Key Points
The mechanism of bone marrow failure (BMF) in PNH is not known.
Novel CD1d-restricted, GPI-specific T cells are present in PNH patients and might be responsible for BMF.
Abstract
The mechanism of bone marrow failure (BMF) in paroxysmal nocturnal hemoglobinuria (PNH) is not yet known. Because in PNH the biosynthesis of the glycolipid molecule glycosylphosphatidylinositol (GPI) is disrupted in hematopoietic stem and progenitor cells by a somatic mutation in the PIG-A gene, BMF might result from an autoimmune attack, whereby T cells target GPI in normal cells, whereas PIG-A mutant GPI-negative cells are spared. In a deliberate test of this hypothesis, we have demonstrated in PNH patients the presence of CD8+ T cells reactive against antigen-presenting cells (APCs) loaded with GPI. These T cells were significantly more abundant in PNH patients than in healthy controls; their reactivity depended on CD1d expression and they increased upon coculture with CD1d-expressing, GPI-positive APCs. In GPI-specific T cells captured by CD1d dimer technology, we identified, through global T-cell receptor α (TCRα) analysis, an invariant TCRVα21 sequence, which was then found at frequencies higher than background in the TCR repertoire of 6 of 11 PNH patients. Thus, a novel, autoreactive, CD1d-restricted, GPI-specific T-cell population, enriched in an invariant TCRα chain, is expanded in PNH patients and may be responsible for BMF in PNH.
Introduction
Paroxysmal nocturnal hemoglobinuria (PNH) is caused by somatic mutations in the X-linked PIG-A gene in hematopoietic stem and progenitor cells (HSPCs).1,2 PIG-A mutations hinder the production of glycosylphosphatidylinositol (GPI), a glycolipid to which 2% of all proteins are posttranslationally attached before they are sorted and expressed on the cell membrane as GPI-linked molecules.3
The clinical phenotype of PNH consists of a triad of intravascular hemolysis, prothrombotic diathesis, and cytopenias due to bone marrow failure (BMF).4,5 Whereas hemolysis and thrombosis are largely due to deficiency of the GPI-linked complement inhibitors CD55 and CD59,4 the pathogenesis of BMF has remained elusive. The close clinical relationship between PNH and idiopathic aplastic anemia (AA), a T-cell–mediated autoimmune BMF disorder characterized by depletion of HSPCs,6 points toward an immune mechanism mediating BMF in PNH as well. In fact, since the formulation of the “escape” hypothesis,7,8 it was suggested that a T-cell–mediated autoimmune process targeting GPI-positive but not GPI-negative HSPCs may underlie BMF in PNH.8-10 Accordingly, whereas GPI-negative HSPCs arise spontaneously,11 they do not contribute significantly to hematopoiesis unless a T-cell–mediated autoimmune process, by selectively targeting GPI-positive HSPCs, favors expansion and differentiation of GPI-negative HSPCs: this gives rise to a large proportion of GPI-negative hematopoiesis involving all blood cell lineages, which becomes manifest as classic hemolytic PNH.
What is not yet clear is how T cells might selectively target GPI-positive HSPCs. An attractive hypothesis is that GPI itself is a target of autoreactive T cells in PNH.12 In support of this hypothesis, analysis of the T-cell receptor (TCR) complementarity determining region (CDR) β sequence showed that the same identical or quasi-identical TCRβ chain was enriched in CD8+CD57+ T cells of a subset of human leukocyte antigen (HLA)–disparate PNH patients but not in normal controls.13 This is consistent with a T-cell response to the same antigen, restricted by a nonpolymorphic HLA-like molecule such as CD1d. CD1d is expressed on both human and murine HSPCs14,15 and has been shown to associate with GPI,16,17 in keeping with the possibility that CD1d-restricted, GPI-specific T cells might be responsible for targeting HSPCs in PNH. If such cells were involved in the pathogenesis of BMF in PNH, their frequency would be expected to be higher in patients than in normal controls and their TCR structure might resemble that of the CD1d-restricted, glycolipid-specific invariant natural killer T (iNKT) cells: that is, it might have an invariant TCRα chain.18-22 To address these hypotheses, we searched for the presence and assessed the frequency of GPI-specific T cells in the blood of PNH patients and we analyzed the primary structure of their TCRα chain.
Patients, materials, and methods
Patients
Peripheral blood samples from patients with PNH and from age- and gender-matched healthy donors were obtained after written informed consent in accordance with the Declaration of Helsinki and appropriate institutional ethics committee approval. Patient characteristics are shown in supplemental Table 1 (available on the Blood website).
Cell lines and cell culture
The Epstein-Barr virus–transformed C1R B-cell line expresses no or low levels of HLA class I molecules, while C1R-CD1d cells were derived by retroviral transduction of CD1d.23 The KC cell line (a GPI-negative derivative of K562) was a gift from Dr R. W. Finberg (University of Massachusetts Medical School, Worcester, MA). KC-CD1d cells were generated by retroviral transduction of human CD1d complementary DNA (cDNA) into KC cells as described.24 All cell lines were maintained in RPMI 1640 supplemented with 10% fetal calf serum, L-glutamine (2mM), penicillin (50 IU/mL), and streptomycin (50 μg/mL). Immunophenotypic analysis of the above cell lines is shown in supplemental Figure 1A.
T-cell and APC coculture for ELISPOT assays
For trypanosomal GPI (t-GPI)25,26 loading of antigen-presenting cells (APCs; C1R cells, KC cells, and their derivatives), 1 × 106 APCs were loaded with 100 ng of either t-GPI (dissolved in 1 μl of isopropanol:water, 3:2 Vehicle) or Vehicle alone (Veh) in 500 μL of RPMI 1640 for 16 hours at 37°C, 5% CO2. For day 1 assays, T cells were placed directly into enzyme-linked immunospot (ELISPOT) assays in the presence of GPI- or Veh-loaded APCs. A similar approach was used for loading human GPI (h-GPI) onto patient-derived dendritic cells (DCs; see supplemental Methods).
