-SCE-sig, and the NOD/SCID engrafting LSC-R and HSC-R gene sets (Figure 7A). However, given the convergence of AML transcriptomes related to these gene sets, we explored the identity of signaling pathways that are significantly overrepresented in each set using the Ingenuity Pathway Analysis suite (Figure 7B). A core set of 106 pathways was significantly enriched across all 3 gene sets. In order to assess the relevance of these core pathways on patient survival, we compared expression of components of these pathways between G1 and G2 survival groups (classified using the Heptad-sig) using multivariate analysis of variance (see supplemental Experimental Procedures for details). The pathways were ranked according to significance (supplemental Table 5); the top ranked pathway is shown in supplemental Figure 8.
Stem cell signatures in AML converge on a core set of signaling pathways. (A) Venn diagram showing the overlap of genes between the Heptad targets and the P-SCE and LSC-R/HSC-R signatures. (B) Venn diagram showing the overlap of pathways in which the P-SCE-sig genes, Heptad target genes,11 and the LSC-R and HSC-R genes3 are significantly represented. The 106 shared pathways were ranked based on whether the component genes in a pathway were differentially expressed between AML patients in the short and long survival groups in Figure 5A; the highest ranked pathways are listed. Also see supplemental Figure 8.
Stem cell signatures in AML converge on a core set of signaling pathways. (A) Venn diagram showing the overlap of genes between the Heptad targets and the P-SCE and LSC-R/HSC-R signatures. (B) Venn diagram showing the overlap of pathways in which the P-SCE-sig genes, Heptad target genes,11 and the LSC-R and HSC-R genes3 are significantly represented. The 106 shared pathways were ranked based on whether the component genes in a pathway were differentially expressed between AML patients in the short and long survival groups in Figure 5A; the highest ranked pathways are listed. Also see supplemental Figure 8.
Discussion
We performed a detailed analysis of ERG transcript type and abundance in AML and report that ERG expression in AML is regulated by a heptad of HSC TFs binding its promoters and +85 stem cell enhancer. The activity of these elements not only reflects ERG expression but also appears to reflect the primitive nature or stemness of the global AML transcriptome. We also demonstrate that gene expression signatures derived from promoter–stem cell enhancer activity in AML or for expression levels of the regulatory heptad were associated with patient survival. The implications of these findings are that activation and/or maintenance of HSC circuits in AML may account for the shared gene expression profiles reported between LSCs and HSCs. This may relate to the initial transformation event occurring in HSC or to the restoration of a stem cell–like transcriptional environment by the transforming mutations.
Aberrantly expressed oncogenic TFs require accessible DNA targets to bind to and concurrently expressed transcriptional coactivators to bind with in order to generate transforming transcriptional programs. Therefore, the epigenetic context of the leukemia-initiating cell is an important collaborator along with the transcriptional oncogenic driver. The assembly of TF complexes at enhancer elements prevents chromatin compaction and loss of enhancer accessibility and activity.24 In the specific example shown here, ERG is a component of a regulatory heptad, which binds the ERG +85 stem cell enhancer. Continued expression of ERG and components of the heptad in a cell would promote persistent enhancer accessibility. However, as the heptad collectively regulates not only the ERG +85 stem cell enhancer, but also a number of other verified hematopoietic enhancers in HSPCs,11 expression levels of the heptad are associated not only with accessibility of the +85 enhancer and expression of ERG but also with the expression of a set of genes that contribute to the relative immaturity or stemness of the AML transcriptome. Moreover, they are reflective of a transcriptional environment that is permissive for stem cell programs. We believe that the P-SCE signature reflects the degree of primitiveness of the AML cell of origin (ie, the closer the epigenetic profile of the ERG locus is between an AML sample and normal HSPCs, the closer the transcriptome of the AML is to normal HSPCs). However, the P-SCE signature is influenced not only by the color-coded AML samples (n = 25) but also by the control CD34 cells (n = 5) that were used as a reference point for the degree of primitiveness of the AML samples.
Our data demonstrate that expression levels of the heptad factors correlate with survival in AML (supplemental Table 3). Although binding of heptad factors to stem cell enhancers in HSPCs is recognized, the exact roles of individual components as positive or negative regulators are not known. Indeed, assembly of the heptad complex at stem cell enhancers may represent a dynamic interaction, where some components are predominantly positive and their activity is modulated by others that are inhibitory. We propose that the poor survival group represents AML samples with more stem cell–like blasts, where the relative high expression of some factors and low expression of others may indicate corresponding permissive and repressive roles for these factors when acting in concert in HSPCs. Direct assessment of chromatin accessibility at regulatory elements and expression levels not only of ERG but of each of the heptad genes in a large cohort of primary AML samples would strengthen our thesis that these genes help maintain stem cell signatures in leukemic cells by coordinating their own expression and those of critical downstream targets.
The grim prognosis seen in CN-AML patients with FLT3-ITD mutations in the context of wild-type NPM1 and expression of the heptad signature implies a degree of coordination between this receptor tyrosine kinase signaling pathway and heptad activity. Acquisition of FLT3 mutations in cells may prime heptad activity and contribute to poorly differentiated AML or may simply cooperate with heptad activity during leukemia evolution. Mutations in another tyrosine kinase, JAK2, which like FLT3 mutations are also acquired relatively late in disease evolution,25,26 promote epigenetic expression of hematopoietic oncogenes by aberrant phosphorylation of Tyr 41 on histone 3H and prevention of heterochromatinization and gene inactivation.27 The role of FLT3-ITD mutations on chromatin modification and the epigenetic environment is unclear, although NPM1 mutations have been associated with hypermethylated gene promoters and better OS in CN-AML.28 Interestingly, the negative impact of FLT3-ITD and the heptad signature on OS was not seen in the NPM1 mutant cohort. It remains to be determined whether FLT3-ITD mutations directly increase expression or stability of heptad factors or are permissive for their access to target sites. Whether specific underlying somatic mutations in NPM1 wild-type AMLs account for the differential expression of ERG, TAL1, LYL1, and GATA2 and whether they constitute a shared transformative signature remain to be established. However, the association of signaling pathways with survival groups defined by different stem cell signatures underscores the convergence of survival pathways irrespective of the molecular heterogeneity of AML cells. Recognizing pathways that are deregulated in poor survival AML may present opportunities for targeted therapies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Health and Medical Research Council of Australia, Biotechnology and Biological Sciences Research Council, Leukaemia and Lymphoma Research and the Kay Kendall Leukaemia Fund.
Authorship
Contribution: E.D., D.B., E.G., J.A.I.T., K.K., C.P., S.F., D.G., W.K.L., L.B., K.H.M., K.O., J.O., and J.W.H.W. performed research, interpreted and analyzed data. G.M., S.K.B., C.B., A.B., G.S.V., B.G. and B.J.H. provided vital reagents, designed experiments, interpreted and analyzed data. J.E.P. designed the study, analyzed data and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: John E. Pimanda, Level 2, Lowy Cancer Research Centre, University of New South Wales, Sydney, NSW 2052, Australia; e-mail: jpimanda@unsw.edu.au; and Brian J. Huntly, Level 6, Cambridge Institute for Medical Research, Cambridge, CB2 0XY, United Kingdom; e-mail: bjph2@cam.ac.uk.
References
Author notes
E.D. and D.B. contributed equally to this study.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal