Key Points
Stat1 deletion in the presence of JAK2-V617F alters phenotypic manifestations by reducing megakaryopoiesis and favoring erythropoiesis.
IFNγ is elevated in serum of mice with thrombocytosis and in patients with essential thrombocythemia and may drive Stat1 activation.
Abstract
The interferon-γ (IFNγ)/signal transducer and activator of transcription 1 (Stat1) pathway shows higher activity in patients with essential thrombocythemia (ET) than in polycythemia vera (PV) and was proposed to be promoting the ET phenotype. We explored the phenotypic consequences of Stat1 deficiency on the effects of Janus kinase 2 (JAK2)–V617F in vivo by crossing mice expressing JAK2-V617F with Stat1 knockout mice. JAK2-V617F;Stat1−/− double transgenic mice showed higher red cell parameters and lower platelet counts compared with JAK2-V617F;Stat1+/+ mice. Bone marrow transplantation reproduced these phenotypic changes in wild-type recipients, demonstrating that the effect of Stat1 is cell-intrinsic and does not require a Stat1-deficient microenvironment. Deletion of Stat1 increased burst-forming unit–erythroid and reduced colony-forming unit–megakaryocyte colony formation driven by JAK2-V617F, but was not sufficient to completely normalize the platelet count. Gata1, a key regulator of megakaryopoiesis and erythropoiesis, was decreased in Stat1-deficient platelets. V617F transgenic mice with thrombocytosis had higher serum levels of IFNγ than normal controls and patients with ET showed higher IFNγ serum levels than patients with PV. Together, these results support the concept that activating Stat1 in the presence of JAK2-V617F, for example, through IFNγ, constrains erythroid differentiation and promotes megakaryocytic development, resulting in ET phenotype.
Introduction
The Janus kinase 2 (JAK2)–V617F mutation is present in ∼80% of patients with myeloproliferative neoplasms (MPNs) and is considered an important driver for the disease.1-4 This G>T point mutation causes a valine-to-phenylalanine substitution in position 617 of the protein (V617F) and provides cytokine hypersensitivity to hematopoietic progenitor and stem cells.5,6 The role of JAK2-V617F in the pathogenesis of MPNs was confirmed in retroviral, transgenic, and knockin mouse models (reviewed in Van Etten et al7 and Li et al8 ). MPNs can manifest in 3 distinct phenotypes, that is, polycythemia vera (PV), essential thrombocythemia (ET), and primary myelofibrosis (PMF). The JAK2-V617F mutation is not only found in almost all patients with PV but also in ∼50% of patients with ET and PMF.5,6 This situation raises the question of how a single mutation results in distinct clinical presentations. Several factors influencing the phenotypic expression have been proposed, including the presence of a subclone homozygous for JAK2-V617F,9 expression levels of the mutant JAK2-V617F allele in mice,10 and the activity of the interferon and signal transducer and activator of transcription 1 (Stat1) signaling pathways.11
Compelling evidence has been published stressing the fact that Stat1 may promote megakaryopoiesis.12 Moreover, the erythropoietin (Epo)–dependent activation of Stat1 may be critical to normal erythroid differentiation.13 Stat1-deficient mice have altered erythropoiesis, delayed erythroid differentiation in bone marrow (BM) and spleen, and a net decrease in total body colony-forming unit–erythroid (CFU-E) in Stat1−/− mice.14 However, it has been recently shown in burst-forming unit–erythroid (BFU-E) colonies from JAK2-V617F–positive MPN patients that the preferential activation of Stat1 may constrain erythroid differentiation.11 Thus, the consequences of Stat1 activation in JAK2-V617F–positive MPNs are not yet completely understood.
Here, we analyzed the role of Stat1 in determining the MPN phenotype in an in vivo model of JAK2-V617F–driven MPNs. For this purpose, we have used our mouse models of PV and ET in combination with a Stat1 knockout.15 The absence of Stat1 protein reduced the number of platelets by affecting megakaryocytic maturation, reducing myelofibrosis, and promoting erythropoiesis. These features were reproduced upon BM transplantation into wild-type animals. Our results support the role of Stat1 in the pathogenesis of JAK2-V617F–positive ET.
Materials and methods
Transgenic mice
Mice carrying a Cre recombinase–inducible human JAK2-V617F transgene (hereafter called V617F) have been described previously.10 To activate the V617F transgene, the mice were crossed with SclCreER16 or VavCre mice.17 Cre expression in SclCreER;V617F mice was induced by intraperitoneal injection of 2 mg of tamoxifen per day for 5 consecutive days. To generate triple transgenic mice, SclCreER;V617F or VavCre;V617F animals were crossed with Stat1 knockout mice.15 Mice were kept under specified pathogen-free conditions with free access to food and water. All experiments were done in strict adherence to Swiss laws for animal welfare and were approved by the Swiss Cantonal Veterinary Office of Basel-Stadt.
Blood analyses
Blood was collected into EDTA-coated microtainers (BD Biosciences) by tail vein sampling and by cardiac puncture during the takedown procedure. Complete blood counts were determined on the ADVIA120 Hematology Analyzer using the Multispecies Software (Bayer).
BM transplantation assay
Total BM cells were harvested from SclCreER;V617F transgenic mice 10 weeks after induction with tamoxifen or from VavCre;V617F mice. BM cells (2 × 106) were transplanted by tail vein injection into C57BL/6 female recipient mice lethally irradiated with 12 Gy. Blood counts were performed every 4 weeks after transplantation.
Flow cytometric analysis
For acquisition and analysis of erythroid precursors, single-cell suspensions from BM and spleen were stained with phycoerythrin (PE)-conjugated anti-mouse monoclonal antibodies against Ter119, biotin-conjugated anti-mouse antibody against CD71, and allophycocyanin (APC)–conjugated streptavidin antibody (BioLegend). For acquisition and analysis of Lin−Sca1+c-kit+ (LSK) cells, Lin−Sca1−c-kit+ (LK) cells, and megakaryocytic progenitors,18 the samples were stained with anti-mouse lineage depletion biotinylated antibody cocktail (Ter119, Gr1, Mac1, CD5, B220/CD45R) (MagCellect cell enrichment kit; R&D Systems), Pacific Blue–conjugated streptavidin antibody, APC-conjugated anti-mouse monoclonal antibodies against c-Kit (BioLegend), PE-conjugated anti-mouse monoclonal antibodies against Sca1 (BioLegend), PE-Cy7–conjugated anti-mouse monoclonal antibodies against CD150 (BioLegend), and fluorescein isothiocyanate (FITC)–conjugated anti-mouse monoclonal antibodies against CD41 (BioLegend). Dead cells were excluded by gating on the cells negative for propidium iodide. For intracellular detection of pStat3 (Y705) and pStat5 (Y694), 2 × 107 cells in suspension were fixed in 3.6% formaldehyde for 10 minutes at 37°C, permeabilized by incubating in ice-cold 90% methanol for 30 minutes on ice. For analysis, a 200-μL aliquot was incubated with rabbit anti-mouse pStat3 or pStat5 (Cell Signaling) for 1 hour at room temperature, and stained with Alexa Fluor 633–conjugated anti-rabbit secondary antibody. The samples were analyzed on a FACSCalibur flow cytometer (BD Biosciences) and CyAn ADP Analyzer (Beckman Coulter).
Histology
Tibias, femurs, and spleens were fixed in 4% phosphate-buffered formalin, embedded in paraffin, and sectioned. Tissue sections were stained with hematoxylin and eosin (H&E) for morphology or with Gömöri for the analysis of reticulin fibers. Images were taken using an Olympus BX43 microscope (numeric aperture of the objective lenses, 0.75/20× and 0.95/63×) and an Olympus DP73 camera (Olympus Schweiz AG).
Hematopoietic progenitor assays
Erythroid progenitors were assayed in M3436 medium (Stemcell Technologies). BM (3 × 104) or spleen cells (2 × 105) were plated in 35-mm dishes in duplicates. BFU-E colonies were scored after 14 days in culture. CFU-megakaryocyte (CFU-MK) colonies were grown in chamber slides with 50 ng/mL recombinant human thrombopoietin (Tpo) in collagen-based medium (MegaCult-C; STEMCELL Technologies) containing 50 ng/mL interleukin 11 (IL-11), 10 ng/mL recombinant mouse IL-3, and 20 ng/mL recombinant human IL-6 (PeproTech). BM cells (1 × 105) were cultured for 8 days. The slides were then fixed with ice-cold acetone for 5 minutes, stained with acetylcholinesterase for 5 to 6 hours in a humid chamber, fixed with 95% ethanol for 10 minutes, and counterstained with Harris hematoxylin solution. For counting, slides were scanned using 5× and 10× objective lens. CFU-MK colonies were defined as having at least 3 megakaryocytes.
Real-time polymerase chain reaction, allelic ratio and gene expression analysis
Gene expression analysis was performed with Power SYBR Green PCR Master mix on a 7500 Fast machine (Applied Biosystems). Primers for human Jak2, mouse Jak2 were described previously.10 Ratios of human and mouse Jak2 were assessed in the “Absolute Quantification” setup with standard curves made from linearized pMSCV-IRES GFP plasmids containing either human JAK2 or mouse Jak2. The messenger RNA (mRNA) expression of mouse Gata1, Gp1βa, NF-E2 were calculated with the Δ cycle threshold (ΔCT) method with 4 wild-type samples as calibrator, and using the following primers: GAAGCGAATGATTGTCAGCA and TTCCTCGTCTGGATTCCATC (Gata1), GTGCAGAGGGCAAGGCAAGT and TGACTCAGAGCTGAGGGTCG (Gp1βa), CAGGTCTCCACAAGCACAAA and CCAGCCTCTCAGGGACACTA (NF-E2). Mouse Gusb was used for normalization and relative expression.10
Protein lysates and western blot analysis
BM cells were lysed in lysis buffer (Tris HCl 20mM, 1% Triton X-100, NaCl 150mM, EDTA 5mM, 200mM Na3VO4, 200mM phenylarsine oxide with protease and phosphatase inhibitors; Thermo Fisher Scientific Inc). Protein lysates (17 μg for Stat1 and 5 μg for Stat3 and Stat5) were run on 7.5% Tris acetate sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels, transferred to nitrocellulose membranes, and blotted with the indicated antibodies: anti-Stat1 (1:2000, Cell Signaling; or 1:2000, p84/91, Santa Cruz Biotechnology) anti-phospho-Stat3 (1:3000; Cell Signaling), anti-β-Actin (1:500; Santa Cruz Biotechnology), anti-phospho-Stat5 (1:3000; Cell Signaling), and Stat3 (1:1000; Santa Cruz Biotechnology), anti-Stat5 (1:1000; Santa Cruz Biotechnology), and peroxidase-conjugated anti-rabbit immunoglobulin (1:20 000).
Immunoassay for mouse and human interferon-γ plasma concentration
The collection of blood samples was performed at the study center in Basel, Switzerland, and was approved by the Ethik Kommission Beider Basel. Written consent was obtained from all patients in accordance with the Declaration of Helsinki. The diagnosis of MPN was established according to the criteria of the World Health Organization.19-21 Plasma from patients and mice were collected using EDTA as an anticoagulant. The samples were centrifuged for 20 minutes at ∼2000 × g within 30 minutes of collection. The assays were performed using the Proteome Profiler Array kit according the manufacturer’s instruction (R&D Systems).
Platelet isolation
For platelet isolation, blood samples collected into EDTA-coated microtainers were mixed 1:1 with NaCl 0.9%, and centrifuged at low speed without brake (87 × g, 10 minutes, room temperature). Upon spinning, platelet-rich plasma was transferred to a clean tube and 1 mL of Tyrode buffer, pH 7.4 (134mM sodium chloride, 12mM sodium bicarbonate, 2.9mM potasium chloride, 0.34mM sodium phosphate monobasic, 1mM magnesium chloride) was added. The sample was centrifuged, and platelet-rich plasma was transferred to a clean tube. Platelets were pellet by centrifugation at 350 × g, 5 minutes, at room temperature. The pellet was resuspended in peqGold TriFast (PEqLAB Biotechnologie GmbH) for further RNA extraction.
Statistical analysis
Results are presented as means ± standard error of the mean (SEM). To assess the statistical significance among individual cohorts, 1-way analysis of variance (ANOVA) with subsequent Bonferroni posttest (Prism Version 4.00 software; GraphPad) or unpaired t test was used. P ≤ .05 was considered significant.
Results
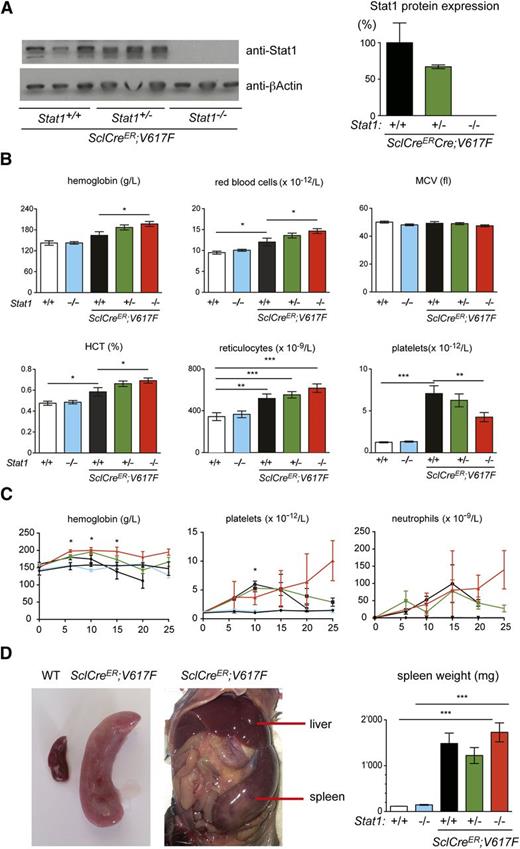
To determine the effects of Stat1 deficiency on the MPN phenotypes, we crossed the Stat1 knockout mice with our Cre-LoxP inducible JAK2-V617F transgenic mice.10,15 Expression of Stat1 was reduced or absent in mice heterozygous and homozygous for the Stat1 knockout, respectively (Figure 1A; supplemental Figure 1A, see supplemental Data available at the Blood Web site). At 10 weeks after injection of tamoxifen, all SclCreER;V617F transgenic mice displayed a PV phenotype with thrombocytosis (Figure 1B). Homozygous loss of Stat1 in V617F transgenic mice increased red cell numbers, whereas platelets were lowered. Marked neutrophilia was observed in V617F transgenic mice in all littermates irrespective of the Stat1 genotype. Loss of Stat1 alone on a wild-type Jak2 background had no effect on blood parameters, as previously reported.14 The time course of blood counts is shown in Figure 1C. The effect of Stat1 deletion on platelet counts disappeared at 15 weeks and a reversed picture was seen at 20 and 25 weeks in SclCre;V617F;Stat1−/− mice, primarily due to a strong decline of platelets in the SclCre;V617F;Stat1+/+ mice. After 10 weeks, the survival of SclCre;V617F;Stat1−/− and also SclCre;V617F;Stat1+/+ mice substantially declined and the platelet counts could be skewed due to selection of the surviving mice. A persistent reduction of thrombocytosis without decreased survival of the transplanted mice was observed in VavCre;V617F;Stat1−/− mice (supplemental Figure 1B). VavCre;V617F mice have lower expression of the V617F transgene than the SclCreER;V617F mice and show a pure ET phenotype (supplemental Figure 1C; Tiedt et al10 ). All V617F transgenic mice also showed marked splenomegaly, irrespective of the Stat1 genotype (Figure 1D and supplemental Figure 1D). Thus, loss of Stat1 accentuated the PV phenotype, whereas thrombocytosis was reduced, but not completely abrogated.
Analysis of SclCreER;V617F transgenic and control mice. (A) Stat1 protein expression in spleen homogenates was assessed by immunoblot analysis. (B) Blood counts 10 weeks after induction with tamoxifen (SclCreER;V617F;Stat1+/+, n = 11; SclCreER;V617F;Stat1+/−, n = 27; SclCreER;V617F;Stat1−/−, n = 22; controls: WT and Stat1−/−, n = 6). One-way ANOVA is shown for comparisons. *P ≤ .05, **P ≤ .01, ***P ≤ .001. (C) Follow-up of blood counts. Genotypes and coloring of the lines is the same as in panel B. The Student t test was used for the comparisons between SclCreER;V617F;Stat1−/− and SclCreER;V617F;Stat1+/+ mice; *P ≤ .05. (D) Spleen weight assessment in transgenic mice 10 weeks after induction with tamoxifen. (SclCreER;V617F;Stat1+/+, n = 3; SclCreER;V617F;Stat1+/−, n = 6; SclCreER;V617F;Stat1−/−, n = 6; WT, n = 4; and Stat1−/−, n = 4). The Student t test was used. Error bars represent SEM. HCT, hematocrit; MCV, mean corpuscular volume; WT, wild type.
Analysis of SclCreER;V617F transgenic and control mice. (A) Stat1 protein expression in spleen homogenates was assessed by immunoblot analysis. (B) Blood counts 10 weeks after induction with tamoxifen (SclCreER;V617F;Stat1+/+, n = 11; SclCreER;V617F;Stat1+/−, n = 27; SclCreER;V617F;Stat1−/−, n = 22; controls: WT and Stat1−/−, n = 6). One-way ANOVA is shown for comparisons. *P ≤ .05, **P ≤ .01, ***P ≤ .001. (C) Follow-up of blood counts. Genotypes and coloring of the lines is the same as in panel B. The Student t test was used for the comparisons between SclCreER;V617F;Stat1−/− and SclCreER;V617F;Stat1+/+ mice; *P ≤ .05. (D) Spleen weight assessment in transgenic mice 10 weeks after induction with tamoxifen. (SclCreER;V617F;Stat1+/+, n = 3; SclCreER;V617F;Stat1+/−, n = 6; SclCreER;V617F;Stat1−/−, n = 6; WT, n = 4; and Stat1−/−, n = 4). The Student t test was used. Error bars represent SEM. HCT, hematocrit; MCV, mean corpuscular volume; WT, wild type.