For 7-day cocultures, T cells were cultured with GPI-loaded C1R-CD1d until 24 hours before performing the ELISPOT assay. For 14-day cocultures, T cells were restimulated on day 7 with GPI-loaded C1R-CD1d cells and then kept in coculture with the GPI-loaded C1R-CD1d until 24 hours before performing the ELISPOT assay. Specifically, after irradiation (6000 Rad), 2.5 × 105 C1R-CD1d were cocultured in 24-well plates with 0.5-1 × 105 CD8+ T cells in 1 mL of RPMI medium (Invitrogen) supplemented with 5% human serum, L-glutamine (2 mM), penicillin (50 IU/mL), and streptomycin (50 μg/mL). Interleukin-2 (IL-2) (100 U/mL; Miltenyi Biotec) was added every 3 days. ELISPOT assay was carried out using an ELISPOT kit (Mabtech) according to the manufacturer’s instructions at days 1, 7, and 14 of the coculture. Briefly, CD8+ T cells were collected and cocultured with freshly prepared GPI- or Veh-loaded irradiated C1R-CD1d at different effector-to-target (E:T) ratios, as indicated in individual experiments, in 96-well polyvinylidene difluoride membrane plates (Whatman) coated with anti-human interferon (IFNγ) monoclonal antibody (mAb). After 16 hours of incubation at 37°C and 5% CO2, secreted IFNγ was detected using biotinylated anti-human IFNγ mAb and revealed by development with streptavidin-alkaline phosphatase and 5-bromo-4-chloro-3-indoyl phosphate-toluidine salt/nitro blue tetrazolium chromogenic substrate. Spots were quantified using an AID ELISPOT reader.
T-cell and APC coculture for intracellular IFNγ staining
A total of 5 × 105 monocyte-depleted peripheral blood mononuclear cells (PBMCs; obtained by collection of nonadherent cells from Ficoll-fractionated PBMCs incubated in serum-free RPMI 1640 medium at 37°C for 2-4 hours) and irradiated 2.5 × 105 APCs were added to wells of a 24-well plate in 1 mL of RPMI medium supplemented with 5% human serum, L-glutamine (2 mM), penicillin (50 IU/mL), and streptomycin (50 μg/mL). IL-2 (100 U/mL; Miltenyi Biotec) was added every 3 days. On day 6, additional APCs (E:T ratio, 2:1) were added to the culture. On day 6, brefeldin A (eBioscience) was added into the coculture at a final concentration of 5 μg/mL, 5 hours after the addition of irradiated APCs. Finally, 16 hours later, intracellular staining for IFNγ was performed using the manufacturer’s reagents and instructions (Fix and Perm; Invitrogen). Briefly, cell suspensions were first stained with the following mouse anti-human mAbs: CD3-phycoerythrin (PE), CD8-PE-Cy7 (BD Pharmingen) for 30 minutes at 4°C, fixed in the dark at room temperature for 15 minutes, and then permeabilized in Medium B permeabilization buffer and stained with anti-human IFNγ Alexa Fluor 647 (BD Pharmingen) in the dark at room temperature for 15 minutes. Finally, cells were analyzed using a FACSCanto II flow cytometer (BD Biosciences) and FlowJo software (Tree Star Inc).
CD1d/GPI dimer generation, staining, and flow cytometric analysis and cell sorting
Recombinant CD1d dimeric complexes (dimer X: soluble dimeric human CD1d:Ig fusion) were from BD Biosciences. To stain 1 × 106 cells, we generated CD1d/GPI dimers according to the following protocol: 2 μg of CD1d dimer X were mixed in a glass vial with 2 μg of t-GPI (dissolved in 2 μL of isopropanol:water, 3:2 vehicle), h-GPI (dissolved in 4 μL of isopropanol:dimethylsulfoxide [DMSO], 10:1 vehicle), or vehicle alone for 16 hours at 37°C. Synthesis of t-GPI and h-GPI were previously described.25,27
For CD1d/GPI or CD1d/Veh dimer staining, cells were first incubated with Fc blocking reagent (Miltenyi Biotec) for 10 minutes at 4°C to reduce nonspecific staining. Loaded CD1d dimer was added for 1 hour at 4°C in phosphate-buffered saline plus 0.5% bovine serum albumin; samples were washed with phosphate-buffered saline and then incubated with 2.5 μL of a 1:100 dilution (5 ng) of PE-conjugated A85-1 anti-mouse immunoglobulin G1 (IgG1) Ab (BD Pharmingen) for 10 minutes a 4°C. After 2 washes, samples were stained with the following mouse anti-human mAb: fluorescein isothiocyanate–CD19, fluorescein isothiocyanate–CD14, Alexa Fluor 405-CD3, allophycocyanin-CD8 (all from BD Pharmingen). Finally, 4′,6-diamidino-2-phenylindole (DAPI) was used in order to exclude dead cells. We performed 6-color flow cytometry for analysis and cell sorting on a FACSAria II cell sorter (BD Biosciences). Doublets were excluded based on FSC-W/FSC-A values. Data were analyzed using the FlowJo software (Tree Star Inc).
T-cell flow-sorting, TCR amplicon library preparation, and GS Junior NGS
Four T-cell subsets, CD3+CD48+TCRVβ19+/− and CD3+CD48−TCRVβ19+/−, were flow-sorted from patient PBMCs. CD3+ T cells from normal donor were flow-sorted in 2 fractions, that is, CD3+TCRVβ19+ and CD3+TCRVβ19− T cells (supplemental Figure 4A). Total RNA was extracted from sorted cells using the RNeasy Plus Micro kit (QIAGEN) and transcribed to cDNA with the RevertAid First Strand cDNA Synthesis Kit (Fermentas).
Polymerase chain reaction (PCR) TCRVα21-Cα amplification using cDNA from each sorted T-cell fraction (supplemental Figure 4A) was performed using fusion primers designed according to the Genome Sequencer (GS) Junior system, amplicon library preparation method (supplemental Figure 4B). The PCR products were run on an agarose 1.6% gel and the band of interest, ∼450 bp, was cut and gel-purified using a Fermentas kit according to the manufacturer’s protocol (supplemental Figure 4C).
Sequencing of the TCRαV21-Cα amplicons was carried out on a 454 GS Junior (Roche) platform targeting >500 reads per amplicon. Reads were processed with the Roche Amplicon Variant Analyzer Version 2.3 software according to the manufacturer’s instructions, and all variant sequences extracted in clusters per amplicon by the corresponding “MID.” All per-amplicon clustered sequences were exported in fasta format to ImMunoGeneTics (IMGT, http://www.imgt.org) and processed by IMGT-HighV-QUEST software.28
Immunomagnetic bead selection, aerolysin selection to generate a C1R-CD1d/GPI-negative B-cell line, in vitro differentiation of GPI-negative monocytes to DCs, and TCR α- and β-gene mRNA repertoire amplification are described in supplemental Materials and Methods.
