Key Points
A novel TESC-NEH1 pathway is involved in FLT3-ITD+ AML pathogenesis.
Inhibition of NHE1 overcomes sorafenib resistance in FLT3-ITD+AML.
Abstract
Internal tandem duplication (ITD) of fms-like tyrosine kinase 3 (FLT3) in acute myeloid leukemia (AML) is associated with inferior clinical prognosis. Sorafenib is effective in clearing leukemic blasts in chemorefractory FLT3-ITD+ AML, but leukemia progression invariably occurs. Mechanisms of drug resistance are not completely understood. We hypothesized that a gene encoding tescalcin (TESC), known to be upregulated at leukemia progression during continuous sorafenib treatment and activate an Na+/H+ exchanger type-1 (NHE1), may underlie tyrosine kinase inhibitor resistance. TESC was highly expressed in FLT3-ITD+ AML lines MOLM-13 and MV4-11, and its knockdown by small-interfering RNA lowered intracellular pH (pHi) and induced apoptosis. The results were recapitulated by treatment with an NHE1 inhibitor, 5-(N,N-hexamethylene) amiloride (HMA). Induction of sorafenib resistance in the MOLM-13 cell line (M13-RE) significantly increased its sensitivity to HMA. The later also enhanced suppression of FLT3 signaling by sorafenib in otherwise resistant cell lines. HMA treatment of MOLM-13 and MV4-11 as well as primary FLT3-ITD+ AML cells significantly reduced leukemia initiation in anti-CD122-primed NOD/SCID mouse xenotransplantation. These observations provided novel information about the pathogenetic role of a TESC-NHE1-pHi axis in mediating sorafenib resistance in AML.
Introduction
Acute myeloid leukemia (AML) is a group of heterogeneous diseases with distinct clinical, biological, cytogenetic, and genetic features. It is characterized by an abnormal increase of myeloblasts in the bone marrow (BM) and peripheral blood (PB). Chemotherapy and allogeneic hematopoietic stem cell (HSC) transplantation are the mainstays of treatment. However, despite an initial remission, leukemia relapses are frequent, with an overall cure rate of only 30% to 40%.1
Internal tandem duplication (ITD) of the fms-like tyrosine kinase 3 (FLT3) gene is one of the commonest gain-of-function mutations in AML, occurring in nearly 30% of cases, particularly in AML with normal cytogenetics, t(6;9) and t(15;17) (acute promyelocytic leukemia). FLT3 is a class III receptor tyrosine kinase highly expressed in human hematopoietic stem and progenitor cells.2 It is activated upon binding to FLT3 ligand, leading to activation of downstream effectors, including RAC-α serine/threonine-protein kinase (AKT) and mitogen-activated protein kinase pathways.3,4 ITD at the juxtamembrane or tyrosine kinase 1 domain disrupts the negative regulatory domain in FLT3, resulting in ligand-independent constitutive activation of FLT35 and hence aberrant activation of signal transducer and activator of transcription (STAT) 5, proto-oncogene tyrosine-protein kinase Src kinase,6 and transcription factor forkhead box O3A.7 Normal and aberrant FLT3 signaling has been extensively reviewed.8,9
FLT3-ITD+ AML has a higher relapse rate and hence inferior disease-free and overall survivals. The prognosis is worse in AML with larger ITD size,10 higher allelic burden,11 and multiple ITD.12 FLT3-ITD as a therapeutic target has been tested in phase 1/2 clinical trials.13-15 In a previous study, we treated FLT3-ITD+ AML with the tyrosine kinase inhibitor (TKI) sorafenib.16 Although there was an initial response, resistance invariably occurred.17 Similar findings were observed in clinical trials of other TKIs, constituting an important limitation to their clinical application.16,18 Genetic analyses of leukemia cells from patients resistant to TKIs including midostaurin,19 sorafenib,16 and quizartinib (AC220)20 have demonstrated evolution of resistant clones carrying tyrosine kinase domain (TKD) mutations in 17% to 75% cases. However, resistant clones without TKD mutation were also frequently encountered, suggesting alternative mechanisms of drug resistance.
Using microarray analysis, we have identified genes that are upregulated when FLT3-ITD+ AML became refractory to sorafenib.16 A working hypothesis is that these upregulated genes are responsible for sorafenib resistance. One of the upregulated genes is tescalcin (TESC). It is an EF-hand Ca2+ binding protein, which is expressed in primary and immortalized human hematopoietic cells21 and is upregulated during differentiation of mouse primary megakaryocytes and human K652 cells. TESC interacts directly with Na+/H+ exchanger type-1 (NHE1) and enhances its maturation, stability, and intracellular transport to the cell surface.22-24 Under normal circumstances, NHE1 takes in 1 extracellular Na+ in exchange for the efflux of an intracellular H+, thereby playing a pivotal role in the regulation of intracellular pH (pHi). Induction of NHE1 activity leads to increased H+ efflux and a subsequent increase in pHi, which has been shown to enhance cell proliferation,25,26 decrease apoptosis,27 increase genome instability,28,29 facilitate cancer metastasis,30 and increase drug resistance in cancer.31-33
In this study, we examined the expression, function, and mechanisms of action of TESC in relationship to changes in pHi and sorafenib resistance in FLT3-ITD+ AML, with a view to defining methods of restoring sorafenib sensitivity in cases that are refractory to it.
Materials and methods
FLT3-ITD+ AML, mobilized PB HSCs, and cord blood cells
In a phase 2 clinical trial, patients with therapy-refractory FLT3-ITD+ AML were treated with sorafenib monotherapy (400 mg twice daily) (supplemental Material 1; see the Blood Web site).16 Paired BM and/or PB samples before sorafenib treatment (sorafenib naïve) and at subsequent leukemia progression (sorafenib resistant) were collected. Normal mobilized PB mononuclear cells (MNCs) were obtained during donation for HSC transplantation. Cord blood (CB) cells were collected immediately after normal cesarean section. Patients and donors gave informed consent, and the procurement of these materials was approved by the institutional review board in accordance with the Declaration of Helsinki.
Cell processing
MNCs from BM and/or PB from AML patients, PB MNCs, and CB cells were collected by density gradient centrifugation (Ficoll-Paque Plus; Amersham Biosciences, Uppsala, Sweden) and stored in liquid nitrogen until use. CD34+ cells were isolated from MNCs immunomagnetically or by fluorescence-activated cell sorting with an anti-human CD34 fluorescein isothiocyanate–conjugated antibody (Beckman Coulter). Leukemia cell lines used in this study included MOLM-13 and MV4-11 (acute monocytic leukemia with FLT3-ITD) and THP-1 (acute monocytic leukemia).
Generation of sorafenib-resistant FLT3-ITD+ AML cell line
The FLT3-ITD+ AML line MOLM-13 was cultured at increasing concentrations of sorafenib, starting from 1 nM to 50 nM, generating sorafenib-resistant MOLM-13 cells (M13-RE). MOLM-13 cultured in parallel with diemethylsulfoxide (DMSO, solvent for sorafenib) was used as control.
In vitro treatment of AML cells
Leukemia cell lines and FLT3-ITD+ primary AML cells were treated with the NHE1 inhibitor 5-(N,N-hexamethylene) amiloride (HMA) (10 nM to 10 μM) (Sigma-Aldrich, St. Louis, MO) and with sorafenib (1 nM to 1 μM) (LC Laboratories, Woburn, MA) for 3 days. DMSO (0.1%) was used as vehicle control. Viable cells after treatment were enumerated by AccuCheck Counting Beads (Life Technologies, Grand Island, NY) based on negative propidium iodide staining.
Transfection
Small-interfering RNAs (siRNAs) targeting TESC (Qiagen, Venlo, The Netherlands) were transfected into MOLM-13 and MV4-11 cells using the Neon Transfection System (Life Technologies) (supplemental Material 2). Scramble siRNA (Thermo Fisher Scientific) was employed as control.
Apoptosis analysis
The proportion of apoptotic cells was evaluated by the PE Annexin V Apoptosis Detection Kit I (BD Bioscience). Briefly, leukemic cells (1 × 105) were suspended in the annexin V binding buffer (1X, 100 μL) and incubated with phycoerythrin-conjugated annexin V and 7-aminoactinomycin D mixture for 15 minutes at room temperature. Binding buffer (1X, 200 μL) was then added, followed by flow cytometric analysis.
Reverse transcription polymerase chain reaction (PCR) and quantitative PCR
Total RNA of mononuclear or CD34+ cells was extracted with TRIZOL reagent (Life Technologies) and reverse transcribed with SuperScript II (Life Technologies). Quantitative PCR was performed with the Power SYBR Green assay (StepOnePlus Real-Time PCR System; Life Technologies) (supplemental Material 2), using the 2[−ΔΔC(T)] method against an internal control gene ACTINB.
Measurement of pHi
Leukemia cells were washed with 1X phosphate-buffered saline (PBS), incubated with 2.5 μM SNARF-1 carboxylic acid, acetate, succinimidyl ester (Life Technologies) at 1 × 106/mL at 37°C for 20 minutes, centrifuged, and resuspended in 1X PBS. Calibration of pHi and its subsequent measurement was described in supplemental Material 3.
Colony-forming assay
The clonogenicity of leukemia cells was evaluated by standard methylcellulose-based culture (MethoCult; Stem Cell Technologies, Vancouver, British Columbia, Canada). Leukemia cells were seeded at 100 cells per mL in triplicates, and colonies were evaluated after 10 days of culture.
Xenotransplantation
MNCs of primary BM or PB FLT3-ITD+ AML samples, leukemia cell lines, and CD34+ cells from normal CB were injected IV into sublethally irradiated (250 cGy) 6- to 8-week-old NOD/SCID mice primed with an anti-CD122 antibody (200 μg, intraperitoneal injection).34 Xenogeneic transplantation was approved by the Committee on the Use of Live Animals for Teaching and Research of the University of Hong Kong.
Flow cytometry analysis of engraftment
Engraftment was defined by the presence of human leukemia/hematopoietic cells in the recipient mouse marrow by 9 weeks. Protocols for its enumeration were described in supplemental Material 3.
Immunoprecipitation and immunoblotting
To examine TESC protein expression, AML cells were lysed in radio-immunoprecipitation assay lysis buffer with protease and phosphatase inhibitors. In the immunoprecipitation experiments, sorafenib-sensitive and sorafenib-resistant MOLM-13 and MV4-11 cells were lysed in Tergitol-Type NP-40 lysis buffer with protease and phosphatase inhibitors and immunoprecipitated with rabbit anti-human FLT3 antibody (S18; Santa Cruz Biotechnology) and recombinant Protein G–Sepharose 4B (Life Technologies). Immuoprecipitated samples and whole cell lysate were separated, transferred, and blotted with primary and horseradish peroxidase–conjugated secondary antibodies (supplemental Material 4). Hybridization signals were visualized with Amersham enhanced chemiluminescence western blot detection reagents (GE Healthcare) or Luminata Forte Western horseradish peroxidase substrate (Millipore) and evaluated by the ChemiDoc XRS+ System (Bio-Rad). Densitometric analysis of the bands was performed by ImageJ 1.47v.
Phosphoflow analysis
Cells (1 × 106) were suspended and fixed in 1 mL Hanks balanced salt solution (GIBCO) containing 2% fetal bovine serum (GIBCO) and 1.45% paraformaldehyde (P6148; Sigma-Aldrich) for 10 minutes at room temperature followed by centrifugation at 500g for 6 minutes at 4°C. Supernatant was decanted, and tubes were blotted dry. The pellet was resuspended in 1 mL cold methanol and stored at −80°C overnight until further analysis.35 The protocols of antibody staining were described in supplemental Material 3.
Sorafenib uptake assay
[3H]sorafenib was custom-made by Moravek Biochemicals (CA).36 MOLM-13 and MV4-11 were either pretreated with HMA (10 μM) or its vehicle control (0.1% DMSO) for 1 hour. Thereafter, [3H]sorafenib (1 and 10 nM) or the vehicle control (0.1% DMSO) was added and incubated for 2 hours. The cells were then washed twice with 1X PBS and centrifuged at 1200 rpm. Supernatant was discarded, and the cell pellets were lysed with 0.1% Triton X-100 PBS. Intracellular accumulation of [3H]sorafenib was measured using a liquid scintillation analyzer TRI-CARB 2900TR (PerkinElmer, MA) and normalized to the amount of protein in the cell pellet as measured by DC Protein Assay (Bio-Rad).
Statistics
All experiments were performed in at least triplicate, and results were expressed as mean ± standard error of the mean. Groups of data were compared by Student t test. A P value of < .05 was considered statistically significant. Excess over highest single agent was obtained by the difference between the percentage inhibition of combined treatment and the higher percentage inhibition of the 2 single agents. Excess over Bliss additivism was obtained by the difference between the observed and the expected (E) percentage inhibition of combined treatment. E is calculated as follows: E = A + B − AB, where A and B are the percentage inhibition of single agent A and B.37
Results
TESC knockdown suppressed growth of FLT3-ITD+ AML cells
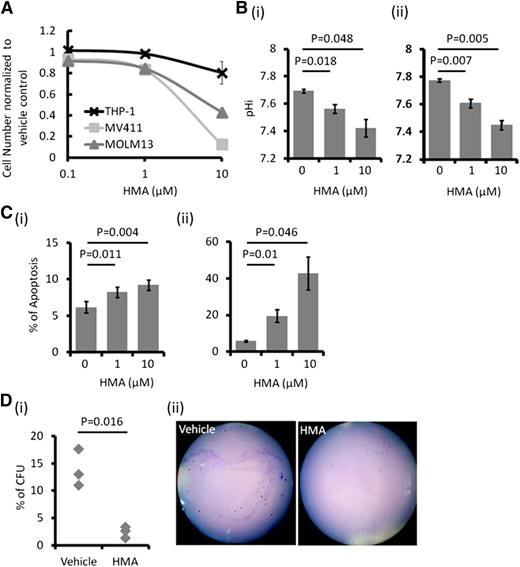
TESC was highly expressed in 2 FLT3-ITD+ AML cell lines, MOLM-13 and MV4-11, as well as in FLT3 wild-type (WT) cell lines, including K562, KG1, and ML2 (supplemental Figure 1A-B). However, THP-1 cells expressed very a low level of TESC. Transfection of MOLM-13 and MV4-11 with siRNA targeting TESC resulted in downregulation of TESC messenger RNA (supplemental Figure 1C) and protein (Figure 1A). TESC knockdown was associated with significant growth inhibition (Figure 1Bi-ii), decrease in pHi (Figure 1Biii-iv), and increase in apoptosis (Figure 1Bv-vi).
Effect of TESC knockdown on AML cell lines. (A) TESC was successfully knocked down by siRNA in (i) MOLM-13 and (ii) MV4-11 as shown by western blot. (B) TESC knockdown significantly reduced the growth of (i) MOLM-13 and (ii) MV4-11 6 days posttransfection, accompanied by intracellular acidification and apoptosis induction in MOLM-13 (iii and v) and MV4-11 (iv and vi). n.s., no significant difference as defined by P > .05; SCR, scrambled siRNA; TC, transfection control, no plasmid; TS3, siRNA3; TS4, siRNA4; U, untransfected.
Effect of TESC knockdown on AML cell lines. (A) TESC was successfully knocked down by siRNA in (i) MOLM-13 and (ii) MV4-11 as shown by western blot. (B) TESC knockdown significantly reduced the growth of (i) MOLM-13 and (ii) MV4-11 6 days posttransfection, accompanied by intracellular acidification and apoptosis induction in MOLM-13 (iii and v) and MV4-11 (iv and vi). n.s., no significant difference as defined by P > .05; SCR, scrambled siRNA; TC, transfection control, no plasmid; TS3, siRNA3; TS4, siRNA4; U, untransfected.
NHE1 inhibition also suppressed FLT3-ITD+ AML cells
Untreated AML cells showed higher pHi than normal hematopoietic cells, which was related to higher NHE1 activity (supplemental Figure 1D). NHE1 inhibition by HMA suppressed growth in different leukemia cell lines (Figure 2A), with the effect being greatest in the FLT3-ITD+ MOLM-13 and MV4-11 cells (50% inhibition concentration [IC50] of 6.38 and 5.33 μM, respectively). Suppression of cellular growth correlated with a dose-dependent decrease in pHi and induction of apoptosis (Figure 2B-C) and a significant reduction of colony formation (Figure 2D). HMA was ineffective in suppressing THP-1 cell growth (Figure 2A). This was correlated with a lack of effect on pHi, apoptosis, and colony formation in this cell line (supplemental Figure 2). These observations supported the proposition that NHE1 activity, and hence the pHi, was mechanistically linked to apoptosis and cellular proliferation.
In vitro effect of HMA on AML cell lines. (A) MOLM-13 and MV4-11 were most sensitive to HMA compared with THP-1. (B) HMA reduced pHi in (i) MOLM-13 and (ii) MV4-11 in a dose-dependent manner. (C) HMA also induced apoptosis in (i) MOLM-13 and (ii) MV4-11 cells. (D) In MV4-11 cells, the number of colony-forming cells was significantly reduced by HMA treatment compared with vehicle control. (ii) Representative photo showing the reduced number of colony in HMA-treated MV4-11 cells.
In vitro effect of HMA on AML cell lines. (A) MOLM-13 and MV4-11 were most sensitive to HMA compared with THP-1. (B) HMA reduced pHi in (i) MOLM-13 and (ii) MV4-11 in a dose-dependent manner. (C) HMA also induced apoptosis in (i) MOLM-13 and (ii) MV4-11 cells. (D) In MV4-11 cells, the number of colony-forming cells was significantly reduced by HMA treatment compared with vehicle control. (ii) Representative photo showing the reduced number of colony in HMA-treated MV4-11 cells.
