Key Points
Epigenetics and in vivo behavior can distinguish MSCs from different sources.
BM-derived MSCs form a hematopoietic niche via a vascularized cartilage intermediate.
Abstract
In the last decade there has been a rapid expansion in clinical trials using mesenchymal stromal cells (MSCs) from a variety of tissues. However, despite similarities in morphology, immunophenotype, and differentiation behavior in vitro, MSCs sourced from distinct tissues do not necessarily have equivalent biological properties. We performed a genome-wide methylation, transcription, and in vivo evaluation of MSCs from human bone marrow (BM), white adipose tissue, umbilical cord, and skin cultured in humanized media. Surprisingly, only BM-derived MSCs spontaneously formed a BM cavity through a vascularized cartilage intermediate in vivo that was progressively replaced by hematopoietic tissue and bone. Only BM-derived MSCs exhibited a chondrogenic transcriptional program with hypomethylation and increased expression of RUNX3, RUNX2, BGLAP, MMP13, and ITGA10 consistent with a latent and primed skeletal developmental potential. The humanized MSC–derived microenvironment permitted homing and maintenance of long-term murine SLAM+ hematopoietic stem cells (HSCs), as well as human CD34+/CD38−/CD90+/CD45RA+ HSCs after cord blood transplantation. These studies underscore the profound differences in developmental potential between MSC sources independent of donor age, with implications for their clinical use. We also demonstrate a tractable human niche model for studying homing and engraftment of human hematopoietic cells in normal and neoplastic states.
Introduction
Human mesenchymal stromal cells (MSCs) from various tissue origins, including bone marrow (BM), umbilical cord (UC), white adipose tissue (WAT), dental pulp, skin, and many others, have entered clinical application as a potential regenerative cell therapy.1 Despite increasing human trials evaluating allogeneic or autologous MSCs, our understanding of cell fate after transplantation in vivo and of the transcription factors that regulate their developmental potential is limited.2 For most MSC sources, differentiation capacity has been studied primarily with in vitro assays using induction reagents, which may not necessarily reflect the full developmental capacity of forming functional tissues in vivo,3 thus creating uncertainty regarding the biological effects of MSCs in ongoing clinical trials. For example, although almost all MSCs can accumulate calcium (Ca2+) on stimulation in vitro, histologic examination revealed dystrophic calcification rather than mature bone formation after transplantation.4 We have previously established that BM-derived MSCs (BM-MSCs) can give rise to a vascularized, extramedullary hematopoietic microenvironment in vivo, but whether this developmental capacity is also shared by MSCs from other sources has not been rigorously tested.5,6 Additionally, recent work studying MSC reprogramming into other cell types has shown differences in plasticity toward certain lineages and epigenome characteristics, depending on the tissue source of the MSC population.7
Almost half a century ago, experiments by Tavassoli and Crosby demonstrated ectopic bone and marrow formation after transplanting unfractionated BM to heterotopic anatomical sides,8 leading to the discovery of nonhematopoietic cells within BM.9 In vertebrates, long bones form through endochondral ossification, a tightly regulated process that generates bone through an intermediate cartilage template, which is finally replaced by mature bone tissue10 and appears to occur in parallel with definitive hematopoiesis. Recent studies have indicated that MSCs derived from sites of endochondral ossification are capable of forming a functional hematopoietic stem cell niche,11,12 but whether other sites can give rise to a functional hematopoietic niche has not been explored.
The idea of a nonhematopoietic “mesenchymal stem cell” was conceptualized following the identification of clonogenic adherent mesenchymal populations that could differentiate along osteogenic, chondrogenic, and adipogenic lineages ex vivo and that could recapitulate parts of these functions after being transplanted in vivo.13,14 Initially, the term MSC was restricted to BM, but in recent years the definition has broadened to now include connective tissue cells from many sources such as UC blood (UCB),15,16 WAT,17 UC,18 dental pulp,3 skin,19 and others.20 However, functional equivalence in vivo and epigenetic characteristics of these cells have not been studied.
Here, we performed a comprehensive study of 4 different MSC populations commonly employed in cell therapy protocols. Unexpectedly, we found that only BM-MSCs underwent spontaneous chondrogenic differentiation with subsequent formation of a functional marrow niche that could support both murine and human hematopoietic stem cell (HSC) homing. In vivo endochondral differentiation was associated with a unique DNA methylation signature and with upregulation of key cartilage/bone developmental regulators. Our model provides an accessible ectopic platform to study normal and malignant hematopoiesis and details of epigenetic determinants for future MSC-based regenerative therapies.
Materials and methods
Collection of human samples and animal ethics
Approval for human sample collection was obtained from the Institutional Review Boards of the Medical University of Graz (protocols 19-252, 18-243, 21-060, and 19-284) and Stanford University (IRB 28853). Samples were collected in accordance with the Declaration of Helsinki after written informed consent. Animal experiments were approved (BMWF-66.010/0082-II/10b/2009) and held in accordance with the Animal Care and Use Committee and National Institutes of Health guidelines.
MSC isolation, expansion, and in vitro differentiation
All cells were isolated and cultured in α-modified minimum essential medium (α-MEM; Sigma-Aldrich) containing 10% pooled human platelet lysate (pHPL).21-23 BM-MSCs were isolated and expanded as previously published.24 Adipose tissue was obtained by liposuction from healthy female donors aged 18 to 39 years as described.17,25 UC-derived MSCs and skin-derived MSC were isolated as previously described.18 Briefly, biopsy pieces were allowed to adhere to culture dishes (Corning) before cell culture medium was carefully added. Outgrowing cells were expanded and passaged at near confluence.
In vitro differentiation
Adipogenic, osteogenic, and chondrogenic differentiation was done following standard protocols (see the supplemental Experimental Procedures, available on the Blood Web site).
Flow cytometry
A panel of antibodies was used for analysis of human MSCs, as well as analysis of human hematopoiesis, including hematopoietic stem and progenitor cells (HSPCs) in ectopic ossicles. Cells were processed on a Navios (Beckman Coulter), AriaII (BD Biosciences), or LSRII (BD Biosciences) instrument and analyzed with FlowJo (Tree Star) or Kaluza 1.1 (Beckman Coulter) software. For HSPC phenotyping, staining was performed according to previously published protocols.26 Detailed information on all antibodies is available in the supplemental Experimental Procedures.
In vivo bone formation
Subcutaneous transplants.
MSCs (2 × 106 per implant) were resuspended in 300 µL of matrigel-equivalent matrix (Millipore; noninductive protocol) and injected subcutaneously into immunodeficient NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice (6-18 weeks old; Jackson Laboratory). For the osteoinductive protocol, 40 µg of tricalcium phosphate/hydroxyapatite (TCP/HA) powder (BoneCeramic; Straumann) was loaded with MSCs (2 × 106 per implant) and slowly rotated (2 hours at 37°C) for cell seeding before transplanting into subcutaneous dorsal pockets. Mice received 40 µg/kg of human parathyroid hormone ([1-34]; R&D Systems) daily for 21 days in the osteoinductive group only.
Kidney capsule transplants.
For kidney capsule transplants, 2 × 105 cells were resuspended in 2 μL of matrigel and injected underneath the renal capsule of adult anesthetized male NSG mice, as previously described.11
DNA methylation and gene expression analysis
Two hundred nanograms of bisulfite-converted DNA per sample were analyzed by HumanMethylation 450 BeadChip array (Illumina). Image processing, initial data quality control, and raw data analysis were performed with GenomeStudio 2010.3 (Methylation Module 1.8.5; Illumina). For further analysis, we used BioConductor’s lumi package with R statistical software.
For gene expression analysis, single-stranded DNA targets generated from 200 ng total RNA were fragmented, biotin-labeled, and hybridized to GeneChip Human Gene 2.0 ST arrays following standard Affymetrix protocols. Raw data were normalized in R using the oligo library. Genes that were differentially expressed in BM arrays (fold change of at least 1.5) were identified. Unsupervised hierarchical clustering for gene expression was done using the hclust function in R. Gene ontology annotation on hypomethylated and overexpressed genes was performed using DAVID bioinformatics resources.27 The raw data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database and are accessible through accession numbers GSE57151 and GSE41933.