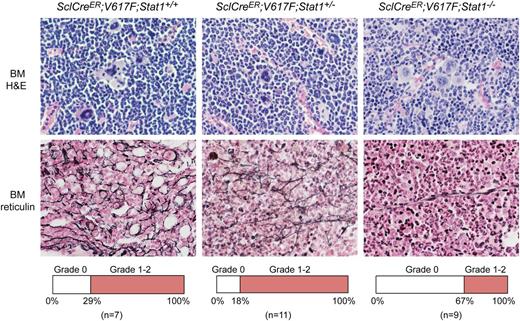
Histopathology of bone sections of V617F transgenic mice with Stat1−/− showed less prominent MPN features and a lower degree of myelofibrosis than V617F transgenic mice with Stat1+/− or Stat1+/+ background (Figure 2). The same result was also observed in VavCre;V617F;Stat1−/− mice (not shown). Sections of the spleen revealed destruction of normal splenic architecture by atypical hematopoiesis in all transgenic mice. Megakaryocytes in spleen of all transgenic mice irrespective of the Stat1 genotype were markedly increased in numbers compared with controls and displayed the same atypical morphology as in the BM (data not shown).
Histopathology of SclCreER;V617F transgenic mice using H&E and Gömöri-stained BM samples (magnification, ×630). Transgenic mice were sacrificed 10 weeks after induction with tamoxifen. SclCreER;V617F;Stat1+/+ mice (n = 6) showed more features of MPN and myelofibrosis compared with SclCreER;V617F;Stat1−/− littermates (n = 3) (hypercellularity with trilineage hyperplasia, markedly increased numbers of megakaryocytes with morphologic abnormalities, hyperchromatic, hyperlobulated nuclei, and bizarre nuclear configuration, often forming clusters). Frequency of mice displaying grade 0 or grade 1-2 myelofibrosis is shown in the lower panel for each genotype.
Histopathology of SclCreER;V617F transgenic mice using H&E and Gömöri-stained BM samples (magnification, ×630). Transgenic mice were sacrificed 10 weeks after induction with tamoxifen. SclCreER;V617F;Stat1+/+ mice (n = 6) showed more features of MPN and myelofibrosis compared with SclCreER;V617F;Stat1−/− littermates (n = 3) (hypercellularity with trilineage hyperplasia, markedly increased numbers of megakaryocytes with morphologic abnormalities, hyperchromatic, hyperlobulated nuclei, and bizarre nuclear configuration, often forming clusters). Frequency of mice displaying grade 0 or grade 1-2 myelofibrosis is shown in the lower panel for each genotype.
To determine whether the observed effects of Stat1 deficiency were entirely mediated by the hematopoietic cells, we transplanted BM cells from SclCreER;V617F;Stat1−/− donors and controls into C57BL/6 wild-type recipients (supplemental Figure 2A). We observed the same effects of Stat1 deficiency on erythropoiesis and megakaryopoiesis as obtained in the nontransplanted mice (supplemental Figure 2B). The decrease of platelets was also confirmed by transplantation of VavCre;V617F;Stat1−/− BM into WT recipients (data not shown).
To assess alterations in the hematopoietic progenitor and stem cell compartment in more detail, we performed flow cytometry analysis and colony-forming assays in SclCreER;V617F;Stat1−/− mice and controls. Loss of Stat1 had a negative effect on the numbers of LSK early progenitor and stem cells (Figure 3A). The same effect was observed in VavCre;V617F;Stat1−/− mice (supplemental Figure 3A). The numbers of erythroid precursors were increased in SclCreER;V617F;Stat1−/− mice by flow cytometric analysis (Figure 3B) and in BFU-E colony assays (Figure 3C). VavCre;V617F;Stat1−/− mice showed normal frequency of erythroid precursors, although all transgenic mice showed a higher number of BFU-E colonies compared with wild-type mice (supplemental Figure 3B-C). Early megakaryocytic progenitors (Lin−/Sca1−/c-Kit+/CD150+/CD41+) showed a trend toward higher numbers in SclCreER;V617F;Stat1−/− mice (Figure 3D). However, the numbers of CFU-MK colonies were decreased in both BM and spleen of SclCreER;V617F;Stat1−/− mice (Figure 3E), suggesting that the combination of V617F and Stat1 deficiency expands early megakaryocytic progenitors, but interferes with their terminal differentiation. The same trend was observed in BM but not spleen of transplanted recipients of both SclCreER;V617F;Stat1−/− (supplemental Figure 2E) and VavCre;V617F;Stat1−/− (supplemental Figure 3D).
Flow cytometric analysis of BM and spleen, and hematopoietic progenitor colony assays. (A) flow cytometric analysis of LSK in BM and spleen and quantification of LSK, (B) flow cytometric analysis of erythroid precursors in region II (CD71+/Ter119+) and (D) flow cytometric analysis of megakaryocytic progenitors (Lin−Sca1−ckit+/CD150+CD41+). (C,E) Number of hematopoietic progenitors assessed by colony assays in methylcellulose or collagen-based media. Error bars represent SEM. One-way ANOVA was used for comparisons. *P ≤ .05; ns, not significant. (SclCreER;V617F;Stat1+/+, n = 5; SclCreER;V617F;Stat1+/−, n = 8 ; SclCreER;V617F;Stat1−/−, n = 9; wild type, n = 8; and Stat1−/−, n = 6). AchE, acetylcholinesterase; FCS, forward scatter; SSC, side scatter.
Flow cytometric analysis of BM and spleen, and hematopoietic progenitor colony assays. (A) flow cytometric analysis of LSK in BM and spleen and quantification of LSK, (B) flow cytometric analysis of erythroid precursors in region II (CD71+/Ter119+) and (D) flow cytometric analysis of megakaryocytic progenitors (Lin−Sca1−ckit+/CD150+CD41+). (C,E) Number of hematopoietic progenitors assessed by colony assays in methylcellulose or collagen-based media. Error bars represent SEM. One-way ANOVA was used for comparisons. *P ≤ .05; ns, not significant. (SclCreER;V617F;Stat1+/+, n = 5; SclCreER;V617F;Stat1+/−, n = 8 ; SclCreER;V617F;Stat1−/−, n = 9; wild type, n = 8; and Stat1−/−, n = 6). AchE, acetylcholinesterase; FCS, forward scatter; SSC, side scatter.
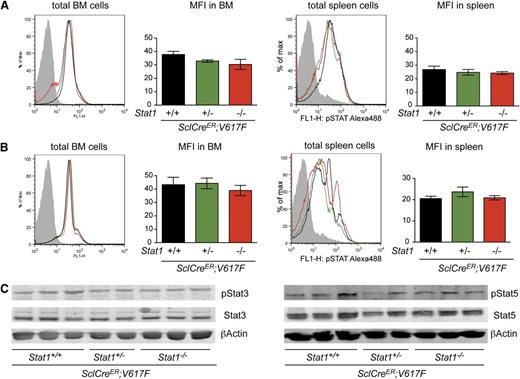
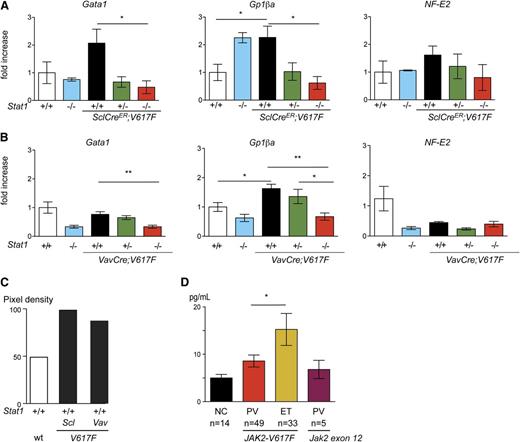
To test whether the absence of Stat1 had an effect on intracellular signaling by other Stat proteins, we examined the in vivo steady-state phosphorylation of Stat3 (pStat3) and Stat5 (pStat5) in hematopoietic tissues. The combination of JAK2-V617F and Stat1−/− did not increase levels of pStat3 or pStat5 in BM or spleen, irrespective of Stat1 status (Figure 4A-B). The same results were observed after assessing BM protein lysates by immunoblotting (Figure 4C) and VavCre;V617F;Stat1−/− mice (supplemental Figure 3F-G). To further characterize the effects of Stat1 deficiency on megakaryocytic differentiation, we analyzed the expression levels of specific genes in mRNA from platelets in SclCreER;V617F;Stat1−/− and VavCre;V617F;Stat1−/− mice. Gata1, a key regulator for megakaryopoiesis, was expressed at lower levels in SclCreER;V617F transgenic mice on the Stat1−/− background, compared with littermates with the same genotype on the Stat1+/+ background (Figure 5A). Gp1βa, a target gene of Gata1, was also expressed at lower levels in platelets of SclCreER;V617F;Stat1−/− mice (Figure 5A) and VavCre;V617F;Stat1−/− mice (Figure 5B). A trend toward lower expression of NF-E2 was observed in SclCreER;V617F;Stat1−/− mice only (Figure 5A). Together, these findings support the concept that Stat1 deficiency reduces thrombocytosis by affecting the maturation of megakaryocytes.
Analysis of Stat3 and Stat5 phosphorylation in BM and spleen of transgenic mice (n = 4, per group). Intracellular detection by flow cytometry of pStat3 (A) and pStat5 (B) in BM and spleen. Data shown are representative of 2 independent experiments (n = 4 per arm). (C) Levels of total Stat3 and pStat3 proteins (left panel), and Stat5 and pStat5 proteins (right panel) in BM homogenates assessed by immunoblot analysis. Error bars represent SEM. One-way ANOVA is shown for comparisons. MFI, mean fluorescent intensity; ns, not significant.
Analysis of Stat3 and Stat5 phosphorylation in BM and spleen of transgenic mice (n = 4, per group). Intracellular detection by flow cytometry of pStat3 (A) and pStat5 (B) in BM and spleen. Data shown are representative of 2 independent experiments (n = 4 per arm). (C) Levels of total Stat3 and pStat3 proteins (left panel), and Stat5 and pStat5 proteins (right panel) in BM homogenates assessed by immunoblot analysis. Error bars represent SEM. One-way ANOVA is shown for comparisons. MFI, mean fluorescent intensity; ns, not significant.
Gene expression analysis in platelets of transgenic mice and controls by quantitative PCR. Fold changes in mRNA expression of selected genes in platelets of SclCreER;V617F;Stat1−/− and controls (A), and VavCre;V617F;Stat1−/− and controls mice (B). Expression of mouse Gusb was used for normalization and relative expression was calculated with the ΔCT method. The mean value of 4 mice per group is shown. Error bars represent SEM. One-way ANOVA is shown for comparisons. (C) Serum levels of IFNγ in transgenic mice and controls. Duplicates of 1 mouse per genotype are shown. (D) Serum levels of IFNγ in patients with JAK2-V617F–positive and JAK2 exon 12–mutated MPN, and healthy controls. The Student t test is shown for the comparisons. *P ≤ .05, **P ≤ .01. NC, normal control; ns, not significant; PCR, polymerase chain reaction; wt, wild type.
Gene expression analysis in platelets of transgenic mice and controls by quantitative PCR. Fold changes in mRNA expression of selected genes in platelets of SclCreER;V617F;Stat1−/− and controls (A), and VavCre;V617F;Stat1−/− and controls mice (B). Expression of mouse Gusb was used for normalization and relative expression was calculated with the ΔCT method. The mean value of 4 mice per group is shown. Error bars represent SEM. One-way ANOVA is shown for comparisons. (C) Serum levels of IFNγ in transgenic mice and controls. Duplicates of 1 mouse per genotype are shown. (D) Serum levels of IFNγ in patients with JAK2-V617F–positive and JAK2 exon 12–mutated MPN, and healthy controls. The Student t test is shown for the comparisons. *P ≤ .05, **P ≤ .01. NC, normal control; ns, not significant; PCR, polymerase chain reaction; wt, wild type.
Because Stat1 is essential for IFNγ receptor signaling, we investigated whether elevated levels of interferon-γ (IFNγ) correlated with thrombocytosis. We found that mice with elevated platelet counts, that is, SclCreER;V617F and VavCre;V617F, showed higher concentrations of IFNγ in serum than wild-type controls (Figure 5C). A similar increase in IFNγ was also found in patients diagnosed with JAK2-V617F–positive ET, whereas patients with PV showed lower levels of IFNγ in serum (Figure 5D). Interestingly, patients with PV due to a JAK2-exon 12 mutation also displayed low levels of IFNγ in serum. The results suggest that IFNγ could be one of the drivers of Stat1 activation that promotes thrombocytosis in ET.
Discussion
The factors determining the phenotypic manifestation of PV vs ET in JAK2-V617F–positive MPNs are only partially understood. Although PV patients in almost all cases carry a subclone homozygous for JAK2-V617F, patients with ET in most cases lack such a homozygous subclone,9 or this subclone is difficult to detect.22 We showed in a mouse model of MPN that a higher ratio of expression of the mutant JAK2-V617F compared with WT Jak2 favored a PV phenotype.10 Stat1 was proposed to be an additional factor that can influence the decision between an ET and PV phenotype by promoting megakaryopoiesis and reducing erythropoiesis.11,12 Here, we examined the role of Stat1 in a mouse knockout model and showed that Stat1 deficiency in combination with JAK2-V617F favors a PV phenotype by augmenting erythropoiesis and repressing megakaryopoiesis. Thus, our findings are compatible with the model proposed by Chen and colleagues that was derived from studying single BFU-E colonies from ET and PV patients.11 Furthermore, we found that IFNγ, a cytokine that signals through Stat1, was increased in serum of mice with thrombocytosis and was higher in patients with ET than PV, suggesting that increased IFNγ could be a driver favoring an ET phenotype in JAK2-V617F–positive MPNs.
Stat1−/− mice have been reported to have normal blood counts, but showed higher numbers of BFU-E in spleen.14 We did not observe significant differences in BFU-E numbers between wild-type and Stat1−/− mice, but Stat1 deficiency combined with JAK2-V617F clearly acted synergistically on erythropoiesis (Figure 3). Thus, it seems that Stat1 loss favors erythropoiesis only in a state where erythropoiesis is already activated. SclCreER;V617F;Stat1−/− mice also showed more pronounced PV phenotype in peripheral blood than SclCreER;V617F;Stat1+/+ littermates (Figure 1). Furthermore, absence of Stat1 accentuated the shift of the main site of erythropoiesis from BM to spleen that we also found in our SclCreER;V617F;Stat1+/+ transgenic mice (Figure 3B). In stress erythropoiesis, this phenomenon was shown to be highly dependent on signals from the EpoR-Jak2 axis.23,24 All phenotypic changes were confirmed in mice that were transplanted with BM from V617F;Stat1−/− mice (supplemental Figure 2). This implies that the observed effects of Stat1 deficiency were cell autonomous and not dependent on the Stat1−/− microenvironment. Our data from VavCre;V617F;Stat1−/− also imply that the effect of Stat1−/− in promoting erythropoiesis relies on sufficient levels of JAK2-V617F expression in the hematopoietic tissues.
Chen et al suggested that the reduction of Stat1 leads to a compensatory increase in Stat5 signaling.11 In this study, we did not observe a compensatory increase in Stat3 or Stat5 phosphorylation in BM and spleen (Figure 4), but our data cannot exclude the possibility that Stat3 or Stat5 phosphorylation is altered in lineage-specific cells (eg, in erythroid cells). The precise role of Stat3 in the JAK2-V617F–driven MPN has not yet been established. However, the knockout of Stat5 was shown to abrogate the manifestations of the MPN phenotype on all 3 hematopoietic lineages in a Jak2-V617F knockin model and in retroviral Jak2-V617F transduction experiments.25,26 We observed changes in mRNA expression patterns of Gata1 and downstream genes. Gata1 is a key regulator for the erythroid and megakaryocytic lineages. Stat1 was reported to promote megakaryopoiesis downstream of Gata1.12 Enforced expression of Stat1 in megakaryoblastic Gata1-null cell line rescued multiple defects in megakaryopoiesis.12 We found that Stat1 deficiency in V617F transgenic mice resulted in a reduction of late megakaryocytic differentiation (Figure 3 and supplemental Figures 2-3). This is supported by the reduction of Gata1 expression in platelets and of its downstream target genes Gp1βa and NF-E2 (Figure 5). Only 1 of the 2 promoters of NF-E2 is Gata1 dependent.27 Nevertheless, Stat1 was not absolutely required for the ET phenotype because V617F;Stat1−/− mice maintained thrombocytosis, albeit at lower levels than V617F;Stat1+/+ mice (Figure 1 and supplemental Figures 1-2). Similar pattern of mRNA expression for these genes was found in CFU-MK colonies from mice deficient for Gata1.28
Stat1 can be activated by a large number of hematopoietic cytokines. Tpo and Epo receptors have been shown to be able to phosphorylate Stat1. The IFNγ receptor signals through Jak1 and Jak2 and induces phosphorylation of Stat1. Thus, the presence of JAK2-V617F could increase the phosphorylation of Stat1 by amplifying the IFNγ-receptor signals. However, it was recently suggested that ET patients with JAK2-V617F, but not PV patients, have an enhanced IFNγ-expression signature in microarray analysis of human BFU-E colonies.11 These results may reflect a differential exposure of the hematopoietic cell to IFNγ in vivo. Here, we found that V617F transgenic mice with thrombocytosis had higher serum levels of IFNγ than normal controls. Furthermore, patients with ET showed higher IFNγ serum levels than patients with PV. IFNγ was shown to enhance megakaryocyte colony formation in mice.29 Thus, the IFNγ pathway appears to be involved in modulating the activity of erythropoiesis and megakaryopoiesis and our data support a role of IFNγ signaling in modifying the phenotypic manifestation of JAK2-V617F–positive MPN. Inhibition of Stat1 may have also effects on improving myelofibrosis. The decrease in thrombocytosis observed in V617F;Stat1−/− mice was paralleled by a reduction of myelofibrosis (Figure 2), consistent with reports that linked the degree of myelofibrosis with the platelet counts.30
In summary, our results provide experimental evidence for the role of Stat1 in increasing the numbers of platelets and lowering erythrocytes in JAK2-V617F–driven MPN. Our data also suggest a possibly prominent role of IFNγ in driving these Stat1 effects.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank David E. Levy for providing the Stat1 knockout mice and Jürg Schwaller for helpful comments on the manuscript.
This work was supported by the Botnar Foundation (A.D.) and by grants 310000-120724/1 and 32003BB_135712/1 from the Swiss National Science Foundation and the Swiss Cancer League (KLS-02398-02-2009) (R.C.S.).
Authorship
Contribution: A.D. performed research, analyzed data, and wrote the paper; P.L., T.S., J.G., A.K., L.K., and H.H-S. performed research and analyzed data; S.D. prepared and analyzed histology samples; and R.C.S. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Radek C. Skoda, Department of Biomedicine, Experimental Hematology, University Hospital Basel, Hebelstrasse 20, 4031 Basel, Switzerland; e-mail: radek.skoda@unibas.ch.