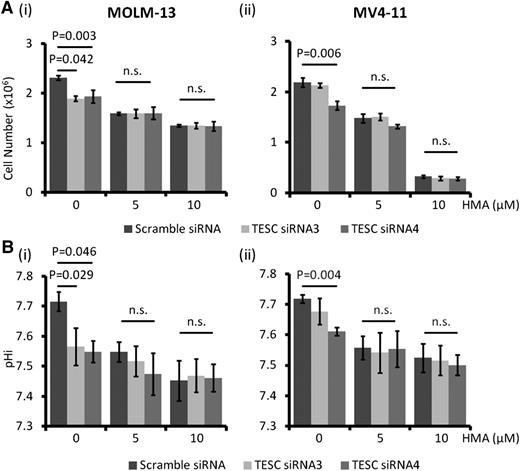
TESC might act through NHE1
TESC knockdown in MOLM-13 and MV4-11 resulted in a decrease in cell number and pHi, and treatment with HMA led to further responses. On the other hand, HMA treatment significantly reduced leukemia growth and pHi in MOLM-13 and MV4-11 and completely abolished the cellular responses to further TESC knockdown (Figure 3A-B), supporting the proposition that both interventions perturbed NHE1 activity as a common target.
NHE1 being the target of TESC in AML. HMA treatment completely abolished the growth inhibitory effects (A) and intracellular acidification (B) of TESC knockdown in MOLM-13 (Ai, Bi) and MV4-11 (Aii, Bii). n.s., no significant difference as defined by P > .05.
NHE1 being the target of TESC in AML. HMA treatment completely abolished the growth inhibitory effects (A) and intracellular acidification (B) of TESC knockdown in MOLM-13 (Ai, Bi) and MV4-11 (Aii, Bii). n.s., no significant difference as defined by P > .05.
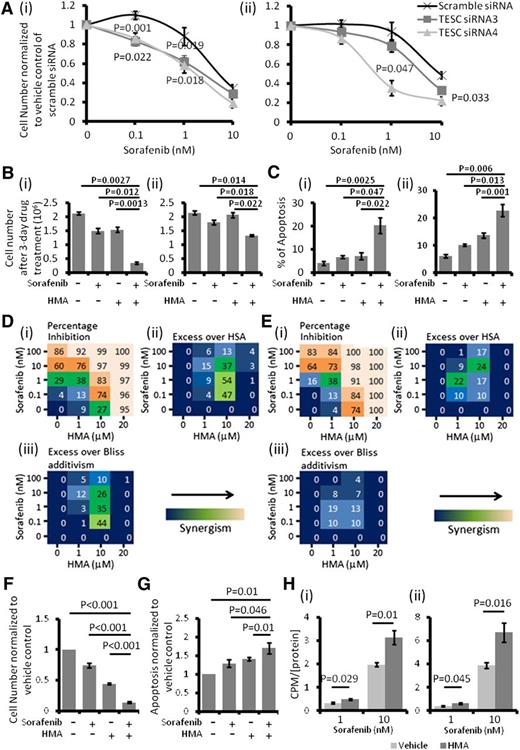
Perturbation of TESC/NHE1 increased sensitivity of leukemia cells to sorafenib
TESC knockdown significantly enhanced the cytotoxicity of sorafenib in MOLM-13 and MV4-11 cells (Figure 4A). Expectedly, HMA treatment in MOLM-13 and MV4-11 cells also significantly increased their sensitivity to sorafenib treatment (Figure 4B-C). Based on inhibition of leukemia growth at different drug doses in combination, synergism between HMA and sorafenib could be demonstrated in both MOLM-13 and MV4-11 cells (Figure 4D-E).37 These effects were recapitulated in primary FLT3-ITD+ AML samples (Figure 4F-G). Importantly, HMA treatment significantly increased the intracellular uptake of [3H]sorafenib into MOLM-13 and MV4-11 cells, providing a potential mechanism for the synergism between HMA and sorafenib (Figure 4H).
NHE1 inhibition resulted in sorafenib sensitivity induction. (A) TESC knockdown significantly potentiated the cytotoxic effect of sorafenib in (i) MOLM-13 and (ii) MV4-11. (B) HMA and sorafenib synergistically inhibited the growth of (i) MOLM-13 and (ii) MV4-11. (C) Combination of HMA and sorafenib also induced more apoptosis in (i) MOLM-13 and (ii) MV4-11 compared with single agent and vehicle control. The concentrations of sorafenib and HMA used in MOLM-13 (Bi and Ci) were 1 nM and 10 μM and in MV4-11 (Bii and Cii) were 1 nM and 1 μM. (Di) Effects of HMA and sorafenib-mediated percentage growth inhibition in MOLM-13. Each number represented the mean of triplicate experiments. (ii) Difference in percentage growth inhibition between combination treatment and either sorafenib or HMA alone, whichever had a stronger effect. HSA, highest single agent. (iii) Difference in percentage growth inhibition between combination treatment and the multiplication product of growth inhibition by each treatment alone (excess over Bliss additivism). The difference reflected the magnitude of the synergism as shown by the scale bar. (Ei-iii) The results obtained from MV4-11. (F) Combination of HMA and sorafenib enhanced the growth inhibitory and (G) apoptosis effects of primary FLT3-ITD+ AML samples. The average results of 5 primary samples were shown. (H) Pretreatment with HMA (10 μM) significantly upregulated the intracellular level of sorafenib in both (i) MOLM-13 and (ii) MV4-11. The count per minute (CPM) was normalized by the amount of protein in the samples.
NHE1 inhibition resulted in sorafenib sensitivity induction. (A) TESC knockdown significantly potentiated the cytotoxic effect of sorafenib in (i) MOLM-13 and (ii) MV4-11. (B) HMA and sorafenib synergistically inhibited the growth of (i) MOLM-13 and (ii) MV4-11. (C) Combination of HMA and sorafenib also induced more apoptosis in (i) MOLM-13 and (ii) MV4-11 compared with single agent and vehicle control. The concentrations of sorafenib and HMA used in MOLM-13 (Bi and Ci) were 1 nM and 10 μM and in MV4-11 (Bii and Cii) were 1 nM and 1 μM. (Di) Effects of HMA and sorafenib-mediated percentage growth inhibition in MOLM-13. Each number represented the mean of triplicate experiments. (ii) Difference in percentage growth inhibition between combination treatment and either sorafenib or HMA alone, whichever had a stronger effect. HSA, highest single agent. (iii) Difference in percentage growth inhibition between combination treatment and the multiplication product of growth inhibition by each treatment alone (excess over Bliss additivism). The difference reflected the magnitude of the synergism as shown by the scale bar. (Ei-iii) The results obtained from MV4-11. (F) Combination of HMA and sorafenib enhanced the growth inhibitory and (G) apoptosis effects of primary FLT3-ITD+ AML samples. The average results of 5 primary samples were shown. (H) Pretreatment with HMA (10 μM) significantly upregulated the intracellular level of sorafenib in both (i) MOLM-13 and (ii) MV4-11. The count per minute (CPM) was normalized by the amount of protein in the samples.
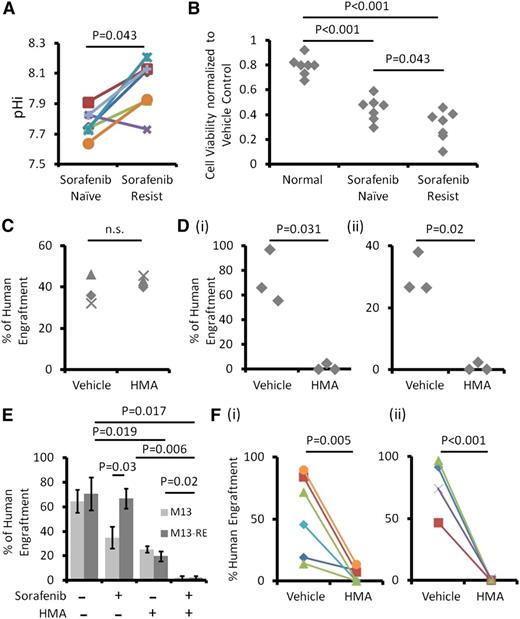
Sorafenib resistance in MOLM-13 cells might be mediated by NHE1
The M13-RE subline of MOLM-13, generated by exposure to increasing concentrations of sorafenib, has an IC50 of sorafenib at 69.9 nM, as compared with 7.8 nM in the parental line (Figure 5A). Sequencing of the entire FLT3 gene by Sanger sequencing did not show acquisition of new FLT3-ITD and TKD mutation in M13-RE (data not shown). Restriction enzyme digestion was also performed, showing the absence of the commonest TKD D835 mutation (Figure 5B). M13-RE was also resistant to ponatinib and quizartinib, both potent inhibitors of FLT3 (supplemental Figure 3A-B). Compared with MOLM-13, M13-RE showed much higher pHi (Figure 5C) and was more sensitive to the growth inhibitory effect of HMA treatment (Figure 5D), with a larger drop in pHi (Figure 5E) and an increase in apoptosis (Figure 5F). Interestingly, although sorafenib was ineffective in M13-RE, it significantly enhanced the growth inhibition and apoptosis induced by HMA (Figure 6A-B). Sorafenib significantly suppressed FLT3 signaling via STAT5, AKT, and extracellular signal-regulated kinases 1/2 phosphorylation in MOLM-13 and MV4-11 but not in M13-RE (Figure 6C-E and supplemental Figure 4A-B). However, in the presence of HMA, sorafenib exerted comparable effects on MOLM-13 and M13-RE (Figure 6C-E; supplemental Figure 3B for quizartinib and ponatinib). These results suggested that NHE1 played an important part in mediating sorafenib resistance in M13-RE, with its inhibition resulting in potentiation of the actions of sorafenib.
Sorafenib-resistant MOLM-13 cells were more sensitive to HMA. (A) Compared with M13, M13-RE showed an increase in IC50 to sorafenib. (B) No novel ITD and D835Y mutation in M13-RE. Detection of D835Y has been described previously.16 M13-RE cells also showed (C) higher pHi, (D) higher sensitivity to HMA in term of growth inhibition, (E) intracellular acidification, and (F) apoptosis.
Sorafenib-resistant MOLM-13 cells were more sensitive to HMA. (A) Compared with M13, M13-RE showed an increase in IC50 to sorafenib. (B) No novel ITD and D835Y mutation in M13-RE. Detection of D835Y has been described previously.16 M13-RE cells also showed (C) higher pHi, (D) higher sensitivity to HMA in term of growth inhibition, (E) intracellular acidification, and (F) apoptosis.
Effects of HMA on sorafenib-resistant cell line M13-RE. (A) Sorafenib (10 nM) and HMA (10 μM) inhibited leukemia growth and (B) induced apoptosis of M13 (fold change normalized to vehicle control) with combined sorafenib and HMA treatment resulting in more pronounced inhibition. Note that HMA but not sorafenib inhibited leukemia growth and induced apoptosis in M13-RE. (C) In M13, HMA modestly inhibited FLT3 signaling and potentiated sorafenib-induced inhibition of downstream signaling. (D) M13-RE was resistant to sorafenib but sensitive to the combination of HMA and sorafenib. Densitometric analysis of the bands was performed by ImageJ. The ratios of phosphorylated to total proteins are shown at the top of each lane. (E) Effects of HMA (10 μM) and sorafenib (10 nM) on FLT3 signaling (i, p-STAT; ii, p-AKT; iii, p-ERK [extracellular signal-regulated kinase]) evaluated quantitatively by phosphoflow. Sorafenib induced significantly stronger inhibition in M13 than M13-RE. Combination of sorafenib with HMA resulted in significant inhibition of FLT3 signaling in both M13 and M13-RE.