Quantitative reverse transcription–polymerase chain reaction was done as described in detail in the supplemental Experimental Procedures.
Western blot analysis
Cells were lysed in radioimmunoprecipitation assay buffer, and 40 µg of protein was resolved with 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (Bio-Rad) and transferred onto a nitrocellulose membrane (Bio-Rad). Membranes were incubated with rabbit anti-human RUNX3 (clone D6E2; Cell Signaling Technology) or mouse anti-human β-actin (clone 8H10D10; Cell Signaling Technology), followed by secondary horseradish peroxidase–conjugated antibodies (Thermo Scientific). Antibody binding was visualized with enhanced chemiluminescence detection reagents (GE Healthcare).
Hematopoietic transplantation studies
CD34+ HSPCs were enriched using MACS Technology (Miltenyi). Pooled CD34+ HSPCs (1 × 104 to 2 × 105 HSPCs per mouse) from 3 to 5 individual donors were injected intravenously into 6- to 8-week-old female NSG mice 24 hours after sublethal (200 rad) irradiation. HSPCs were pooled to minimize interexperimental variations due to engraftment differences of individual UCB donors. Stably engrafted mice (human chimerism above 10% detected by flow cytometry of peripheral blood) were used to create human ossicles (2-4 transplants per mouse). Eight to 10 weeks later, additional mice were euthanized, and engrafted ossicles (n = 14) were either analyzed for human hematopoietic engraftment or 5 × 106 ossicle-derived mononuclear cells were transplanted into irradiated secondary recipients (n = 3).
Statistical analysis
Student t test and analysis of variance were used as statistical methods in SPSS 19.0 software (SPSS), Prism (GraphPad), or Microsoft Excel. For correlation analysis, a Pearson correlation coefficient was calculated. All values are presented as mean ± standard deviation. P values <.05 were considered significant.
Results
MSCs isolated from diverse tissues do not reveal phenotypic differences in vitro
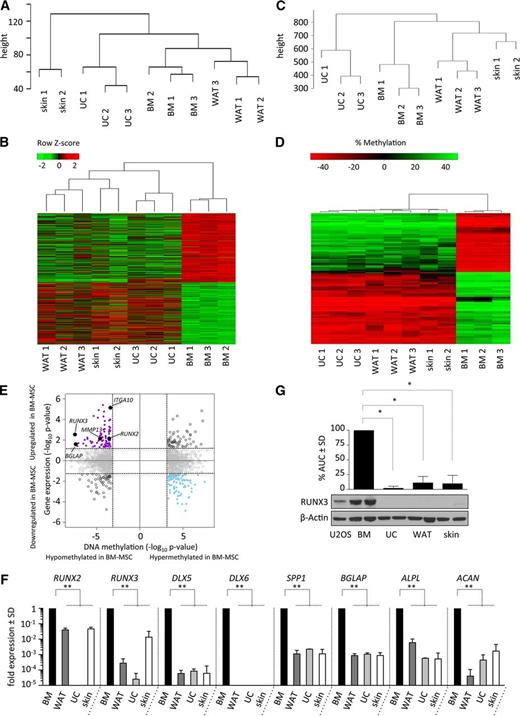
MSCs from healthy normal human BM, WAT, UC, and skin were isolated and expanded in vitro with pHPL, a humanized cell culture protocol used in current MSC-based clinical proposals submitted to the Food and Drug Administration.1 MSC donor characteristics are summarized in supplemental Table 1. All cells were seeded at a low seeding density (50-100 cells per cm2) and expanded for a maximum of 4 passages. Irrespective of the tissue of origin, all MSCs displayed a spindle-shaped, elongated morphology (Figure 1A). Similarly, MSCs derived from all sources revealed an identical immunophenotype consistent with standard criteria agreed by the International Society of Cellular Therapy expert panel (Figure 1B).28 An extended panel of positive MSC markers, including CD44, CD140a, CD140b, CD166, CD63, CD49a, CD10, CD29, CD13 and CD146, could also not clearly distinguish between tissue sources (supplemental Figure 1). Furthermore, in vitro adipogenic, osteogenic, and chondrogenic differentiation assays performed at passage 2 to 3, showed trilineage differentiation potential for each expanded MSCs population obtained from BM, WAT, UC, and skin (Figure 1C). Although all MSCs could clearly form osteocytes, adipocytes, and chondrocytes, BM-MSCs reproducibly displayed enhanced chondrocyte formation with increased deposition of toluidine blue–positive extracellular matrix (Figure 1C).
BM-MSCs, but not WAT-, UC-, or skin-derived MSCs form bone, cartilage, and marrow tissue in vivo. (A) Phase-contrast images depicting spindle-shaped morphology of BM-MSCs and WAT-, UC-, and skin-derived MSCs. (B) Flow cytometric profiles of the same respective cell populations as in panel A. Histograms show fluorescent cell surface staining intensity of anti-CD90, anti-CD73, anti-CD105, anti-CD45, anti-HLA-DR, anti-CD14, anti-CD19, and anti-CD34 monoclonal antibodies conjugated to fluorophores (gray shading) and corresponding isotype control (no shading). (C) Trilineage in vitro differentiation of BM-MSCs and WAT-, UC-, and skin-derived MSCs (shown left to right). MSCs were induced to differentiate along osteogenic, adipogenic, and chondrogenic lineage in vitro according to standard protocols. After 21 days of osteogenic induction, cells were stained with alizarin red for visualization of Ca2+ accumulation (upper left image for each). Adipogenic differentiation capacity was visualized after 14 days of induction by staining lipid droplets with fluorescent nile red (upper right image for each). A 3-dimensional chondrogenic differentiation assay in Transwell plates was used for evaluation of chondrogenic differentiation potential. Tissue was fixed and further processed for histologic evaluation of chondrotypic glucosaminoglycans with toluidine blue after 28 days of induction. (D) BM-MSCs form bone, cartilage, and marrow tissue in vivo. Upper image shows schematic of the in vivo protocol. After expansion in pHPL-supplemented α-modified minimum essential medium (α-MEM), 2 × 106 MSCs were resuspended in matrigel and subcutaneously injected into NSG mice. Spontaneous in vivo tissue formation was evaluated 8 to 12 weeks postimplantation by histology. Representative photomicrographs of hematoxylin and eosin ([H&E], upper row) and pentachrome-stained sections (lower row) are shown. Inserts show low-magnification overview photographs. ECM, extracellular matrix.
BM-MSCs, but not WAT-, UC-, or skin-derived MSCs form bone, cartilage, and marrow tissue in vivo. (A) Phase-contrast images depicting spindle-shaped morphology of BM-MSCs and WAT-, UC-, and skin-derived MSCs. (B) Flow cytometric profiles of the same respective cell populations as in panel A. Histograms show fluorescent cell surface staining intensity of anti-CD90, anti-CD73, anti-CD105, anti-CD45, anti-HLA-DR, anti-CD14, anti-CD19, and anti-CD34 monoclonal antibodies conjugated to fluorophores (gray shading) and corresponding isotype control (no shading). (C) Trilineage in vitro differentiation of BM-MSCs and WAT-, UC-, and skin-derived MSCs (shown left to right). MSCs were induced to differentiate along osteogenic, adipogenic, and chondrogenic lineage in vitro according to standard protocols. After 21 days of osteogenic induction, cells were stained with alizarin red for visualization of Ca2+ accumulation (upper left image for each). Adipogenic differentiation capacity was visualized after 14 days of induction by staining lipid droplets with fluorescent nile red (upper right image for each). A 3-dimensional chondrogenic differentiation assay in Transwell plates was used for evaluation of chondrogenic differentiation potential. Tissue was fixed and further processed for histologic evaluation of chondrotypic glucosaminoglycans with toluidine blue after 28 days of induction. (D) BM-MSCs form bone, cartilage, and marrow tissue in vivo. Upper image shows schematic of the in vivo protocol. After expansion in pHPL-supplemented α-modified minimum essential medium (α-MEM), 2 × 106 MSCs were resuspended in matrigel and subcutaneously injected into NSG mice. Spontaneous in vivo tissue formation was evaluated 8 to 12 weeks postimplantation by histology. Representative photomicrographs of hematoxylin and eosin ([H&E], upper row) and pentachrome-stained sections (lower row) are shown. Inserts show low-magnification overview photographs. ECM, extracellular matrix.