Data and statistical analysis were performed using GraphPad Prism software.
Results
CD1d-dependent T-cell reactivity in response to exogenous GPI
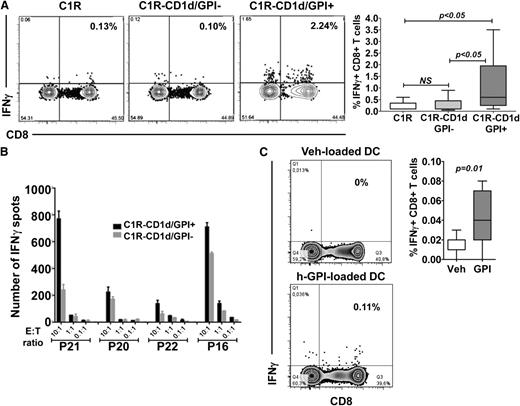
To test whether GPI-specific T-cell responses could be identified in patients with the classic, hemolytic form of PNH (supplemental Table 1), we cultured CD8+ T cells with CD1d-expressing C1R (C1R-CD1d) B cells (supplemental Figure 1A) as APCs loaded with chemically synthesized t-GPI25,26 (supplemental Figure 1B); 24 hours later, we carried out IFNγ ELISPOT assays. Because C1R cells express HLA class II but not class I molecules,29 to minimize CD4+ T-cell–mediated alloreactive responses, we used CD8+ but not CD4+ T cells. In pilot experiments (supplemental Figure 1C), we found that the number of CD8+ T cells able to form IFNγ spots after exposure to GPI-loaded APCs was higher in patients than in controls.
Similar results were obtained when CD8+ T cells were cocultured with GPI-loaded C1R-CD1d cells for 1, 7, and 14 days: at each time point, T cells from PNH patients formed 4.9-, 4-, and 5.2-fold, respectively, more IFNγ spots than similarly treated T cells from normal donors (Figure 1A; P < .005 at each time point).
CD1d-dependent T-cell reactivity in response to exogenous GPI. (A) IFNγ spot formation by CD8+ T cells from PNH patients and normal donor controls assessed in ELISPOT assays against t-GPI– or Veh-loaded C1R-CD1d cells. Assays were performed on days 1, 7, or 14 at a fixed E:T ratio 3:1 (number of plated T cells varied between 5 × 104 and 105 per well) as described in “Patients, materials, and methods.” The median frequency of IFNγ spot-forming CD8+ T cells as a percentage of total CD8+ T cells was derived after the mean number of spots of triplicate or quadruplicate assays was calculated. In the box-and-whisker plots, the horizontal line inside the box represents the median, the top and bottom of the box represent the interquartile range, and the vertical bars the range. Number of samples used in each time point and group are also shown. Significance P values were estimated by Mann-Whitney test. (B) IFNγ ELISPOT assay performed with CD8+ T cells from 6 PNH patients (P) and 3 normal controls (N) cultured for 24 hours with the parental CD1d-negative C1R cell line or its C1R-CD1d derivative loaded with Veh or t-GPI (E:T ratio of 1:1; 5 × 104 T cells per well plated in triplicate assays). Data are shown as mean ± SEM. (C) IFNγ ELISPOT assay performed with CD8+ T cells from 9 PNH patients and 3 controls cultured for 24 hours with the GPI-negative K562 cell line KC or its derivative KC-CD1d loaded with GPI or Veh (E:T ratio of 1:1; 5 × 104 T cells per well plated in triplicate assays). Data are shown as mean ± SEM. *Not performed.
CD1d-dependent T-cell reactivity in response to exogenous GPI. (A) IFNγ spot formation by CD8+ T cells from PNH patients and normal donor controls assessed in ELISPOT assays against t-GPI– or Veh-loaded C1R-CD1d cells. Assays were performed on days 1, 7, or 14 at a fixed E:T ratio 3:1 (number of plated T cells varied between 5 × 104 and 105 per well) as described in “Patients, materials, and methods.” The median frequency of IFNγ spot-forming CD8+ T cells as a percentage of total CD8+ T cells was derived after the mean number of spots of triplicate or quadruplicate assays was calculated. In the box-and-whisker plots, the horizontal line inside the box represents the median, the top and bottom of the box represent the interquartile range, and the vertical bars the range. Number of samples used in each time point and group are also shown. Significance P values were estimated by Mann-Whitney test. (B) IFNγ ELISPOT assay performed with CD8+ T cells from 6 PNH patients (P) and 3 normal controls (N) cultured for 24 hours with the parental CD1d-negative C1R cell line or its C1R-CD1d derivative loaded with Veh or t-GPI (E:T ratio of 1:1; 5 × 104 T cells per well plated in triplicate assays). Data are shown as mean ± SEM. (C) IFNγ ELISPOT assay performed with CD8+ T cells from 9 PNH patients and 3 controls cultured for 24 hours with the GPI-negative K562 cell line KC or its derivative KC-CD1d loaded with GPI or Veh (E:T ratio of 1:1; 5 × 104 T cells per well plated in triplicate assays). Data are shown as mean ± SEM. *Not performed.
To test whether IFNγ reactivity in response to GPI is dependent on CD1d expression on APCs, we used, along with its C1R-CD1d derivative, also the parental CD1d-negative C1R cell line (supplemental Figure 1A), with and without t-GPI loading. We found that CD8+ T cells from 4 of 6 patients tested formed the highest number of IFNγ spots in the presence of t-GPI–loaded C1R-CD1d cells. By contrast, no such effect was observed with T cells from 3 normal donors (Figure 1B). In another set of similar experiments, reactivity of T cells was tested against the CD1d-negative, GPI-negative K562 cell line KC and its derivative KC-CD1d (supplemental Figure 1A). In 7 of 9 PNH patients, the IFNγ spot-forming activity of T cells was highest against KC-CD1d cells loaded with GPI than in other conditions (Figure 1C). This effect was not observed in any of the 4 normal donor controls tested. Overall, in 7 of 8 PNH patients, the highest T-cell reactivity was observed against CD1d-expressing APCs loaded with t-GPI, while none of the 7 controls showed this pattern (P = .001, Fisher exact test).