Effects of HMA on sorafenib-resistant cell line M13-RE. (A) Sorafenib (10 nM) and HMA (10 μM) inhibited leukemia growth and (B) induced apoptosis of M13 (fold change normalized to vehicle control) with combined sorafenib and HMA treatment resulting in more pronounced inhibition. Note that HMA but not sorafenib inhibited leukemia growth and induced apoptosis in M13-RE. (C) In M13, HMA modestly inhibited FLT3 signaling and potentiated sorafenib-induced inhibition of downstream signaling. (D) M13-RE was resistant to sorafenib but sensitive to the combination of HMA and sorafenib. Densitometric analysis of the bands was performed by ImageJ. The ratios of phosphorylated to total proteins are shown at the top of each lane. (E) Effects of HMA (10 μM) and sorafenib (10 nM) on FLT3 signaling (i, p-STAT; ii, p-AKT; iii, p-ERK [extracellular signal-regulated kinase]) evaluated quantitatively by phosphoflow. Sorafenib induced significantly stronger inhibition in M13 than M13-RE. Combination of sorafenib with HMA resulted in significant inhibition of FLT3 signaling in both M13 and M13-RE.
Sorafenib-resistant primary AML cells were also dependent on NHE1-mediated increase in pHi for proliferation and survival
In FLT3-ITD+ AML patients, sorafenib-resistant cells showed significantly higher pHi than the corresponding sorafenib-naïve ones (Figure 7A). NHE1 inhibition by HMA (10 μM) also suppressed the growth of sorafenib-resistant AML cells to a significantly larger extent than sorafenib-naïve cells, whereas normal hematopoietic cells were unaffected by HMA at this concentration (Figure 7B). Moreover, HMA induced more apoptosis in sorafenib-resistant samples (supplemental Figure 4C). These results suggested that the proliferation and survival of sorafenib-resistant FLT3-ITD+ AML cells, as compared with sorafenib-naïve AML and normal hematopoietic cells, were more dependent on NHE1.
Primary sorafenib-resistant AML samples were dependent on NHE1 for survival. (A) Six out of 7 sorafenib-resistant AML samples exhibited higher pHi compared with their sorafenib-naïve counterparts. (B) HMA had minimal suppressive effects on normal hematopoietic cells but significantly suppressed sorafenib-resistant myeloblasts and to a lesser extent sorafenib-naïve AML cells. (C) HMA treatment had no effect on the engrafting potential of HSCs from umbilical CB. (D) HMA treatment at the same dose in (i) MOLM-13 and (ii) MV4-11 nearly abolished leukemia engraftment in NOD/SCID mice. (E) Both HMA and sorafenib reduced leukemia engraftment by MOLM-13 (blue column), and their combination completely abolished the engraftment. For M13-RE (red column), sorafenib treatment had no effect on human engraftment. HMA reduced leukemia engraftment and when combined with sorafenib completely abolished leukemia engraftment. (F) HMA significantly reduced human engraftment from (i) sorafenib-naïve and (ii) sorafenib-resistant primary FLT3-ITD+ AML compared with the vehicle control.
Primary sorafenib-resistant AML samples were dependent on NHE1 for survival. (A) Six out of 7 sorafenib-resistant AML samples exhibited higher pHi compared with their sorafenib-naïve counterparts. (B) HMA had minimal suppressive effects on normal hematopoietic cells but significantly suppressed sorafenib-resistant myeloblasts and to a lesser extent sorafenib-naïve AML cells. (C) HMA treatment had no effect on the engrafting potential of HSCs from umbilical CB. (D) HMA treatment at the same dose in (i) MOLM-13 and (ii) MV4-11 nearly abolished leukemia engraftment in NOD/SCID mice. (E) Both HMA and sorafenib reduced leukemia engraftment by MOLM-13 (blue column), and their combination completely abolished the engraftment. For M13-RE (red column), sorafenib treatment had no effect on human engraftment. HMA reduced leukemia engraftment and when combined with sorafenib completely abolished leukemia engraftment. (F) HMA significantly reduced human engraftment from (i) sorafenib-naïve and (ii) sorafenib-resistant primary FLT3-ITD+ AML compared with the vehicle control.
The effects of NHE1 inhibition on leukemia initiation
HMA treatment (10 μM) for 3 days had no significant effects on engraftment and multilineage differentiation of normal hematopoietic cells in NOD/SCID mice (Figure 7C and supplemental Figure 5A). However, HMA treatment at the same dose and treatment duration abolished the engraftment of MOLM-13 and MV4-11 cells (Figure 7D and supplemental Figure 5B-C). Sorafenib also reduced the engraftment by the M13 but not the M13-RE cell line (Figure 7E). In both cell lines, HMA (5 μM) also significantly reduced their engraftment. Importantly, when combined with sorafenib, engraftment was completely abolished, suggesting that HMA treatment had restored sorafenib sensitivity even in the M13-RE cell line. HMA (10 μM) nearly abolished leukemia engraftment by both sorafenib-naïve (Figure 7Fi) and sorafenib-resistant (Figure 7Fii) FLT3-ITD+ AML samples.
Discussion
In this study, we demonstrated a link between TESC expression, NHE1 activity, pHi, and leukemia progression in FLT3-ITD+ AML during sorafenib resistance. The results were consistent with emerging evidence that NHE1 plays an important role in cancer progression and metastasis by maintaining a higher pHi permissive for cellular proliferation, antiapoptosis, and drug resistance. The cytoplasmic domain of NHE1 provides the docking site for various signaling and cytoskeletal proteins, resulting in its constitutive activation and hence a higher set point of pHi in cancer cells.38 An increased pH gradient across the plasma membrane may also activate extracellular proteases and enhance cellular motility. In addition, our observations have provided unique insights into the roles of TESC and NHE1 in FLT3-ITD+ AML, which may have important implications for the design of therapeutic strategies to overcome resistance against TKIs.