BM-MSCs but not WAT-, UC-, or skin-derived MSCs, form BM tissue in vivo
To test the spontaneous in vivo differentiation potential, 2 × 106 MSCs at passage 2 to 3 were resuspended in matrigel (without induction protocols) and transplanted subcutaneously into immunocompromised NSG mice. Strikingly, histologic examination of MSC-explants at 8 weeks after transplant revealed areas of cartilage, bone, and hematopoietic tissue (as visualized by hematoxylin and eosin and pentachrome stains) in transplants from BM-MSCs but not in transplants from WAT-, UC-, or skin-derived MSCs (Figure 1D). These data are consistent with previous reports indicating that despite obvious similarities in morphology, immunophenotype, and in vitro differentiation assays, the in vivo behavior of transplanted BM-MSCs differs substantially from MSCs derived from other tissue sources.3,29,30
BM-MSCs have a distinct gene expression and DNA methylation signature
To investigate underlying differences in epigenetic or transcriptional programs that may contribute to the striking in vivo phenotype observed, we performed gene expression (Figure 2A-B) and DNA methylome studies (Figure 2C-D) of BM-MSCs compared with WAT-, UC- and skin-derived MSCs expanded for 2 to 3 passages (3 healthy donors for each source) using Affymetrix GeneChip Human Gene 2.0 ST and Illumina HumanMethylome450 BeadChip arrays. Unsupervised hierarchical clustering of both platforms revealed similarity within MSCs of the same tissue source consistent with an underlying tissue-specific gene expression and DNA methylation (Figure 2A, C). Interestingly, BM-MSCs were more closely related to WAT than to skin or UC according to both gene expression and methylation. Supervised analysis of the top 1000 most differentially methylated regions in BM-MSCs compared with the other three MSC groups revealed genomic regions that were differentially hypomethylated and differentially hypermethylated (Figure 2D). Integrating the 2 analyses revealed a number of transcription factors and differentiation-marker genes noted to be both hypomethylated and upregulated in only BM-MSCs (Figure 2E). These genes included RUNX2, RUNX3, BGLAP, MMP13, and ITGA10. Functional annotation of all hypomethylated and upregulated genes to gene ontology terms revealed enrichment for skeletal and mesodermal development genes (DAVID; corrected P value of .02; supplemental Table 2) suggesting that BM-MSCs in culture, prior to transplant, are already primed for skeletal development. A complete list of upregulated genes with associated hypomethylated differentially methylated regions is available in supplemental Table 3. To validate these findings, we performed quantitative gene expression of known skeletal developmental regulators at baseline, without adding osteocyte- or chondrocyte-inducing agents, on all MSC populations. We found increased expression of RUNX2, RUNX3, DLX5, DLX6, SPP1, BGLAP, ALPL, and ACAN in BM-MSCs compared with WAT-, UC-, and skin-derived MSCs (Figure 2F), including genes that were differentially hypomethylated in BM-MSCs (RUNX3, RUNX2, and BGLAP; Figure 2E). Only BM-MSCs expressed detectable levels of DLX6, a homeobox transcription factor critically regulating chondrocyte hypertrophy and subsequent endochondral ossification in mouse studies (Figure 2F).31 RUNX3 has recently been identified as a critical factor in the coordination of chondrocyte development, maturation, and hypertrophy during endochondral ossification.32 To further examine RUNX3 expression in BM-MSCs, we performed quantitation of RUNX3 protein levels, which confirmed high expression only in BM-MSCs, consistent with the gene expression and methylation data (Figure 2G). We also noted that in vivo BM-formation did not correlate with younger donor age (age vs percent marrow in histologic cross-sections; Pearson correlation r = 0.245, P = .46). DNA methylation of our samples based on 353 previously defined “clock” CpG sites showed high correlation with chronological donor age (Pearson r = 0.9211, P < .0001) (supplemental Figure 2A).33 These data revealed latent changes in developmental transcription factors and DNA methylation in MSC populations that are not apparent with cell surface marker studies but are consistent with developmental fates observed in vivo.
BM-MSCs have a distinct gene expression and DNA methylation signature. (A) Unsupervised hierarchical clustering dendrograms of gene expression analysis performed using Affymetrix GeneChip Human Gene 2.0 ST arrays of early-passage BM-MSCs and WAT-, UC-, and skin-derived MSCs cultured in pHPL. (B) Heat map representing all significant differentially expressed genes, with red indicating high gene expression and green indicating decreased gene expression of BM-MSCs compared with non-BM-derived (WAT/UC/skin) MSCs. (C) Unsupervised hierarchical clustering dendrograms of differentially methylated regions analyzed using HumanMethylation450 BeadChip arrays performed on the same BM-MSCs and WAT-, UC-, and skin-derived MSCs cultured in pHPL as in panel A. (D) Heat map showing the top 100 differentially methylated regions in BM-MSCs compared with non-BM-derived (WAT/UC/skin) MSCs, with red indicating hypomethylation and green indicating hypermethylation. (E) Identification of genes with concordant changes in messenger RNA expression and DNA methylation. Quadrant plot showing differentially methylated CpGs (only CpGs with the most significant P values are depicted) and expression of associated genes. The x-axis depicts the −log10P value for differentially methylated CpGs; the y-axis depicts the −log10P value of differential expression for associated genes. Vertical dashed lines indicate a false discovery rate of 5%; horizontal dashed lines indicate a P value of .05. The 4 quadrants are (i) hypomethylated and upregulated in BM-MSCs (solid purple circles and solid black circles); (ii) hypermethylated and downregulated in BM-MSCs (solid blue circles); (iii) hypomethylated and downregulated in BM-MSCs (open black circles); and (iv) hypermethylated and upregulated in BM-MSCs (open black circles). Hypomethylated and upregulated genes with known roles in chondrogenesis and osteogenesis (solid black circles) are labeled with arrows. (F) Quantitative reverse transcription–polymerase chain reaction–based validation of the baseline expression in key genes found to be hypomethylated and overexpressed, as well as other transcription factors and lineage-specific genes associated with cartilage and bone formation in various MSC populations. Runt-related transcription factor 2 and 3 (RUNX2 and RUNX3), distal-less homeobox 5 and 6 (DLX5 and DLX6), osteopontin (SPP1), osteocalcin (BGLAP), bone-specific alkaline phosphatase (ALPL), and aggrecan (ACAN) expression was analyzed in MSCs derived from BM (black bars) compared with WAT (dark gray bars), UC (light gray bars), and skin (white bars). Expression differences for each gene compared with the GAPDH housekeeping gene are shown relative to BM levels. (G) Western blot analysis of RUNX3 protein levels in the positive-control U2OS osteosarcoma cell line, as well as in BM-MSCs and WAT-, UC-, and skin-derived MSCs. Bar graphs depict densitometry-derived mean areas under the curve (AUC) ± standard deviation (SD) normalized to β-actin. All densitometry values are in relation to BM-MSCs (n = 3 per MSC source; *P < .0001).
BM-MSCs have a distinct gene expression and DNA methylation signature. (A) Unsupervised hierarchical clustering dendrograms of gene expression analysis performed using Affymetrix GeneChip Human Gene 2.0 ST arrays of early-passage BM-MSCs and WAT-, UC-, and skin-derived MSCs cultured in pHPL. (B) Heat map representing all significant differentially expressed genes, with red indicating high gene expression and green indicating decreased gene expression of BM-MSCs compared with non-BM-derived (WAT/UC/skin) MSCs. (C) Unsupervised hierarchical clustering dendrograms of differentially methylated regions analyzed using HumanMethylation450 BeadChip arrays performed on the same BM-MSCs and WAT-, UC-, and skin-derived MSCs cultured in pHPL as in panel A. (D) Heat map showing the top 100 differentially methylated regions in BM-MSCs compared with non-BM-derived (WAT/UC/skin) MSCs, with red indicating hypomethylation and green indicating hypermethylation. (E) Identification of genes with concordant changes in messenger RNA expression and DNA methylation. Quadrant plot showing differentially methylated CpGs (only CpGs with the most significant P values are depicted) and expression of associated genes. The x-axis depicts the −log10P value for differentially methylated CpGs; the y-axis depicts the −log10P value of differential expression for associated genes. Vertical dashed lines indicate a false discovery rate of 5%; horizontal dashed lines indicate a P value of .05. The 4 quadrants are (i) hypomethylated and upregulated in BM-MSCs (solid purple circles and solid black circles); (ii) hypermethylated and downregulated in BM-MSCs (solid blue circles); (iii) hypomethylated and downregulated in BM-MSCs (open black circles); and (iv) hypermethylated and upregulated in BM-MSCs (open black circles). Hypomethylated and upregulated genes with known roles in chondrogenesis and osteogenesis (solid black circles) are labeled with arrows. (F) Quantitative reverse transcription–polymerase chain reaction–based validation of the baseline expression in key genes found to be hypomethylated and overexpressed, as well as other transcription factors and lineage-specific genes associated with cartilage and bone formation in various MSC populations. Runt-related transcription factor 2 and 3 (RUNX2 and RUNX3), distal-less homeobox 5 and 6 (DLX5 and DLX6), osteopontin (SPP1), osteocalcin (BGLAP), bone-specific alkaline phosphatase (ALPL), and aggrecan (ACAN) expression was analyzed in MSCs derived from BM (black bars) compared with WAT (dark gray bars), UC (light gray bars), and skin (white bars). Expression differences for each gene compared with the GAPDH housekeeping gene are shown relative to BM levels. (G) Western blot analysis of RUNX3 protein levels in the positive-control U2OS osteosarcoma cell line, as well as in BM-MSCs and WAT-, UC-, and skin-derived MSCs. Bar graphs depict densitometry-derived mean areas under the curve (AUC) ± standard deviation (SD) normalized to β-actin. All densitometry values are in relation to BM-MSCs (n = 3 per MSC source; *P < .0001).
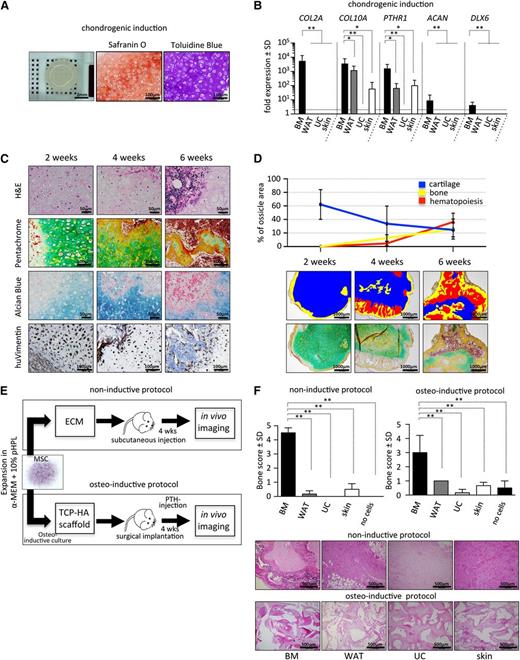
BM-derived MSCs exhibit superior chondrogenic potential in vitro
To further examine the endochondral developmental potential of BM-MSCs, we tested the ability of these cells to form chondrocytes in vitro. We found that BM-MSCs formed a disclike structure with evidence of hypertrophic chondrocytes surrounded by thick extracellular matrix that stained highly positive for Safranin O and toludine blue (Figure 3A). Consistent with this observation, we found strong upregulation of COL2A, COL10A, ACAN, PTHR1, and DLX6 in BM-MSCs compared with WAT-, UC-, and skin-derived MSCs after chondrogenic induction (Figure 3B).
BM-derived MSCs exhibit superior chondrogenic and osteogenic potential. (A) BM-MSCs were differentiated in vitro on Transwell membranes for 28 days in chondrogenic induction medium and then fixed, paraffin-embedded, and analyzed. Macroscopic view (left) of a representative cartilage tissue piece generated by 5 × 105 MSCs. Safranin O (middle) and toluidine blue staining (right) was used for visualization of glycosaminoglycans produced by differentiated hypertrophic chondrocytes. (B) Gene expression analysis of collagen 2a (COL2A), collagen 10a (COL10A), parathyroid hormone receptor 1 (PTHR1), aggrecan (ACAN), and distal-less homeobox 6 (DLX6) by quantitative reverse transcription–polymerase chain reaction after 28 days of chondrogenic induction comparing BM (black bars) to non-BM-derived cells (dark gray, light gray, and white bars). Bar graphs depict fold expression (mean ± SD) of differentiated MSCs over the respective cells in the uninduced state. (C) Endochondral ossification of BM-MSCs precedes marrow development in vivo. Time course analysis (2, 4, and 6 weeks posttranspant) of BM-MSCs transplanted in NSG mice (noninductive protocol), removed and sectioned for histology. Representative images show hematoxylin and eosin (H&E; upper row), pentachrome (second row), and alcian blue staining (third row). Immunohistochemical staining for human vimentin (huVimentin, bottom row) delineates human origin of the developed tissue. (D) Top: quantification of cartilage, bone, and hematopoietic tissue at 2-, 4-, and 6-week time points following transplant (noninductive protocol). Bottom: pentachrome-stained tissue sections (n = 4 ossicles per time point from 4 different human donors) were evaluated for relative percentage of respective tissue using the area calculation tool in ImageJ software. Representative pseudocolored images illustrating areas used for quantification are shown. (E) Bone formation in vivo is restricted to BM-MSCs. Two protocols (noninductive and osteoinductive) for testing bone and marrow niche formation capacity of MSCs are illustrated. Cells (2 × 106) were either directly injected subcutaneously in ECM (matrigel, noninductive protocol) or, to achieve maximal osteogenic induction, were cultured for 72 hours in a medium inducing osteogenic differentiation before being seeded onto (TCP-HA) particles. Thereafter, TCP-HA with attached MSCs was surgically implanted. Transplanted NSG mice received a daily anabolic dose of parathyroid hormone (PTH) for the first 21 days of the experiment to further enhance osteogenesis in vivo (osteoinductive protocol). After 4 weeks, in vivo imaging was performed to evaluate bone formation in both protocols used. Active bone growth and resorption was semiquantitatively evaluated in vivo using the bisphosphonate imaging agent OsteoSense on a Maestro imaging instrument. (F) Top: bar graphs represent bone scores from transplants containing BM-MSCs and WAT-, UC-, and skin-derived MSCs or control implants without cells (n = 3 per source). Cells were applied following either a noninductive (left) or an osteoinductive (right) transplantation protocol. Eight weeks posttransplant, mice were euthanized, and explants were analyzed to confirm in vivo imaging results. Bottom: representative photographs of H&E-stained tissues sections derived from all 4 different MSC-sources are shown as indicated. Student t test *P < .05, **P < .00001.
BM-derived MSCs exhibit superior chondrogenic and osteogenic potential. (A) BM-MSCs were differentiated in vitro on Transwell membranes for 28 days in chondrogenic induction medium and then fixed, paraffin-embedded, and analyzed. Macroscopic view (left) of a representative cartilage tissue piece generated by 5 × 105 MSCs. Safranin O (middle) and toluidine blue staining (right) was used for visualization of glycosaminoglycans produced by differentiated hypertrophic chondrocytes. (B) Gene expression analysis of collagen 2a (COL2A), collagen 10a (COL10A), parathyroid hormone receptor 1 (PTHR1), aggrecan (ACAN), and distal-less homeobox 6 (DLX6) by quantitative reverse transcription–polymerase chain reaction after 28 days of chondrogenic induction comparing BM (black bars) to non-BM-derived cells (dark gray, light gray, and white bars). Bar graphs depict fold expression (mean ± SD) of differentiated MSCs over the respective cells in the uninduced state. (C) Endochondral ossification of BM-MSCs precedes marrow development in vivo. Time course analysis (2, 4, and 6 weeks posttranspant) of BM-MSCs transplanted in NSG mice (noninductive protocol), removed and sectioned for histology. Representative images show hematoxylin and eosin (H&E; upper row), pentachrome (second row), and alcian blue staining (third row). Immunohistochemical staining for human vimentin (huVimentin, bottom row) delineates human origin of the developed tissue. (D) Top: quantification of cartilage, bone, and hematopoietic tissue at 2-, 4-, and 6-week time points following transplant (noninductive protocol). Bottom: pentachrome-stained tissue sections (n = 4 ossicles per time point from 4 different human donors) were evaluated for relative percentage of respective tissue using the area calculation tool in ImageJ software. Representative pseudocolored images illustrating areas used for quantification are shown. (E) Bone formation in vivo is restricted to BM-MSCs. Two protocols (noninductive and osteoinductive) for testing bone and marrow niche formation capacity of MSCs are illustrated. Cells (2 × 106) were either directly injected subcutaneously in ECM (matrigel, noninductive protocol) or, to achieve maximal osteogenic induction, were cultured for 72 hours in a medium inducing osteogenic differentiation before being seeded onto (TCP-HA) particles. Thereafter, TCP-HA with attached MSCs was surgically implanted. Transplanted NSG mice received a daily anabolic dose of parathyroid hormone (PTH) for the first 21 days of the experiment to further enhance osteogenesis in vivo (osteoinductive protocol). After 4 weeks, in vivo imaging was performed to evaluate bone formation in both protocols used. Active bone growth and resorption was semiquantitatively evaluated in vivo using the bisphosphonate imaging agent OsteoSense on a Maestro imaging instrument. (F) Top: bar graphs represent bone scores from transplants containing BM-MSCs and WAT-, UC-, and skin-derived MSCs or control implants without cells (n = 3 per source). Cells were applied following either a noninductive (left) or an osteoinductive (right) transplantation protocol. Eight weeks posttransplant, mice were euthanized, and explants were analyzed to confirm in vivo imaging results. Bottom: representative photographs of H&E-stained tissues sections derived from all 4 different MSC-sources are shown as indicated. Student t test *P < .05, **P < .00001.
Chondrogenic differentiation and vascularization precedes BM formation in vivo
Development of long bones and regenerative bone formation is dependent on the capacity to form bone tissue through an intermediate chondrogenic state.10 To quantify time-dependent chondrogenic differentiation and BM formation, we performed serial histologic analyses of transplanted BM-MSCs. At 2 weeks, we observed graft vascularization associated with differentiation of BM-MCSs into hypertrophic chondrocytes and formation of cartilage-associated extracellular matrix as evidenced by formation of lacunar structures, as well as strong blue and green signals in alcian blue and pentachrome staining, respectively. We did not observe signs of graft vascularization in WAT-, UC-, and skin-derived MSCs (data not shown). Consistent with this observation, we also found significantly higher transcript levels for proangiogenic molecules, including VEGFA, FZD1, IL6, and ANGPT, in BM-MSCs compared with WAT-, UC-, and skin-derived MSCs (supplemental Figure 5). At this 2-week time point, no discernible hematopoietic tissue and little osteocyte formation were present (Figure 3C-D). Cartilage-rich areas were then increasingly replaced by hematopoietic tissue, with a concomitant increase of bone matrix at 4 and 6 weeks as evidenced by formation of eosinophilic areas within cartilage, overlapping with yellow/red areas in pentachrome stainings. These areas corresponded to bone formation with increased collagen and osteoid deposition, composed almost entirely of human cells as evidenced by human-specific vimentin staining. By 6 weeks, areas of presumably murine hematopoietic tissue negative for human vimentin could be observed within osteocyte-rich tissue, resembling a trabecular bone structure (Figure 3C-D). Time-dependent chondrogenic differentiation followed by hematopoietic invasion and bone formation was consistently observed in transplanted BM-MSCs from multiple donors.
To further quantify the extent of calcified bone formed in mice transplanted with either BM-MSCs, WAT-, UC-, or skin-derived MSCs, we performed in vivo near-infrared imaging using the bisphosphonate imaging agent OsteoSense, which integrates into areas of active bone growth and resorption after both noninductive and osteoinductive protocols (Figure 3E). BM-MSCs exhibited superior levels of detectable calcified bone, scored relative to the animal’s knee (supplemental Figure 3A), in transplanted tissues (Figure 3F). Importantly, obvious bone formation from WAT-, UC-, and skin-derived MSCs was missing not only after subcutaneous injection in matrigel (noninductive protocol) but also after maximal osteogenic stimulation, which includes osteogenic cell culture, cell seeding on osteoconductive TCP/HA scaffolds, and daily subcutaneous anabolic parathyroid hormone treatment (osteoinductive protocol, Figure 3E).34 Bone formation in BM-MSC-containing transplants generated exclusively under noninductive conditions was additionally verified by flat-panel volume computed tomography, microcomputed tomography, and magnetic resonance imaging. Depicted dense areas in the flanks of transplanted mice resemble normal bone tissue (supplemental Figure 3B-D; supplemental Video 1).
BM niche develops from sites that undergo endochondral ossification
Pelvic and long bones ossify via a cartilage intermediate (endochondral ossification), whereas skull bones undergo intramembranous ossification. To determine whether in vivo BM niche formation is linked exclusively to MSCs derived from sites of endochondral ossification, we transplanted iliac crest–derived BM-MSCs and human skull–derived (parietal bone) mesenchymal cells into murine kidney capsules in a previously established model.11 Similar to findings using the subcutaneous model, histologic analysis of transplants from iliac crest–derived BM-MSCs revealed evidence of time-dependent cartilage and bone formation accompanied by vascularization and sinusoidal structure development resembling a BM cavity. However, despite an identical immunophenotype of both cell types in vitro (supplemental Figure 4), mesenchymal cells derived from human parietal bone did not form cartilage or bone and showed only limited vascularization compared with BM-MSC-derived transplants. Reduced graft vascularization might in part explain the lack of hematopoietic engraftment in non-BM-MSC transplants.
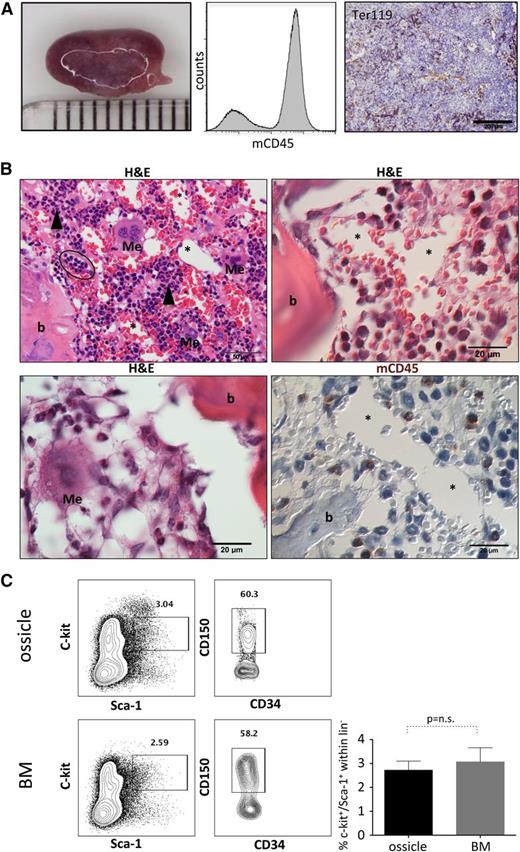
Homing of murine hematopoietic stem cells to human BM-MSC-derived transplants
Previous work has noted areas of hematopoietic tissue within bone and cartilaginous structures in ectopic transplants of human BM-MSCs, including different hematopoietic lineages and HSCs.12,35,36 We also noted that BM-MSC-derived transplants (hereby referred to as “ossicles”) adopted a dark purplish hue over 4 to 6 weeks (Figure 4A), which correlated with the presence of mouse hematopoietic tissue evidenced by murine CD45-positive cells directly aspirated from each ossicle by flow cytometry and evidenced by histology and murine Ter119 immunostaining (Figure 4A-B). Areas of myeloid precursor cells adjacent to both endosteal vascular structures, frequent megakaryocytes with multiple nuclei, erythroid islands, and vascular sinusoids lined with endothelial cells surrounding luminal erythrocytes could be discerned under higher power in keeping with formation of a near-complete but ectopic BM niche structure that can attract and support murine hematopoiesis (Figure 4B). Furthermore, we observed a similar frequency of lineage−sca1+c-kit+ (LSK) progenitors (2.7 ± 0.2% vs 3.1 ± 0.3%, P value not significant) and LSK/flk-2−/CD34−/CD150+ long-term HSCs (35.9 ± 17.3% vs 35 ± 16.4% within LSK/flk-2−CD34−, P value not significant) aspirated from ossicle cavities (12 ossicles from a total of 3 transplanted mice) compared with the normal BM of the same animals (Figure 4C).
Homing and maintenance of murine hematopoiesis in ectopic niches. (A) Left: representative macroscopic photograph of an ossicle generated by 2 × 106 BM-MSCs at 10 weeks posttransplant (millimeter scale is shown). Abundant hematopoietic tissue within the transplant is reflected by purple coloring. Middle: flow cytometric assessment of the hematopoietic tissue after crushing the ossicle confirmed the presence of mouse CD45+ cells. Right: immunohistochemical staining for TER119 marks erythroid cells within the invaded marrow space. (B) Upper left, upper right, and lower left: hematoxylin and eosin staining illustrating invasion of all three major blood cell lineages, including leukocytes and myeloid precursors (black arrowheads), erythrocytes with foci of immature red cells (black oval), as well as multinucleated megakaryocytes (Me) adjacent to bone (b) and/or BM sinusoids (asterisks). Lower right: immunohistochemistry for murine CD45 (mCD45) showing murine and hematopoietic origin of migrated cells within the ectopic marrow niche. Megakaryocytes (Me) and BM sinusoids (asterisks) are shown. (C) BM-MSC-derived marrow niches are colonized by immature murine HSPCs. Representative flow cytometric contour plots showing analysis of c-kit and Sca-1 surface expression on lineage− cells (left plots) within BM-MSC-derived ossicles (upper row) and the respective mouse BMs (lower row). Lineage−, c-kit+, Sca-1+ cells (rectangular gate) were used for further analysis of murine long-term HSCs. Representative contour plots depict gating on CD150+ LT-HSCs within LSK/CD34−/CD135− cells (right plots). Bar graph summarizes percentage (mean ± SD) of c-kit+/Sca-1+ cells within the lineage− (lin−) population (n = 4; Student t test P value not significant).
Homing and maintenance of murine hematopoiesis in ectopic niches. (A) Left: representative macroscopic photograph of an ossicle generated by 2 × 106 BM-MSCs at 10 weeks posttransplant (millimeter scale is shown). Abundant hematopoietic tissue within the transplant is reflected by purple coloring. Middle: flow cytometric assessment of the hematopoietic tissue after crushing the ossicle confirmed the presence of mouse CD45+ cells. Right: immunohistochemical staining for TER119 marks erythroid cells within the invaded marrow space. (B) Upper left, upper right, and lower left: hematoxylin and eosin staining illustrating invasion of all three major blood cell lineages, including leukocytes and myeloid precursors (black arrowheads), erythrocytes with foci of immature red cells (black oval), as well as multinucleated megakaryocytes (Me) adjacent to bone (b) and/or BM sinusoids (asterisks). Lower right: immunohistochemistry for murine CD45 (mCD45) showing murine and hematopoietic origin of migrated cells within the ectopic marrow niche. Megakaryocytes (Me) and BM sinusoids (asterisks) are shown. (C) BM-MSC-derived marrow niches are colonized by immature murine HSPCs. Representative flow cytometric contour plots showing analysis of c-kit and Sca-1 surface expression on lineage− cells (left plots) within BM-MSC-derived ossicles (upper row) and the respective mouse BMs (lower row). Lineage−, c-kit+, Sca-1+ cells (rectangular gate) were used for further analysis of murine long-term HSCs. Representative contour plots depict gating on CD150+ LT-HSCs within LSK/CD34−/CD135− cells (right plots). Bar graph summarizes percentage (mean ± SD) of c-kit+/Sca-1+ cells within the lineage− (lin−) population (n = 4; Student t test P value not significant).
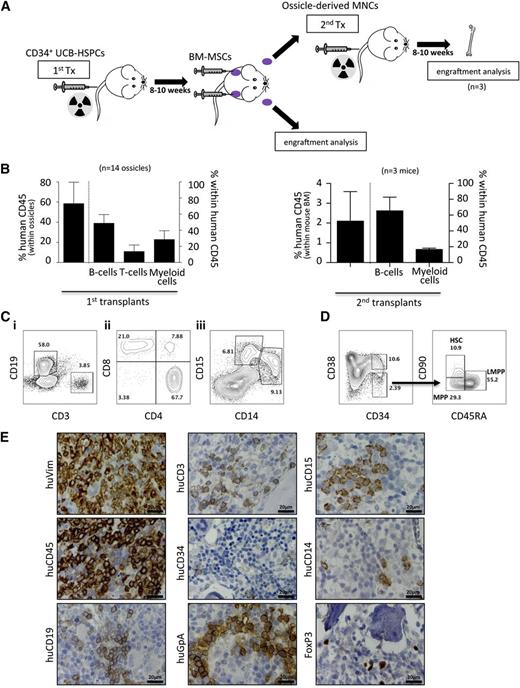
BMMSC ossicles support human hematopoiesis
Gene expression analysis of BM-MSCs showed significantly higher expression of several human hematopoietic regulators, including INHBA, FZD1, TLR4, PDGFC, ANGPT1, VEGFA, CTGF, and VEGFC compared with MSCs from WAT, UC, and skin (supplemental Figure 5). To test whether BM-MSCs could also support human hematopoiesis in vivo, irradiated NSG mice were first transplanted with UCB-derived CD34+ HSPCs 8 weeks prior to subcutaneous implantation of BM-MSCs to generate human/mouse hematopoietic chimeras (n = 5 UCB donors; Figure 5A). After >10% blood chimerism was confirmed by flow cytometry, BM-MSCs were subsequently transplanted into dorsal subcutaneous tissue as previously, allowing them to develop a BM niche over 8 to 10 weeks. We then assessed these established ossicles (n = 14) for engraftment of both human and mouse hematopoietic elements. We observed up to 80% human chimerism within ossicle marrow spaces by flow cytometry (Figure 5B) comprising human B-lymphoid (CD19), T-lymphoid (CD3, CD8, CD4, and FoxP3), and myeloid (CD14 and CD15) cells (Figure 5B-D). Furthermore, human hematopoietic stem and progenitor subcompartments, including HSCs (CD34+/CD38−/CD90+/CD45RA−), multipotent progenitor cells (CD34+/CD38−/CD90−/CD45RA−), and lymphoid primed multipotent progenitor cells (CD34+/CD38−/CD90−/CD45RA+) were robustly engrafted in all ossicles analyzed (Figure 5D). Human glycophorin A+ erythroid cells were discernible by immunostaining of ossicle marrow with a species-specific monoclonal antibody (Figure 5E). Most importantly, mononuclear cells derived from ossicles engrafted with 3 different UCB-derived CD34+ HSPCs gave rise to human hematopoiesis in mouse BM when transplanted by tail vein injection into secondary recipients (n = 3; Figure 5B). Together, these results indicate that human BM-MSC-derived ossicles facilitate homing and establishment of trilineage human hematopoiesis and hematopoietic stem cells.
Homing and maintenance of multilineage human hematopoiesis and functional HSPCs in ectopic niches. (A) Schematic illustration of the experimental procedure for studying homing and engraftment of human hematopoiesis. Hematopoietic chimerism (human/mouse) was induced by transplanting UCB-derived CD34+ HSPCs in advance (8-10 weeks) of BM-MSC transplants (1st Tx). After formation of ectopic niches by BM-MSC (indicated by purple color), hematopoietic cells were recovered and either subjected to flow cytometric engraftment analysis or transplanted into irradiated secondary recipients (2nd Tx). (B) Bar graphs show percentage of human chimerism and corresponding leukocyte subsets, including B cells, T cells, and myeloid cells within human CD45-positive cells within ossicles (n = 14; 5 different UCB transplants) of primary recipients (left graph) and within BM (n = 3; 3 different primary UCB transplants) of secondary recipients (right graph). (C) Representative flow-cytometric contour plots depicting (i) representative gating of human B cells (CD19) and T-cells (CD3) within human CD45-positive cells; (ii) T-cell subpopulation analysis revealing CD4+ T-helper cells, CD8+ cytotoxic T-cells, and a minor proportion of double-positive T cells; and (iii) CD33-positive myeloid cells comprising substantial amounts of CD14+ monocytes and CD15+ granulocytes. (D) Human cells within ossicles were analyzed for CD34, CD38, CD90, and CD45RA by flow cytometry. The left plot is gated on lineage negative live cells, whereas the right plots are gated on lineage-negative CD34+CD38− cells. The latter cells contain human HSCs (CD90+/CD45RA−), multipotent progenitor cells (MPPs, CD90−/CD45RA−), and lymphoid primed multipotent progenitor cells (LMPPs, CD90−/CD45RA+). Data shown are representative of multiple samples. Numbers within the plots indicate percentages of the respective parent population. (E) Immunohistochemistry of explanted ossicles confirms flow cytometric results. Representative photographs of immunohistochemical stainings using antibodies specifically binding human vimentin (huVim), huCD45, huCD19, huCD3, huCD34, human glycophorin A (huGpA), huCD15, huCD14, and huFoxP3. MNCs, mononuclear cells.
Homing and maintenance of multilineage human hematopoiesis and functional HSPCs in ectopic niches. (A) Schematic illustration of the experimental procedure for studying homing and engraftment of human hematopoiesis. Hematopoietic chimerism (human/mouse) was induced by transplanting UCB-derived CD34+ HSPCs in advance (8-10 weeks) of BM-MSC transplants (1st Tx). After formation of ectopic niches by BM-MSC (indicated by purple color), hematopoietic cells were recovered and either subjected to flow cytometric engraftment analysis or transplanted into irradiated secondary recipients (2nd Tx). (B) Bar graphs show percentage of human chimerism and corresponding leukocyte subsets, including B cells, T cells, and myeloid cells within human CD45-positive cells within ossicles (n = 14; 5 different UCB transplants) of primary recipients (left graph) and within BM (n = 3; 3 different primary UCB transplants) of secondary recipients (right graph). (C) Representative flow-cytometric contour plots depicting (i) representative gating of human B cells (CD19) and T-cells (CD3) within human CD45-positive cells; (ii) T-cell subpopulation analysis revealing CD4+ T-helper cells, CD8+ cytotoxic T-cells, and a minor proportion of double-positive T cells; and (iii) CD33-positive myeloid cells comprising substantial amounts of CD14+ monocytes and CD15+ granulocytes. (D) Human cells within ossicles were analyzed for CD34, CD38, CD90, and CD45RA by flow cytometry. The left plot is gated on lineage negative live cells, whereas the right plots are gated on lineage-negative CD34+CD38− cells. The latter cells contain human HSCs (CD90+/CD45RA−), multipotent progenitor cells (MPPs, CD90−/CD45RA−), and lymphoid primed multipotent progenitor cells (LMPPs, CD90−/CD45RA+). Data shown are representative of multiple samples. Numbers within the plots indicate percentages of the respective parent population. (E) Immunohistochemistry of explanted ossicles confirms flow cytometric results. Representative photographs of immunohistochemical stainings using antibodies specifically binding human vimentin (huVim), huCD45, huCD19, huCD3, huCD34, human glycophorin A (huGpA), huCD15, huCD14, and huFoxP3. MNCs, mononuclear cells.
Discussion
Despite the surge in clinical interest, little is known about the in vivo biology of MSCs when transplanted from different tissue origins, and such differences may be underappreciated and clinically relevant. We chose to use a standard humanized expansion protocol (pHPL) to study clinical grade–expanded MSCs, a method common to a number of current proposals submitted to the Food and Drug Administration.1 We found in vivo differences between 4 commonly used sources of MSCs (BM, WAT, UC, and skin) cultured with this method when transplanted as a pool of cells into subcutaneous tissue in mice. Most apparent was the unique ability of BM-MSCs to form functional BM supportive of both murine and human hematopoietic tissue, which was chronologically preceded by extensive cartilage tissue formation. In our study, immunohistochemical detection of human vimentin allowed us to identify the human origin of the newly formed cartilage and bone, proving their derivation from transplanted human MSCs.
Endochondral ossification and ectopic hematopoietic niche formation by BM-MSCs has been reported previously,36-38 and ectopic bone formation was shown to be superior in BM-MSCs compared with adipose tissue–derived MSCs.30 Nevertheless, a more comprehensive comparison with other MSC sources—in humanized culture conditions and in the absence of specific induction protocols—has not been shown before. Although various MSC sources show similarities in cell surface markers and morphology, our in vivo results inspired a search for underlying differences in these cells at an early time point during in vitro expansion. Here, although applying an extended antibody panel including previously suggested MSC-defining epitopes,28 pericyte-associated epitopes,39 and surface proteins expressed on primary BM-colony forming unit fibroblastoid–generating cells,40,41 we could not distinguish between MSC sources simply by flow cytometry. However, we found significant differences in the gene expression pattern and genome-wide CpG methylation of BM-MSCs versus other sources. Integrating the transcriptome and methylome differences revealed a BM-MSC-specific signature consisting of 85 hypomethylated and transcriptionally upregulated genes, including RUNX2, RUNX3, BGLAP, ITGA10, and MMP13 (Figure 2E). Intriguingly, this relatively small epigenetically determined gene set was significantly enriched for known regulators of skeletal and mesodermal development (DAVID; P < .02). This finding is consistent with a latent epigenetic program for endochondral differentiation present in BM-MSCs that is independent of donor age. Interestingly, we did not see significant upregulation of the histone-lysine demethylases KDM4B and KDM6B, previously reported to be upregulated during osteogenic differention of MSCs (supplemental Figure 2B).42 Differences in epigenetic marks between MSC sources and expression levels of transcription factors such as RUNX3 may therefore be more informative regarding in vivo developmental potential than analysis of cell surface markers by flow cytometry.
In vitro differentiation assays are another method commonly used to evaluate the potency of MSCs. Although somewhat informative, these assays can be misleading.43 Although similarly capable of Ca2+ accumulation and lipid uptake, MSCs from BM displayed higher baseline chondrogenic gene expression (Figure 2F) and robust formation of hypertrophic chondrocytes in a 3-dimensional differentiation assay (Figure 3A).44 It will be interesting to test other sources of MSCs for 3-dimensional chondrocyte differentiation potential in vitro and to determine whether this correlates with BM niche formation in vivo as a potential surrogate biological assay.
Recent studies show that BM-MSCs can provide a supportive microenvironment by establishing a physiological stem cell niche through ectopic endochondral ossification12,36,38 or “endochondral myogenesis.”45 Our time course analysis suggests that a cartilage intermediate arising from BM-MSCs may be required for proper ectopic BM formation, because it demonstrated a close dependence of the marrow niche on a specific endochondral differentiation program attributed selectively to BM-MSCs. Whether ossification per se is absolutely required for marrow cavity formation in our model remains to be tested.
Furthermore, MSCs that undergo chondrogenic differentiation show increased graft vascularization, which might contribute to the observed distinct hematopoietic content. Endochondral bone formation is a tightly regulated process in the skeletal development and is dependent on regulatory mechanisms precisely controlling the balance between proliferation and differentiation. We now provide additional evidence that epigenetic events control MSC commitment toward cartilage and bone fate, and found a number of transcription factors including RUNX3 to be hypomethylated and overexpressed in BM-MSCs compared with other sources. RUNX3 is a Runt-domain family transcription factor that controls craniofacial skeletal development,46 cartilage formation, and the process of endochondral bone formation.32 Future studies using short hairpin RNA or gene knockout techniques with chromatin-immunoprecipitation approaches will be required to determine the biological role of this gene in hematopoietic niche formation.
Our finding that BM-MSCs robustly build a functional human marrow niche in NSG mice also allowed us to directly study human HSC engraftment and maintenance in a structurally similar human microenvironment. Human HSCs gave rise to multipotent hematopoietic progenitors (multipotent progenitor cells and lymphoid primed multipotent progenitor cells) in our model, which was also consistent with local production of niche hematopoietic factors (Figure 5D). Similar findings in a murine system have been reported previously.36 The fact that we could also detect high numbers of human erythroid lineage cells as well as robust differentiation of mature myeloid cells suggests a local cytokine milieu resembling human marrow conditions. Potentially, this model circumvents the need to use mice expressing human cytokines, which have been shown to improve engraftment rates of some hematopoietic malignancies and are capable of forming innate human immune cells.47-49 Using a similar model, we previously showed feasibility of genetic modifications in niche cells.5 Here, we provide further evidence that humanized marrow models can facilitate elucidation of critical niche stem cell components for the study of both healthy and malignant hematopoiesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Daniela Thaler, Birgit Freilhauer, Claudia Url, Daniela Blattl, and Anna Ortner for excellent technical assistance; Alexander Hofmeister for assistance with microcomputed tomography imaging; Clemens Diwoky for assistance with magnetic resonance imaging; Alexander Deutsch for help with analyzing quantitative reverse transcription–polymerase chain reaction data; Monica Farrell for editorial assistance; and Matthias Schick (German Cancer Research Center, Heidelberg, Germany) for hybridization and preliminary analysis of DNA-methylation data. Krisztina Szöke and Jan E. Brinchmann provided WAT-derived SVF.
This work was supported by Austrian Research Foundation grant N211-NAN (D.S.); the Adult Stem Cell Research Foundation and a Young Investigator starting grant of the Medical University of Graz (A.R.); German Research Foundation grants KFO-183 and TP6 (U.F.H.) and WA 1706/2-1 (W.W.); the Stem Cell Network North Rhine-Westphalia (W.W.); National Institutes of Health, National Institute of Dental and Craniofacial Research grants R01 DE021683-01 (M.A.) and R21 DE02423001 (M.T.L.), National Heart, Lung, and Blood Institute grants U01 HL099776 (M.T.L.) and U01 HL099999 (I.L.W. and R.M.), and National Cancer Institute grants AML P01 CA55164, CA016672, CA143805, CA049639, CA136411, and CA100632 (M.A.) and R01 86065 (I.L.W.); the Virginia and D.K. Ludwig Fund for Cancer Research (I.L.W.); a Paul and Mary Haas Chair in Genetics grant (M.A.); California Institute for Regenerative Medicine grant TR1-01249 (M.T.L.); the Oak Foundation, the Hagey Laboratory for Pediatric Regenerative Medicine, and the Gunn/Olivier Research Fund (M.T.L); and a CJ Martin Overseas Biomedical Research Fellowship (D.T.). A.R. is currently supported by an Erwin Schroedinger fellowship from the Austrian Science Fund.
Authorship
Contribution: A.R. designed and performed the research, analyzed the data, and wrote the manuscript; N.E. designed and performed the research, analyzed the data, and reviewed the manuscript; D.T. designed and performed the research, analyzed the data, and wrote the manuscript; N.A.H., M.F., C.K.C., K.S.-Y., E.-Y.S., T.W., U.F.H., C.D., F.A., and K.S. performed the research; S.S., C.B.-S., S.T., Q.L., and W.W. analyzed the data; M.A., I.L.W., M.T.L., and R.M. contributed to the research design and critically reviewed the manuscript; and D.S. designed the research, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dirk Strunk, Experimental and Clinical Cell Therapy Institute, Spinal Cord and Tissue Regeneration Center Salzburg, Paracelsus Medical University, Strubergasse 22, A-5020 Salzburg, Austria; e-mail: dirk.strunk@pmu.ac.at; and Andreas Reinisch, Institute for Stem Cell Biology and Regenerative Medicine, Stanford University, 265 Campus Dr, Stanford, CA 94305; e-mail: reinisch@stanford.edu.

![Figure 1. BM-MSCs, but not WAT-, UC-, or skin-derived MSCs form bone, cartilage, and marrow tissue in vivo. (A) Phase-contrast images depicting spindle-shaped morphology of BM-MSCs and WAT-, UC-, and skin-derived MSCs. (B) Flow cytometric profiles of the same respective cell populations as in panel A. Histograms show fluorescent cell surface staining intensity of anti-CD90, anti-CD73, anti-CD105, anti-CD45, anti-HLA-DR, anti-CD14, anti-CD19, and anti-CD34 monoclonal antibodies conjugated to fluorophores (gray shading) and corresponding isotype control (no shading). (C) Trilineage in vitro differentiation of BM-MSCs and WAT-, UC-, and skin-derived MSCs (shown left to right). MSCs were induced to differentiate along osteogenic, adipogenic, and chondrogenic lineage in vitro according to standard protocols. After 21 days of osteogenic induction, cells were stained with alizarin red for visualization of Ca2+ accumulation (upper left image for each). Adipogenic differentiation capacity was visualized after 14 days of induction by staining lipid droplets with fluorescent nile red (upper right image for each). A 3-dimensional chondrogenic differentiation assay in Transwell plates was used for evaluation of chondrogenic differentiation potential. Tissue was fixed and further processed for histologic evaluation of chondrotypic glucosaminoglycans with toluidine blue after 28 days of induction. (D) BM-MSCs form bone, cartilage, and marrow tissue in vivo. Upper image shows schematic of the in vivo protocol. After expansion in pHPL-supplemented α-modified minimum essential medium (α-MEM), 2 × 106 MSCs were resuspended in matrigel and subcutaneously injected into NSG mice. Spontaneous in vivo tissue formation was evaluated 8 to 12 weeks postimplantation by histology. Representative photomicrographs of hematoxylin and eosin ([H&E], upper row) and pentachrome-stained sections (lower row) are shown. Inserts show low-magnification overview photographs. ECM, extracellular matrix.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/2/10.1182_blood-2014-04-572255/4/m_249f1.jpeg?Expires=1767728544&Signature=pjFoex3RvtyKhtc6aAmuce-gnL2~f~c95W7ltagw-lBVrJq9iqBZ-w0lWQ4ebEYYMRtkSr2jUzjDf5g5taLdvbEWInm1Dhd1jVVrtodY4ot6govgnvXNTJUncn7QLGLhYQwNtVHLm2q4IFklUb4SGrH6tX4tOvlHADfbDO52Js2oQA30YSeeCOg2Ztcd2~AszVEblELUnLnjTASVDs97eDo7EiRuslghH5KDQXXN5nB3ml~kazWQKTPWnBxp5G5vVkMhTRuNl8ZfrhLyrC65BOuAiUdrEsKXMQfoinfdrZK5-GeD-4WVo8tn9Hrf2IyxFB1QN2UAmKoglDQQwDi6fQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)