Together, these results show that CD1d is required for induction of optimal T-cell responses elicited by exogenous GPI in PNH patients.
CD1d-dependent T-cell reactivity in response to endogenous GPI
Because a trend for higher T-cell reactivity was observed even in the absence of exogenous GPI (Figure 1A) and, similarly, T cells from patients P16 and P22 (Figure 1B) formed a high number of spots even when GPI had not been added to C1R-CD1d cells, we surmised that patient T cells might be responding to endogenous GPI. Indeed, after a 7-day coculture of monocyte-depleted PBMCs with APC, the number of reactive T cells, as determined by IFNγ-intracellular staining and flow cytometry, was 6-fold higher when APCs were C1R-CD1d/GPI-positive rather than when they were either C1R-CD1d/GPI-negative or C1R (ie, CD1d-negative; n = 9; P < .05; Figure 2A). Also consistent with reactivity elicited by endogenous GPI, in 3 of 4 patients tested, a higher number of IFNγ ELISPOTs formed after exposure of T cells to C1R-CD1d/GPI-positive than to C1R-CD1d/GPI-negative cells (Figure 2B). It should be noted that 6 of 9 patients shown in Figure 2A, and 3 of 4 patients in Figure 2B, had not received any blood transfusions at the time of testing or at any time prior to this (supplemental Table 1), suggesting that T-cell GPI reactivity is not influenced by blood transfusion. This is in line with our previous observation that blood transfusion does not skew the TCRβ repertoire.30
CD1d-dependent T-cell reactivity in response to endogenous GPI. (A) (Left) Representative flow cytometry plots showing intracellular IFNγ-producing CD8+ T cells from a PNH patient after monocyte-depleted PBMCs were cocultured with C1R, C1R-CD1d/GPI-positive, or C1R-CD1d/GPI-negative B cells for 7 days, followed by the addition of the same number of APCs. Brefeldin A was added 16 hours before harvesting the culture. Cells were stained with mAbs against human CD3, CD8, and intracellular IFNγ. Flow cytometry plots shown are gated on CD3+ cells. (Right) Cumulative data from 9 patients showing frequency of CD8+ T cells expressing intracellular IFNγ (Wilcoxon signed rank test). (B) IFNγ ELISPOT assays performed with CD8+ T cells from 4 PNH patients cultured for 24 hours with C1R-CD1d/GPI-positive cells and its GPI-negative derivative. The APC number was 2 × 104 in each well; E:T ratios as shown. (C) (Left) Representative flow cytometry plots showing IFNγ-producing, patient CD8+ T cells after a 3-day coculture of monocyte-depleted PBMCs with autologous, GPI-negative, mature DCs loaded with h-GPI or Veh. Flow cytometry plots shown are gated on CD3+ cells. (Right) Cumulative data from 11 PNH patients (Wilcoxon signed rank test).
CD1d-dependent T-cell reactivity in response to endogenous GPI. (A) (Left) Representative flow cytometry plots showing intracellular IFNγ-producing CD8+ T cells from a PNH patient after monocyte-depleted PBMCs were cocultured with C1R, C1R-CD1d/GPI-positive, or C1R-CD1d/GPI-negative B cells for 7 days, followed by the addition of the same number of APCs. Brefeldin A was added 16 hours before harvesting the culture. Cells were stained with mAbs against human CD3, CD8, and intracellular IFNγ. Flow cytometry plots shown are gated on CD3+ cells. (Right) Cumulative data from 9 patients showing frequency of CD8+ T cells expressing intracellular IFNγ (Wilcoxon signed rank test). (B) IFNγ ELISPOT assays performed with CD8+ T cells from 4 PNH patients cultured for 24 hours with C1R-CD1d/GPI-positive cells and its GPI-negative derivative. The APC number was 2 × 104 in each well; E:T ratios as shown. (C) (Left) Representative flow cytometry plots showing IFNγ-producing, patient CD8+ T cells after a 3-day coculture of monocyte-depleted PBMCs with autologous, GPI-negative, mature DCs loaded with h-GPI or Veh. Flow cytometry plots shown are gated on CD3+ cells. (Right) Cumulative data from 11 PNH patients (Wilcoxon signed rank test).
Finally, we tested the ability of a form of h-GPI obtained by organic synthesis27 (supplemental Figure 1B) to elicit patient T-cell reactivity. We used as APCs mature myeloid DCs obtained from autologous, GPI-negative monocytes (supplemental Figure 2). We took advantage of the fact that in PNH patients the large majority of monocytes are CD14-negative. In the absence of added GPI, we could not demonstrate autologous reactive T cells; however, when h-GPI was added there was a sixfold increment in reactive autologous T cells, (n = 11; P = .005; Figure 2C), thus providing direct evidence that primary, professional APCs, when GPI-loaded, are also able to elicit IFNγ T-cell responses.
Thus, endogenous as well as exogenous GPI activates CD1d-dependent, GPI-reactive T cells, and such reactive cells are increased in patients with PNH.
CD1d/GPI dimer staining of T cells
To prospectively identify, enumerate, and physically isolate GPI-reactive, CD1d-restricted T cells ex vivo, we stained patient or control PBMCs with fluorescently labeled CD1d-dimer complexes loaded with t-GPI. To minimize nonspecific staining, we used a stringent protocol that excludes monocytes, B cells, and dead cells.31 The frequency of CD8+ CD1d/t-GPI dimer+ T cells was 10-fold higher in patients than in controls (P = .02; Figure 3A-B). In vitro, after 14 days of coculture of CD8+ T cells with C1R-CD1d cells loaded with t-GPI, the frequency of CD1d/t-GPI dimer+ T cells was fourfold higher in patients than in controls (P < .01), and significantly higher than the background staining frequencies of patient and control CD1d/Veh dimer+ T cells (P < .01; Figure 3C-D). Similar results were obtained using CD1d dimer loaded with h-GPI (Figure 3E-F). Therefore, CD1d/GPI dimer staining provides independent evidence that compared with controls, the frequency of CD1d-restricted, GPI-specific T cells is increased in patients with PNH.
CD1d/GPI dimer staining of T cells. (A) Representative flow cytometry plots showing ex vivo staining of patient and control PBMCs with anti-CD8 and CD1d/t-GPI dimer followed by analysis by multiparameter flow cytometry. Plots shown are gated on DAPI−CD14−CD19−CD3+ cells. (B) Cumulative data showing frequency of CD8+CD1d/t-GPI dimer+ T cells from 4 PNH patients and 4 healthy controls after ex vivo staining of PBMCs as described in panel A (staining with CD1d/Veh dimer was not performed in this set of experiments). (C) Representative flow cytometry plots showing staining of patient CD8+ T cells with (left) CD1d/t-GPI or (right) unloaded dimer after 14 days of coculture with C1R-CD1d cells loaded with t-GPI. Gating strategy as described in panel A. (D) Cumulative data showing frequency of CD8+ CD1d/t-GPI dimer+ vs CD1d/Veh dimer+ T cells from patients (n = 7) and controls (n = 11), after 14 days of coculture of CD8+ T cells with C1R-CD1d cells loaded with t-GPI (1-way analysis of variance with Tukey multiple comparison test; NS, not significant). (E) Staining of CD8+ T cells with CD1d/h-GPI dimer or CD1d/Veh dimer after 14 days of coculture with C1R-CD1d cells loaded with h-GPI. Gating strategy as described in panel A. Staining of T cells from a PNH patient is shown. (F) Cumulative data from 10 PNH patients and 7 normal controls.
CD1d/GPI dimer staining of T cells. (A) Representative flow cytometry plots showing ex vivo staining of patient and control PBMCs with anti-CD8 and CD1d/t-GPI dimer followed by analysis by multiparameter flow cytometry. Plots shown are gated on DAPI−CD14−CD19−CD3+ cells. (B) Cumulative data showing frequency of CD8+CD1d/t-GPI dimer+ T cells from 4 PNH patients and 4 healthy controls after ex vivo staining of PBMCs as described in panel A (staining with CD1d/Veh dimer was not performed in this set of experiments). (C) Representative flow cytometry plots showing staining of patient CD8+ T cells with (left) CD1d/t-GPI or (right) unloaded dimer after 14 days of coculture with C1R-CD1d cells loaded with t-GPI. Gating strategy as described in panel A. (D) Cumulative data showing frequency of CD8+ CD1d/t-GPI dimer+ vs CD1d/Veh dimer+ T cells from patients (n = 7) and controls (n = 11), after 14 days of coculture of CD8+ T cells with C1R-CD1d cells loaded with t-GPI (1-way analysis of variance with Tukey multiple comparison test; NS, not significant). (E) Staining of CD8+ T cells with CD1d/h-GPI dimer or CD1d/Veh dimer after 14 days of coculture with C1R-CD1d cells loaded with h-GPI. Gating strategy as described in panel A. Staining of T cells from a PNH patient is shown. (F) Cumulative data from 10 PNH patients and 7 normal controls.
Structural characterization of the TCR in CD1d/GPI dimer+ T cells
Although T-cell clones from flow-sorted CD1d/GPI dimer− cells were obtained at high frequency in response to mitogens, IL-2, IL-7, or IL-15 in parallel experiments, several attempts to generate clones or cell lines from CD1d/GPI dimer+ T cells were unsuccessful. We therefore sought insights into their function by analyzing the primary structure of their TCR. For this purpose, after flow-sorting CD8+CD1d/GPI dimer+ T cells from 3 patients, the whole TCR α- and β-chain mRNA repertoire was analyzed using an approach32 successfully tested previously in amplifying TCR mRNA from as few as 3 T cells (supplemental Figure 3).
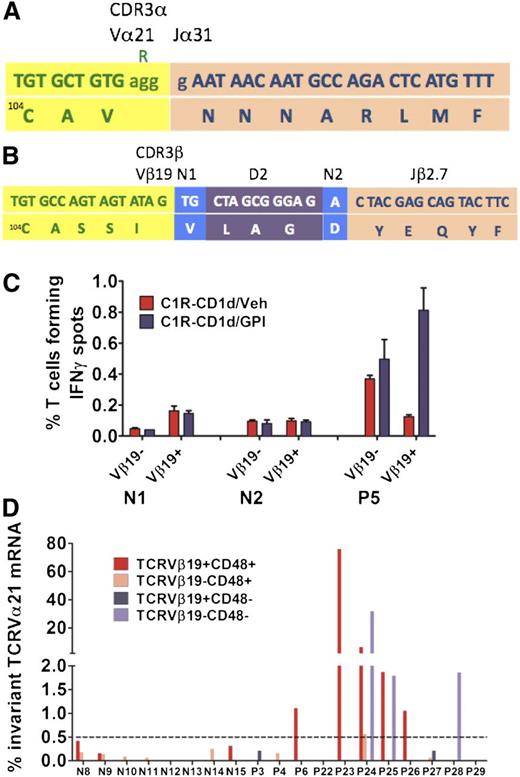
A novel invariant TCRVα21Jα31-1 sequence was identified in 9 of 9 clones in 1 of the 3 patients (P5; supplemental Table 2). Unlike in the majority of TCRα chains, the CDR3 of an invariant TCRα chain is defined by not having any J segment N insertions or deletions: indeed, our Jα31-1 segment is germline and therefore invariant (Figure 4A). Interestingly, analysis of the CDR3β sequence in the same population of flow-sorted CD8+CD1d/GPI dimer+ T cells also revealed the same variant TCRVβ19-D2-Jβ2-7 sequence in all 8 clones sequenced (Figure 4B).
Characterization of the TCR α and β chains of CD1d/GPI dimer+ T cells. Flow-sorted CD1d/h-GPI dimer+ T-cell fractions were subjected to a 2-step reverse transcription PCR to amplify the whole TCR α- and β-chain mRNA repertoire followed by plasmid cloning and sequencing of individual clones (see also supplemental Figure 4). (A) Nucleotide (top) and amino acid (bottom) sequence of the TCRVα21-Jα31-1 CDR3α chain identified in the CD8+ CD1d/GPI dimer+ T cells of P5. The last nucleotide triplet in the Vα21 gene, corresponding to arginine, is deleted; there are no Jα segment N insertions or deletions, that is, the TCRVα21Jα31 amino acid sequence is invariant. (B) Nucleotide (top) and amino acid (bottom) sequence of the TCRVβ19-D2-Jβ2.7 chain identified in the CD8+ CD1d/GPI dimer+ T cells of P5 with a typically diverse CDR3β region as shown. (C) Flow-sorted TCRVβ19+ and TCRVβ19− T cells from P5 and 2 healthy controls were placed immediately after sorting into an IFNγ ELISPOT assay against C1R-CD1d cells loaded with Veh or GPI at an E:T ratio of 1:1 (20 × 103 T cells per well). Data are shown as mean ± SEM of triplicate assays. (D) Bar diagram showing the frequency of the invariant TCRVα21Jα31 mRNA within the whole TCRVα21-Cα repertoire of the flow-sorted T-cell fractions shown (see also Table 1).
Characterization of the TCR α and β chains of CD1d/GPI dimer+ T cells. Flow-sorted CD1d/h-GPI dimer+ T-cell fractions were subjected to a 2-step reverse transcription PCR to amplify the whole TCR α- and β-chain mRNA repertoire followed by plasmid cloning and sequencing of individual clones (see also supplemental Figure 4). (A) Nucleotide (top) and amino acid (bottom) sequence of the TCRVα21-Jα31-1 CDR3α chain identified in the CD8+ CD1d/GPI dimer+ T cells of P5. The last nucleotide triplet in the Vα21 gene, corresponding to arginine, is deleted; there are no Jα segment N insertions or deletions, that is, the TCRVα21Jα31 amino acid sequence is invariant. (B) Nucleotide (top) and amino acid (bottom) sequence of the TCRVβ19-D2-Jβ2.7 chain identified in the CD8+ CD1d/GPI dimer+ T cells of P5 with a typically diverse CDR3β region as shown. (C) Flow-sorted TCRVβ19+ and TCRVβ19− T cells from P5 and 2 healthy controls were placed immediately after sorting into an IFNγ ELISPOT assay against C1R-CD1d cells loaded with Veh or GPI at an E:T ratio of 1:1 (20 × 103 T cells per well). Data are shown as mean ± SEM of triplicate assays. (D) Bar diagram showing the frequency of the invariant TCRVα21Jα31 mRNA within the whole TCRVα21-Cα repertoire of the flow-sorted T-cell fractions shown (see also Table 1).
Furthermore, purified TCRVβ19+ T cells from P5 were 6.5 times more reactive after coculture with GPI-loaded than with unloaded C1R-CD1d cells (Figure 4C). By contrast, TCRVβ19+ T cells from the 2 normal controls showed no increase in IFNγ spots in the presence of GPI.
NGS analysis of the TCRVα21 repertoire
We postulated that if the invariant TCRVα21Jα31-1 chain played a pathogenic role in PNH, its frequency might be increased within the repertoire of the whole TCRVα21 family. Because a TCRVα21-specific mAb is not available, and in patient P5, the TCRVα21Jα31-1 chain seemed to be paired with the TCRVβ19-D2-Jβ2-7 chain, we used a TCRVβ19 family-specific mAb to first flow-sort and then quantify, by next-generation sequencing (NGS), the frequency of invariant TCRVα21 chain mRNA in TCRVβ19+ and in TCRVβ19− T cells. Patient T cells were further sorted into GPI-positive and GPI-negative fractions on the basis of expression of CD48 (supplemental Figure 4) .
Overall, invariant TCRVα21Jα31-1 mRNA was identified in 6 of 8 (75%) controls in both TCRVβ19+ and TCRVβ19− fractions, suggesting that the invariant TCRVα21Jα31-1 chain is part of the normal TCR repertoire (Table 1). In all 6 cases, and in both T-cell fractions, the frequency of the invariant TCRVα21Jα31-1 chain was <0.5% of the total family TCRVα21 mRNA repertoire, thereby providing a measure of the background frequency of the invariant TCRVα21Jα31-1 chain within the normal repertoire (Figure 4D; Table 1).
NGS analysis of the TCRVα21-Cα mRNA repertoire
| . | TCRVβ19+CD48+ . | TCRVβ19−CD48+ . | TCRVβ19+CD48− . | TCRVβ19−CD48− . | ||||
|---|---|---|---|---|---|---|---|---|
| Sorted cells, ×103 . | % TCRVα21Jα31 (no. of reads*) . | Sorted cells, ×103 . | % TCRVα21Jα31 (no. of reads*) . | Sorted cells, ×103 . | % TCRVα21Jα31 (no. of reads*) . | Sorted cells, ×103 . | % TCRVα21Jα31 (no. of reads*) . | |
| Patients†‡ | ||||||||
| P3 | 43.3 | 0 (2 999) | 767.3 | 0 (5 495) | 17.6 | 0.21 (2763) | 711.4 | 0 (5627) |
| P4 | 306.8 | 0 (3 514) | 1300 | 0.16 (2 406) | 5.5 | 0 (4538) | ||
| P6 | 2.2 | 1.11 (499) | 598 | 0 (807) | 0 (921) | |||
| P22 | 93.4 | 0 (2 468) | 1100 | 0 (920) | 32.2 | 0 (2355) | ||
| P23 | 28.4 | 75.8 (2 970) | 359 | 0 (5 643) | 8.9 | 0 (4389) | 2.89 | 0 (1016) |
| P24 | 21.8 | 6.34 (4 457) | 414 | 0.56 (4 243) | 0.96 | 31.79 (585) | ||
| P25 | 96.9 | 1.87 (1 229) | 1150 | 0 (3 232) | 328 | 1.79 (782) | ||
| P26 | 21.6 | 1.05 (2 185) | 592 | 0 (874) | 18.6 | 0 (1176) | ||
| P27 | 46.7 | 0 (1 359) | 1100 | 0.06 (6 209) | 17 | 0.21 (910) | 306 | 0 (1973) |
| P28 | 86.6 | 0 (3 911) | 8.7 | 1.86 (5585) | ||||
| P29 | 54.3 | 0 (3 389) | 1370 | 0 (4 386) | 35.9 | 0 (3845) | ||
| Controls† | ||||||||
| N8 | 77.6 | 0.42 (10 669) | 840 | 0.18 (10 778) | ||||
| N9 | 160 | 0.16 (10 914) | 1250 | 0.14 (10 281) | ||||
| N10 | 150 | 0 (3 532) | 1800 | 0.09 (4 103) | ||||
| N11 | 128 | 0 (3 722) | 1200 | 0.06 (4 558) | ||||
| N12 | 90 | 0 (779) | 800 | 0 (715) | ||||
| N13 | 60 | 0 (6 976) | 1040 | 0 (11 929) | ||||
| N14 | 60 | 0 (5 704) | 1500 | 0.25 (79 369) | ||||
| N15 | 80 | 0.31 (2 248) | 1420 | 0.025 (3 945) | ||||
| . | TCRVβ19+CD48+ . | TCRVβ19−CD48+ . | TCRVβ19+CD48− . | TCRVβ19−CD48− . | ||||
|---|---|---|---|---|---|---|---|---|
| Sorted cells, ×103 . | % TCRVα21Jα31 (no. of reads*) . | Sorted cells, ×103 . | % TCRVα21Jα31 (no. of reads*) . | Sorted cells, ×103 . | % TCRVα21Jα31 (no. of reads*) . | Sorted cells, ×103 . | % TCRVα21Jα31 (no. of reads*) . | |
| Patients†‡ | ||||||||
| P3 | 43.3 | 0 (2 999) | 767.3 | 0 (5 495) | 17.6 | 0.21 (2763) | 711.4 | 0 (5627) |
| P4 | 306.8 | 0 (3 514) | 1300 | 0.16 (2 406) | 5.5 | 0 (4538) | ||
| P6 | 2.2 | 1.11 (499) | 598 | 0 (807) | 0 (921) | |||
| P22 | 93.4 | 0 (2 468) | 1100 | 0 (920) | 32.2 | 0 (2355) | ||
| P23 | 28.4 | 75.8 (2 970) | 359 | 0 (5 643) | 8.9 | 0 (4389) | 2.89 | 0 (1016) |
| P24 | 21.8 | 6.34 (4 457) | 414 | 0.56 (4 243) | 0.96 | 31.79 (585) | ||
| P25 | 96.9 | 1.87 (1 229) | 1150 | 0 (3 232) | 328 | 1.79 (782) | ||
| P26 | 21.6 | 1.05 (2 185) | 592 | 0 (874) | 18.6 | 0 (1176) | ||
| P27 | 46.7 | 0 (1 359) | 1100 | 0.06 (6 209) | 17 | 0.21 (910) | 306 | 0 (1973) |
| P28 | 86.6 | 0 (3 911) | 8.7 | 1.86 (5585) | ||||
| P29 | 54.3 | 0 (3 389) | 1370 | 0 (4 386) | 35.9 | 0 (3845) | ||
| Controls† | ||||||||
| N8 | 77.6 | 0.42 (10 669) | 840 | 0.18 (10 778) | ||||
| N9 | 160 | 0.16 (10 914) | 1250 | 0.14 (10 281) | ||||
| N10 | 150 | 0 (3 532) | 1800 | 0.09 (4 103) | ||||
| N11 | 128 | 0 (3 722) | 1200 | 0.06 (4 558) | ||||
| N12 | 90 | 0 (779) | 800 | 0 (715) | ||||
| N13 | 60 | 0 (6 976) | 1040 | 0 (11 929) | ||||
| N14 | 60 | 0 (5 704) | 1500 | 0.25 (79 369) | ||||
| N15 | 80 | 0.31 (2 248) | 1420 | 0.025 (3 945) | ||||
NGS analysis of the TCRVα21-Cα mRNA repertoire in flow-sorted TCRVβ19+ and TCRVβ19− T cells from PNH patients and normal controls (see also supplemental Figure 4).
mRNA, messenger RNA; NGS, next-generation sequencing.
Median age (range) patients vs controls: 36 years (28-59) vs 36 years (27-52).
Due to very low frequencies of GPI-negative T-cell fractions and small sample sizes, deep sequencing of TCRVα21-Cα amplicons was not possible for all 4 populations in all patients with PNH.
Represent productive TCR reads.
Although the proportion of cases in which we could detect invariant TCRVα21Jα31-1 mRNA was similar in PNH patients and in normal controls (9 of 11 patients; 81%), it was only in PNH patients (6 of 11) that the frequencies were higher than background in either or both of the TCRVβ19+ and TCRVβ19− T-cell fractions (median, 1.86%; range, 1.05%-75%). Interestingly, expanded invariant TCRVα21Jα31-1 T cells were identified in both GPI-positive and GPI-negative T-cell fractions (Figure 4D; Table 1).
Taken together, these data suggest that invariant TCRVα21Jα31-1 cells are expanded within the TCRVα21 family of both TCRVβ19+ and TCRVβ19− T cells in a substantial proportion of patients with PNH.
Discussion
The notion that PNH develops thanks to the escape of GPI-negative hematopoiesis from T-cell–mediated autoimmune attack against normal (GPI-positive) hematopoietic cells was first outlined over 20 years ago, and has been supported by several studies.30,33 It has been further suggested that the target of the autoimmune attack may be, rather than a GPI-linked protein, the GPI molecule itself.12 Here, we provide direct evidence for this hypothesis. We have used a number of complementary methodologies (ie, ELISPOT and intracellular staining of IFNγ-producing cells and CD1d/GPI dimer staining), different types of APCs (B cells, DCs, and K562 cells) and 2 different GPI molecules (t-GPI and h-GPI). All of these approaches converged in enabling us to identify and quantify a novel population of GPI-specific, CD1d-restricted T cells that is expanded in a substantial proportion of PNH patients. Given that GPI has been previously demonstrated in the presentation groove of CD1d16,17,34 and given that that CD1d is expressed on HSPCs,14,15 these findings provide strong support for the pathogenetic model which posits that CD1d-restricted, GPI-specific T cells target and deplete GPI-positive but not GPI-negative HSPCs in PNH.12
We found small numbers of GPI-specific T cells in normal controls, suggesting that they are part of the normal T-cell repertoire. However, their frequency does not increase in response to in vitro–added exogenous GPI, consistent with a state of functional anergy. By contrast, in PNH patients, the frequency of GPI-reactive T cells was not only higher at baseline, but also increased in the course of cocultures carried out for up to 14 days with GPI-loaded APCs. Our results with 2 different GPI-deficient cell lines and their isogenic GPI-positive counterparts, as well as with patient-derived autologous DCs, are consistent with the notion that GPI-specific T cells are autoreactive.
The numbers of GPI-reactive T cells, although higher than in controls, comprised a small percentage of all T cells in PNH patients. There could be several reasons for these low numbers. First, GPI anchors undergo several structural modifications3 and we do not know which form of GPI is most commonly presented by CD1d: therefore, although we tested 2 different GPI preparations, these may not be optimal for detecting T-cell reactivity, resulting in a rather weak pattern of CD1d/GPI dimer staining. Second, GPI-reactive T cells might be more numerous in bone marrow than in peripheral blood; third, their frequency may change in the course of the disease. Despite these complexities, by applying a stringent protocol we were able to demonstrate a significantly higher frequency of CD8+CD1d/GPI dimer+ T cells in PNH patients compared with controls. Finally, we note that the frequency of GPI-specific, CD1d-restricted T cells in PNH reported here is similar to the frequency of pathogenic autoreactive T cells in other organ-specific autoimmune disorders such as vitiligo,35 or to that of CD1d-restricted iNKT cells in normal individuals.36,37
In analogy with activation of iNKT cells by exogenous glycolipid antigens,38 exogenous GPI might activate T cells either directly via their TCR or indirectly through pattern recognition receptors and cytokine secretion. Because GPI has been found in the CD1d antigen-presentation groove and because we have shown that GPI is required for T-cell activation, we favor the notion of direct interaction between GPI as a self-antigen and the invariant Vα21 TCR.
Global analysis of the TCR α- and β-chain repertoire of CD1d-GPI dimer+ T cells in P5 revealed a novel invariant TCRVα21Jα31-1 CDR3α and a diverse TCRVβ19-D2-Jβ2-7 CDR3β sequence. Because both chains were identified at clonal frequencies in the same CD1d/GPI dimer+ T cells, it is very likely that they form a TCR heterodimer in a configuration that is similar to that of the TCR of iNKT cells.39 In support of this, CD8+TCRVβ19+ T cells from the same patient displayed their highest reactivity in the presence of CD1d-expressing APCs pulsed with GPI: this establishes a link between TCRVα21Jα31-1–expressing T cells with GPI-specific reactivity. The frequency distribution of the invariant TCRVα21Jα31-1 chain in normal controls is consistent with the low frequency of GPI T-cell reactivity identified in healthy individuals. A higher frequency pattern was seen exclusively in 6 of 11 patients with PNH: in patient P22, the invariant TCRVα21Jα31-1 chain was as high as 75% of the total TCRVα21 repertoire in TCRVβ19+ T cells, consistent with a process of GPI-dependent expansion of the TCRVα21Jα31-1 T cells. A TCRVα21-specific mAb and TCR transfer experiments will be needed to further define the functional, phenotypic, and structural features of these T cells in health and in disease, including AA. These will include characterization of the complete repertoire of TCRVβ chains that can pair with the invariant TCRVα21Jα31-1 chain, functional validation of the CD1d dimers in complex with different structural variants of GPI, and testing the effect of the GPI-specific T cells on GPI-positive and GPI-negative HSPCs. In the meantime, the higher frequency of CD1d-restricted, GPI-specific T cells in patients than in controls strongly supports the potential of these cells to target CD1d-expressing GPI-positive HSPCs and thus contribute to the pathogenesis of BMF in PNH. This notion offers a rather straightforward explanation as to why PNH patients have often been previously diagnosed with AA. If AA was mediated by T cells that recognize a “conventional” (non-GPI) antigen, then this expansion would not take place. Future work will focus on a group of AA patients who do not have evidence of a PNH clone: we expect that in this group, the GPI-reactive T cells will not be above background. Because PIG-A mutations arise in multipotent hematopoietic stem cells (HSC) spontaneously,11 it is possible, however, that rare, GPI-negative invariant TCRVα21Jα31-1 T cells, progeny of the mutant HSCs, may exist at very low frequencies both before and after the expansion of the PNH hematopoietic clone. Once an autoimmune process has been triggered, both GPI-positive and GPI-negative TCRVα21Jα31-1 T cells may become autoreactive against GPI-positive HSCs and expand in an antigen-dependent manner, either concurrently or sequentially.
In conclusion, we have demonstrated, in patients with PNH, expansion of a novel, autoreactive, CD1d-restricted, GPI-specific T-cell population enriched in an invariant TCRα chain. These T cells may be responsible for BMF in PNH.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients and their families for their collaboration and support, and Kostas Tsatalas, Alexandroupolis, Kostas Stamatopoulos, Thessaloniki, John Apostolidis, and Nikos Anagnostopoulos (all in Athens, Greece) and Despina Kyriakou (in Larissa, Greece) for kindly providing patient samples. The authors thank Laurence Game and Tim Aitman (Imperial National Institute for Health Research Biomedical Research Centre [NIHR BRC] Clinical Genome Laboratory) for facilitating the NGS aspect of the work.
This work was supported by grants from Leukaemia and Lymphoma Research, Imperial Healthcare Charity and the NIHR BRC (L.G., A.C., I.R., J.d.l.F., M.L., and A.K.), and from the Associazione Italiana per la Ricerca sul Cancro (AIRC), the Italian Ministry of Education, University and Research (PRIN 2007HX43y2_002), and the Regione Toscana (M.S., G.T., R.N., L.L.). M.P. was on sabbatical leave from Aristotelion University (Thessaloniki, Greece).
Authorship
Contribution: L.G. and M.P. performed research, analyzed data, and wrote the manuscript; M.S. performed research and analyzed data; G.T. and A.C. performed research; B.R., C.N., and A.V.N. contributed vital new reagents; M.L. and J.d.l.F. contributed to the writing of the manuscript; I.R. designed and supervised research and wrote the manuscript; and L.L., R.N., and A.K. were in charge of the clinical management of some of the patients, designed and supervised research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Anastasios Karadimitris, Centre for Haematology, Department of Medicine, Imperial College London, Hammersmith Hospital, Du Cane Rd, London W12 0NN, United Kingdom; e-mail: a.karadimitris@imperial.ac.uk.
References
Author notes
M.P. and M.S. contributed equally to this study.