First, we demonstrated for the first time a pathogenetic role of TESC in leukemogenesis. The investigation on TESC was based on its preferential expression in primary FLT3-ITD+ myeloblasts resistant to sorafenib. TESC knockdown in FLT3-ITD+ cell lines MOLM-13 and MV4-11 suppressed leukemia growth, lowered pHi, and induced apoptosis. The results suggested that TESC might play a role in leukemogenesis by maintaining high NHE1 activity, consistent with previous studies showing NHE1 to be the direct target of TESC.22-24 We also demonstrated an additive effect of chemical inhibition of NHE1 activity and gene knockdown of TESC. TESC knockdown in MOLM-13 and MV4-11 resulted in a decrease in cell number and pHi, and treatment with HMA led to further responses. In contrast, direct inhibition of NHE1 abolished the effect of TESC knockdown. The observations supported the proposition that mechanisms in addition to TESC might play a role in the regulation of NHE1. The pathogenetic role of TESC in AML progression might be specific to FLT3-ITD+ AML because its knockdown in the FLT3-WT HEL cell line paradoxically increased cellular proliferation and impaired megakaryocytic differentiation.21 The role of TESC in solid organ tumors is entirely unknown.
Second, we also demonstrated a hitherto undescribed mechanistic link between the TESC-NHE1 pathway and FLT3 signaling in FLT3-ITD+ AML. In particular, TESC knockdown and HMA treatment exhibited synergism with sorafenib in their antileukemia effects in FLT3-ITD+ cell lines and primary AML cells. HMA treatment also enhanced inhibition of FLT3 signaling by sorafenib. Previous studies showed that high pHi might modulate sensitivity to chemotherapeutic agents via changes in intracellular drug concentration.32,39,40 Similar mechanisms might underlie sorafenib resistance in FLT3-ITD+ AML. The proposition was supported by the observation that HMA restored the ability of sorafenib to inhibit FLT3 signaling in the resistant M13-RE line. In fact, based on the acid dissociation constant (pKa) of sorafenib, HMA at 10 and 20 μM lowered pHi in MOLM-13 to an extent that might result in 1.45- and 1.86-fold increase in intracellular sorafenib concentration (supplementary Material 5),32 providing the mechanistic basis for its ability to overcome sorafenib resistance in FLT3-ITD+ AML. Importantly, this was demonstrated in the sorafenib uptake assay where HMA treatment (10 μM) increased [3H]sorafenib uptake by 1.6-fold (Figure 4H) in MOLM-13. Whether overexpression of TESC or NHE1 could induce sorafenib resistance in otherwise sensitive FLT3-ITD+ AML should be evaluated.
Third, data from xenotransplantation suggested that the TESC-NHE1 pathway might operate at the level of leukemia-initiating cells (LICs). In fact, HMA treatment at 10 μM for 3 days totally abolished LIC activity in both MOLM-13 and MV4-11 cell lines as well as both sorafenib-naïve and sorafenib-resistant primary AML samples. The effects were specific to leukemia cells because the engrafting potential of hematopoietic cells from CB when treated with the same dose of HMA were unaffected. Synergism between NHE1 and FLT3 inhibition was also observed, and HMA at 5 μM when combined with sorafenib nearly abolished LIC activity in both sorafenib-sensitive and sorafenib-resistant MOLM-13 lines.
These results might provide clues for the design of a therapeutic strategy based on NHE1 inhibition to overcome sorafenib resistance. Although clinical trials on the use of NHE1 inhibitor in cancer treatment have yet to emerge at present, the pathogenic roles of NHE1 in solid organ malignancies41-43 and sorafenib-resistant FLT3-ITD+ AML as reported in the present study imply that NHE1 inhibition as adjunctive or monotherapy could be explored. HMA was 10 times more potent than its prototype amiloride, which is commonly used as a potassium-sparing diuretic. Whether amiloride at therapeutic doses would exhibit antileukemia effects remains to be determined. HMA was 100-fold more specific to NHE1 than the other 5 members of the NHE family.44 Furthermore, HMA in micromolar concentrations in vivo has been shown to protect heart and brain injury against ischemia or reperfusion injury in rats.38 We also demonstrated that HMA at 10 μM had no effect on the stem cell activity of normal CB CD34+ cells (Figure 7C). Therefore, we believe that micromolar doses of HMA would be pharmacologically relevant for it (or its analog) to be exploited for future therapeutic consideration. More potent and specific NHE1 inhibitors that can be applied clinically should also be evaluated.45
A number of outstanding issues remain to be resolved. First, siRNA3 did not work well in MV4-11 cells, despite our effort to optimize the transfection protocol. The less effective siRNA could be because of its instability and low transfection efficiency in a sequence- and cell-specific manner. As a result, we designed siRNA4, which showed superior knockdown efficiency and more significant cellular effects in both MOLM-13 and MV4-11 cell lines. Second, the regulation of NHE1 in AML has not been fully defined. An increase in pHi was shown in all AML cell lines compared with normal hematopoietic cells, including THP-1, which exhibited a very low level of TESC. Furthermore, in contrast to primary sorafenib-resistant FLT3-ITD+ AML samples, TESC was not upregulated in M13-RE (supplemental Figure 6; the latter nevertheless exhibited a significant increase in NHE1 activity. The apparent discrepancy was not unexpected because the regulation of TESC expression might depend on leukemia cell types. In fact, the cell line model has facilitated evaluation of the mechanistic link between sorafenib resistance, NHE1 activity, and pHi, as well as the synergism between sorafenib and HMA. The difference between primary AML samples and cell lines also highlighted the multifaceted and context-dependent mechanism whereby NHE1 might be regulated. Constitutively active NHE1 activity could be maintained by multiple signaling pathways involved in cancer progression and metastasis.30,43 Moreover, HMA also reduced leukemia growth, albeit less significantly, in AML cell lines carrying WT FLT3 (C.H.M. and A.Y.H.L., unpublished), and inhibition of NHE1 has also been shown to induce differentiation in K562 cells that carry the BCR-ABL1 fusion gene.46 Interestingly, NHE1 transcript expression was upregulated in imatinib-resistant K562 cells.31 To our knowledge, association between sorafenib and TESC-NHE1 beyond its link to FLT3-ITD has not been described. Therefore, the potential of NHE1 inhibitor as adjunctive therapy to conventional chemotherapy in FLT3-WT AML should be explored. In addition, the mechanisms underlying TESC upregulation in FLT3-ITD+ AML are currently unclear. In mice, the ZF5 motif was identified in the TESC promoter, which was critical for its activation.47 The ZF5 motif was found to colocalize with BRCA1 in humans and c-myc in mice.48,49 To date, the relationship between TESC and BRCA1 or C-MYC in humans is still unknown. It has also remained unclear if TESC upregulation might occur in patients who developed drug resistance to newer and more specific FLT3 inhibitors like midostaurin and quizartinib.
These limitations notwithstanding, the present study provided an important mechanistic model (Figure 8) whereby novel therapeutic strategies could be further evaluated. In sorafenib-resistant AML, TESC upregulation resulted in activation of NHE1 and increase in pHi, which led to a significant decrease in intracellular sorafenib availability. Increase in pHi might also increase cellular proliferation and cell-cycle progression via cyclin-dependent kinase 1–cyclin B150 and enhanced glycolysis.51 NHE1 might also cross-talk with the canonical Wnt pathway, underscoring its multifaceted actions on leukemia progression.52 HMA inhibited NHE1 and resulted in intracellular acidification. The latter might increase the intracellular sorafenib level and hence FLT3 inhibition. Lowering pHi might also suppress FLT3 activity directly and induce apoptosis by activation of caspase,27 the proapoptotic protein BCL2-associated X protein,53 and endonuclease DNase II associated with DNA fragmentation.54
Proposed role of TESC/NHE1/pH axis in sorafenib-resistant AML. (A) In sorafenib-resistant AML, TESC was significantly upregulated, leading to increased activity of NHE1, more efflux of H+, and hence higher pHi, increased cellular proliferation and antiapoptosis, reduced intracellular sorafenib level, and unopposed FLT3 activity. The question mark represents a yet to be identified mechanism of NHE1 activity in addition to TESC. (B) With HMA, NHE1 activity was reduced, resulting in lower pHi, inhibition of proliferation and apoptosis induction, increased intracellular sorafenib level, and more FLT3 inhibition. Lower pHi also reduced FLT3 signaling directly.
Proposed role of TESC/NHE1/pH axis in sorafenib-resistant AML. (A) In sorafenib-resistant AML, TESC was significantly upregulated, leading to increased activity of NHE1, more efflux of H+, and hence higher pHi, increased cellular proliferation and antiapoptosis, reduced intracellular sorafenib level, and unopposed FLT3 activity. The question mark represents a yet to be identified mechanism of NHE1 activity in addition to TESC. (B) With HMA, NHE1 activity was reduced, resulting in lower pHi, inhibition of proliferation and apoptosis induction, increased intracellular sorafenib level, and more FLT3 inhibition. Lower pHi also reduced FLT3 signaling directly.
In conclusion, this present study demonstrated a novel mechanistic link between TESC, NHE1, pHi and sorafenib resistance in FLT3-ITD+ AML. NHE1 may be a potential target in the treatment of this disease.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr T. Tanaka, Hyogo, University of Health Sciences, Kobe, Japan, for the generous gift of the anti-CD122 antibody.
This work was supported by grants from the S. K. Yee Medical Foundation, the Hong Kong Blood Cancer Foundation, and the General Research Fund (HKU771611). A.Y.H.L. is the Li Shu Fan Medical Foundation Professor in Haematology and received funding from the endowment.
Authorship
Contribution: C.H.M. conducted experiments, analyzed data, and wrote the manuscript; S.S.Y.L., M.K.H.S., H.C.H.C., and H.G. conducted experiments; Y.L.K. analyzed data and wrote the manuscript; and A.Y.H.L. designed the research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Anskar Y. H. Leung, Room K418, K Block, Department of Medicine, Queen Mary Hospital, Pokfulam Rd, Hong Kong, Special Administrative Region, China; e-mail: ayhleung@hku.hk.



![Figure 6. Effects of HMA on sorafenib-resistant cell line M13-RE. (A) Sorafenib (10 nM) and HMA (10 μM) inhibited leukemia growth and (B) induced apoptosis of M13 (fold change normalized to vehicle control) with combined sorafenib and HMA treatment resulting in more pronounced inhibition. Note that HMA but not sorafenib inhibited leukemia growth and induced apoptosis in M13-RE. (C) In M13, HMA modestly inhibited FLT3 signaling and potentiated sorafenib-induced inhibition of downstream signaling. (D) M13-RE was resistant to sorafenib but sensitive to the combination of HMA and sorafenib. Densitometric analysis of the bands was performed by ImageJ. The ratios of phosphorylated to total proteins are shown at the top of each lane. (E) Effects of HMA (10 μM) and sorafenib (10 nM) on FLT3 signaling (i, p-STAT; ii, p-AKT; iii, p-ERK [extracellular signal-regulated kinase]) evaluated quantitatively by phosphoflow. Sorafenib induced significantly stronger inhibition in M13 than M13-RE. Combination of sorafenib with HMA resulted in significant inhibition of FLT3 signaling in both M13 and M13-RE.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/16/10.1182_blood-2013-07-512194/4/m_2530f6.jpeg?Expires=1769124394&Signature=AltG-jmZJLn2XQrMANlalDPF4CcuEu~RsxDv7cMWw~UIcXHGoZLRjrJ0fKltFd5G~TV3gDlGDsLEWc8x03nNbuzSyKhPLO4nzefMujx6J9Wc36fCXkjh~T7wLxta7FiaCbF4ppHES5uNYHlsVdC-cKLkWJ~C6XEP7icPpHQu9gZwvzDsaOykjE6eU~CfSZ9rip~5h9GAGGbclqKl1t4sn9KGm2bqdaNFsk3TW251o2hwBKg~iDFAiMrrlrzpxEP0xu2r4-PLHiuYNPPsa5ghjAQqDv-h2hJi~0ERXJeCFTy1IXPKfw-fIbAujNF4eGLyagnj8eItxsN6uWb-gXNa2Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal