Key Points
IDH2 R140Q expression in TF-1 cells can induce DNA and histone hypermethylation that mirrors human IDH2 mutant acute myeloid leukemia.
The hypermethylation can be reversed on treatment with AGI-6780, an IDH2 mutant-specific small-molecule inhibitor.
Abstract
Mutations of IDH1 and IDH2, which produce the oncometabolite 2-hydroxyglutarate (2HG), have been identified in several tumors, including acute myeloid leukemia. Recent studies have shown that expression of the IDH mutant enzymes results in high levels of 2HG and a block in cellular differentiation that can be reversed with IDH mutant-specific small-molecule inhibitors. To further understand the role of IDH mutations in cancer, we conducted mechanistic studies in the TF-1 IDH2 R140Q erythroleukemia model system and found that IDH2 mutant expression caused both histone and genomic DNA methylation changes that can be reversed when IDH2 mutant activity is inhibited. Specifically, histone hypermethylation is rapidly reversed within days, whereas reversal of DNA hypermethylation proceeds in a progressive manner over the course of weeks. We identified several gene signatures implicated in tumorigenesis of leukemia and lymphoma, indicating a selective modulation of relevant cancer genes by IDH mutations. As methylation of DNA and histones is closely linked to mRNA expression and differentiation, these results indicate that IDH2 mutant inhibition may function as a cancer therapy via histone and DNA demethylation at genes involved in differentiation and tumorigenesis.
Introduction
Active site mutations in IDH1 (R132) and IDH2 (R172 and R140) that produce high levels of 2-hydroxyglutarate (2HG) have been identified in several human cancers.1-3 IDH mutations have been shown to cause DNA hypermethylation in both gliomas and leukemias via inhibition of methylcytosine dioxygenase TET2.4,5 Mutant IDH can also promote histone hypermethylation through competitive inhibition of α-ketoglutarate (aKG)-dependent Jumonji-C histone demethylases, thereby activating or deactivating expression of associated genes.4,6,7
We have shown that mutant IDH1 and IDH2 can affect cell differentiation in solid and liquid tumors.8-10 An IDH1 R132H inhibitor, AGI-5198, delayed growth and promoted differentiation of glioma cells while reducing histone H3K9 trimethylation.8 Leukemic cell differentiation was also induced in primary human patient samples harboring an IDH2 R140Q mutation when they were treated ex vivo with AGI-6780, an IDH2 R140Q allosteric inhibitor.9 However, the mechanism by which IDH2 mutant activity and 2HG levels contribute to cellular differentiation and tumorigenesis is not fully understood. High levels of 2HG have been shown to competitively inhibit aKG-dependent dioxygenases, leading to broad epigenetic changes.11 Therefore, we sought to investigate the global and gene-specific effects of mutant IDH2 inhibition in TF-1 cells expressing IDH2 R140Q. Probing the effects of IDH2 R140Q expression on histone and DNA methylation and gene expression on a genome-wide scale allowed us to identify gene signatures that are affected by IDH2 mutations and that may subsequently function to regulate erythrocyte differentiation.
Materials and methods
Reagents and antibodies
The tri-methyl H3K4, H3K27, and H3K36 total H3 antibodies were from Cell Signaling Technology, Inc. (Danvers, MA). The tri-methyl H3K9 antibody for western blots was from Abcam (Boston, MA). Recombinant human erythropoietin (EPO) was from R&D Systems (catalog no. rhEPO 287-TC). RIPA lysis and extraction buffer and halt protease inhibitor cocktail were from Thermo Scientific (Rockford, IL). IDH2 R140Q inhibitor AGI-6780 compound synthesis, TF-1 cell culturing and single-clone generation, and EPO-induced differentiation were all performed as described previously.9
2HG measurement
Labeled 13C5-2HG was obtained from Agios Pharmaceuticals, and 2HG was obtained from Toronto Research Chemicals (Toronto, Canada). Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis was performed using an AB Sciex 4000 (Framingham, MA) operating in negative electrospray mode. Multiple reaction monitoring (MRM) data were acquired for each compound, using the following transitions: 2HG (146.9/128.8 amu), 13C5-2HG (151.9/133.8 amu), and 3HMG (160.9/98.9 amu). Chromatographic separation was performed using an ion-exchange column (Fast Acid analysis, 9 μm, 7.8 × 100 mm; BioRad, Waltham, MA). The flow rate was 1 mL/min of 0.1% formic acid in water, with a total run time of 4 minutes.
Cell pellets were resuspended in specified volumes of 80:20 MeOH:water, centrifuged for 10 minutes at 14 000 rpm. Next, 30 μL supernatant was extracted by adding 170 μL methanol with 200 ng/mL 13C5-2HG as an internal standard. Samples were then vortexed and centrifuged at 4000 rpm at 5°C, and 150 μL supernatant was transferred to a clean 96-well plate. The samples were dried and reconstituted in 200 μL 0.1% formic acid in water, and 10 μL was injected on column.
Methylation data
Methylation data generated using the Illumina Methylaton450 platform were normalized using Illumina software and the MethyLumi package.12 Differential methylation analysis of replicate methylation samples was done in R, using the Minfi package.13,14 The analysis process includes executing methylumIDAT to normalize raw Illumina (San Diego, CA) IDAT files, forming the result into MethyLumiSets with phenotypic data, and then identifying differentially methylated probes (DMPs) by executing dmpFinder in categorical mode for appropriate contrasts and calculating q-values and β-value differences. Minfi performs an F-test on the methylation values in this mode and then uses the false discovery method to adjust for multiple hypothesis testing and to produce a q-value.13,15 Methylation changes were considered significant at a q-value of 0.05 and minimum β-value difference of 0.1, consistent with criteria suggested in Du et al.16 2HG-specific methylation changes were calculated by subtracting the methylation in the given contrast (eg, ±compound) in TF-1 R140Q (high 2HG) from the equivalent methylation change in TF-1 pLVX (basal 2HG). This controls for any background differential effects of drug vs dimethylsulfoxide that are not related to 2HG. Methylation data sets have been deposited in Gene Expression Omnibus (GEO) as GSE51352. Significant overlap in differentially methylated probes was assessed by the χ-squared test.
Principal component analysis
To assess the similarity between TF-1 IDH2 R140Q cells and acute myeloid leukemia (AML) patient samples, principal component analysis was performed. For both the cell and patient samples, differential methylation analysis (Methylation data) of the TF-1 IDH2 R140Q vs TF-1 pLVX (empty vector) transformed cells identified the most DMPs in each data set. The DMPs present in both data sets were then used to map the samples onto a 2-dimensional grid via principal component analysis.17
Gene signature over-representation analysis
To infer gene signatures that may be commonly affected by a methylation change, significantly differentially methylated probes were mapped to their annotated genes. Because methylation measurements and gene expression values come from different distributions, we did not use quantitative methylation level changes as inputs to pathway enrichment methods that have been designed with gene and protein expression in mind. Instead, we created a list of differentially methylated genes by taking all genes with 1 or more probes exhibiting significant methylation change and excluding all other genes (ie, with no differentially methylated probes). This provides a binary gene scoring metric suitable for over-representation analysis. An over-representation analysis for each gene signature in MSigDB curated gene signatures/chemical and genetic perturbations was performed by calculating the probability of selecting the number of differentially methylated genes from the gene signature by chance via binomial sampling.18 Multiple testing correction was performed via the false-discovery control method.19 Calculations were performed in the R language.
Genomic annotation of methylation regions
Annotation of the gene component to which a methylation site corresponds (ie, 5′ UTR, exon, etc.) was done by mapping the Illumina-provided chromosome number and position to the human assembly hg19 in the University of California, Santa Cruz, Genome Browser.20
DNA methylation assay
Ten million cells were harvested, and DNA was isolated using the DNeasy Blood and Tissues kit (Qiagen, Venlo, The Netherlands). To determine the amount of methylated DNA, DNA was either not digested (M0) or was digested with a methylation-dependent enzyme (Md; Qiagen, Epitect DNA methylation restriction kit), following the manufacturer’s protocol. The amount of completely digested DNA was determined by digesting with the methylation-insensitive enzyme MspI in NEB buffer 4 (MMsp). The amount of remaining DNA was determined by quantitative polymerase chain reaction (qPCR), using the Applied Biosystems 7900 Real-time PCR system. The HBG −51/−54 bp CpG site was detected using the following probes: FAM-CC CCT GGC CTC ACT GGA TAC TCT A-TAM; forward, TGC CTT GAC CAA TAG CCT TG; reverse, AGT GTG TGG AAC TGC TGA AG. The DNA concentrations were calculated as CM0 = 2−CT(M0), CMd = 2−CT(Md), and CMMsp = 2−CT(MMsp). The percentage of methylated HBG (−51/−54 bp) DNA was calculated by the following equation: % meth = 1 − [CMd/(CM0 − CMMsp)] × 100.
Chromatin immunoprecipitation assay
TF-1 IDH2 R140Q and TF-1 pLVX cells were grown for 10 days in the presence of 5 ng/mL granulocyte macrophage-colony-stimulating factor (GM-CSF) and either dimethylsulfoxide, 0.2 μM, or 1 μM AGI-6780. The cells were washed 4 times in PBS and grown in GM-CSF or 2 IU/mL EPO + dimethylsulfoxide, 0.2 μM, or 1 μM AGI-6780 for 7 additional days. Analysis of H3K9me3 association with the HBG promoter via chromatin immunoprecipitation (ChIP) was carried out as recommended by the manufacturer (Upstate Biotechnology, Lake Placid, NY), with the following modifications: 25 million cells were used for protein–DNA cross-linking by incubating at 37°C for 10 minutes in 1% formaldehyde. The cross-linking reaction was quenched by the addition of 0.125 M glycine. Cells were spun down, washed twice in ice-cold PBS, and lysed in 1 mL lysis buffer. The lysate was sonicated (Bioruptor UCD-200, Seraing, Belgium) to produce DNA fragments ranging from 0.2 to 1.0 KB in size, and the efficiency of DNA sonication was evaluated on a 2% agarose gel. DNA was immunoprecipitated from 200 μL of the original lysate (1:10 diluted in dilution buffer) with 3 μg of the anti-H3K9me3 antibody or 3 μg of immunoglobulin G (IgG) control. Immunoprecipitates were washed following manufacturer’s instructions, and the histone–DNA complex was eluted twice with 150 μL elution buffer. Cross-linking of combined eluates was reversed by the addition of 12 μL of 5 M NaCl and boiling for 30 minutes. The eluates were treated with RNAse for 10 minutes, and proteinase K for 1 hour. The DNA was purified using the Qiaquick PCR purification kit and eluted with 50 μL elution buffer (Qiagen, Venlo, The Netherlands). At the same time, 20-μL aliquots of the sonicated lysate (input DNA) were directly subjected to 5 M NaCl, RNase, and proteinase K treatment. DNA was purified with a Qiaquick PCR purification kit and eluted with 50 μL elution buffer. For the ChIP assay, the anti-H3K9me3 antibody (Active Motif, Carlsbad, CA) was used to enrich for H3K9me3 DNA binding regions, whereas nonspecific IgG was used as a negative control. Fold enrichment of H3K9me3 methylation at the HBG −51/−54 bp CpG promoter was determined by qPCR and compared with input DNA, using the Applied Biosystems 7900 Real-time PCR system (Life Technologies, Carlsbad, CA). Reverse transcription PCR for the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene promoter (forward, caattccccatctcagtcgt; reverse, tagtagccgggccctacttt) was used as a negative control.
Results
TF-1 cells with IDH2 mutant expression produce physiologically relevant levels of 2HG
To study the biological effect of AGI-6780, we expressed mutant IDH2 R140Q protein in GM-CSF-dependent TF-1 erythroleukemia cells,9 which have a progenitor-like phenotype and have been used to study the biological consequence of TET2 mutations.21 These cells (hereafter referred to as TF-1 R140Q cells) produce high levels of 2HG (5-30 mM) that correlate with mutant IDH2 expression and grow in the absence of GM-CSF. We selected multiple single-cell clones that produced levels of 2HG observed in human tumors (10-20 mM).3,22 All the clones shared similar biological features in terms of 2HG production and growth in the absence of GM-CSF, allowing us to focus on representative clones 11 and 17 for genome-wide methylation chip studies.9
Expression of IDH2 R140Q in TF-1 cells models the methylation changes seen in patients with IDH2 mutant AML
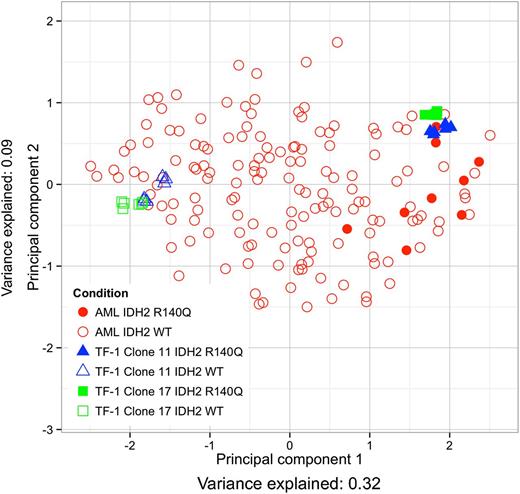
Because IDH2 mutant AML patient samples have been shown to exhibit a DNA hypermethylation phenotype,5 we sought to assess the clinical relevance of using TF-1 R140Q cells as a model system by comparing their DNA methylation pattern with the DNA methylation of primary patient AML samples in The Cancer Genome Atlas.23 The similarity between TF-1 R140Q cells and patient samples was assessed by mapping the probes that were most differentially methylated between IDH2 mutant and IDH2 wild-type (WT) AML patients (while excluding IDH1 mutant AML patients) and TF-1 R140Q and TF-1 pLVX vector control cells onto 2 dimensions, using principal component analysis.17 Replicates of 2 different TF-1 R140Q clones (clones 11 and 17) cluster with the methylation patterns observed in the IDH2 mutant AML patient samples (Figure 1). The greatest amount of methylation change is explained by the first principal component (x-axis) and aligns with IDH2 mutant vs IDH2 WT status of both the TF-1 cells and the AML patient samples (Figure 1 and component mappings in supplemental Table 2, available on the Blood Web site). Other mutations in methylation-related genes did not cluster with IDH2 status (supplemental Figure 1). The number of overlapping DMPs (unless otherwise specified, DMPs are probes with q-value <0.05 and a minimum methylation difference of 0.116 ) between the 2 TF-1 clones and the AML samples is significant in all 3 comparisons and is greatest between the TF-1 clones (supplemental Figure 2A, P-value <1e−16).
TF-1 cells expressing IDH2 R140Q show methylation patterns similar to IDH2 mutant AML patients. Similarity of the most differentially methylated probes is projected onto 2-dimensions by principal component analysis. AML patient samples (red) are either IDH2 WT (open circles) or IDH2 mutant (closed circles). TF-1 samples (clone 11 blue and 17 green) are either IDH2 WT (open triangle/square) or IDH2 R140Q (closed triangle/square).
TF-1 cells expressing IDH2 R140Q show methylation patterns similar to IDH2 mutant AML patients. Similarity of the most differentially methylated probes is projected onto 2-dimensions by principal component analysis. AML patient samples (red) are either IDH2 WT (open circles) or IDH2 mutant (closed circles). TF-1 samples (clone 11 blue and 17 green) are either IDH2 WT (open triangle/square) or IDH2 R140Q (closed triangle/square).
Hierarchical clustering of the most differentially methylated probes between IDH2 mutant and IDH2 WT AML patients and TF-1 R140Q and TF-1 pLVX vector control cells shows that samples cluster first by IDH2 mutation status and then by sample type, AML or TF-1, confirming that larger methylation changes are similar between AML and TF-1 (supplemental Figure 3A). However, if we use a random subset of probes without prefiltering on the most differentially methylated probes, hierarchical clustering segregates by sample type, rather than IDH2 mutant status (supplemental Figure 3B). Altogether, these data suggest that IDH2 R140Q expression that produces high levels of 2HG in TF-1 cells can affect site-specific DNA methylation in a pattern similar to that found in IDH-mutant AML patient samples.
IDH2 mutation leads to increased methylation that is decreased on drug treatment
Further characterization of the DNA methylation pattern showed that TF-1 R140Q cells have an increased number of significantly hypermethylated probes compared with TF-1 pLVX (empty vector control) cells in both clones jointly analyzed (Figure 2A), as well as in each individual clone (Figure 2B-C). No DMPs were detected between TF-1 pLVX cells and TF-1 WT cells expressing IDH2 WT, indicating either would serve as a suitable control (probes with nonsignificant methylation differences are shown in supplemental Figure 4A). Although the majority of the DMPs in each clone show hypermethylation, a nontrivial number of probes are hypomethylated (28% in clone 11 in Figure 2B and 36% in clone 17 in Figure 2C). Joint analysis of the concordance of clones 11 and 17 shows a decrease in hypomethylated probes to 21% (Figure 2A), indicating that DMPs concordant between both clones are largely hypermethylated. Similarly, plotting both clones’ DMPs on 2 axes shows a lower fraction of concordant hypomethylated probes than concordant hypermethylated probes, suggesting that the methylation changes specific to IDH2 mutation are largely hypermethylation (supplemental Figure 5).
IDH2 R140Q inhibitor AGI-6780 reverses DNA hypermethylation. (A-C) R140Q expression results in a DNA hypermethylation phenotype (A, combined clones 11 and 17; B, clone 11; C, clone 17) that can be reversed by 1 μM AGI-6780 treatment of 7 or 8 days (D, left; E) or 28 days (D, middle), with a concomitant decrease in 2HG from 21.44 mM (TF-1 R140Q) to 0.44 mM (TF-1 R140Q + AGI-6780). Venn diagram (inset) shows overlap between methylation sites increased with IDH2 R140Q expression and decreased on AGI-6780 treatment (red circle). A smaller number of DMPs are induced by AGI-6780 treatment of pLVX cells (D, right). Significance, q-value <.05; minimum β-value difference, 0.1.
IDH2 R140Q inhibitor AGI-6780 reverses DNA hypermethylation. (A-C) R140Q expression results in a DNA hypermethylation phenotype (A, combined clones 11 and 17; B, clone 11; C, clone 17) that can be reversed by 1 μM AGI-6780 treatment of 7 or 8 days (D, left; E) or 28 days (D, middle), with a concomitant decrease in 2HG from 21.44 mM (TF-1 R140Q) to 0.44 mM (TF-1 R140Q + AGI-6780). Venn diagram (inset) shows overlap between methylation sites increased with IDH2 R140Q expression and decreased on AGI-6780 treatment (red circle). A smaller number of DMPs are induced by AGI-6780 treatment of pLVX cells (D, right). Significance, q-value <.05; minimum β-value difference, 0.1.
Treatment of TF-1 R140Q clone 11 with 1 μM AGI-6780 decreased intracellular 2HG levels from 21.44 mM (without AGI-6780) to 0.44 mM (supplemental Figure 4B; further experiments showing 2HG reduction in supplemental Figure 4C-D) and, after 7 days, reversed the methylation of 271 probes (Figure 2D, left). Extending the treatment to 28 days further increased the number of demethylated probes to 3107 (Figure 2D, middle). These results suggest that inhibition of mutant IDH2 R140Q has a progressive effect on DNA hypermethylation and are consistent with the idea that 2HG may exert its effects through time-dependent epigenetic rewiring. The magnitude of demethylation at days 7 and 28 of AGI-6780 treatment were significantly correlated; at 7 days, 52% of the 28-day demethylation level was present (supplemental Figure 6A). Similarly, a large fraction of the 3107 probes (3003/3107) demethylated by AGI-6780 at 28 days were the same probes that were hypermethylated by expression of IDH2 R140Q (Figure 2C, Venn diagram). Markedly fewer DMPs were observed in control TF-1 pLVX cells (383 DMPs) compared with TF-1 R140Q cells (3552 DMPs) after 28 days of AGI-6780 treatment (with 1 overlapping probe between the 2 sets), implying a specific effect of AGI-6780 on TF-1 R140Q cells (Figure 2D, right; supplemental Figure 2C; supplemental Table 3).
The methylation changes observed with IDH2 R140Q expression in clone 17 (Figure 2C), after 8 days of treatment with AGI-6780 (Figure 2E), correlate with the magnitude of changes seen in clone 11 (supplemental Figure 6B), whereas the same treatment of control pLVX cells produced no significant methylation changes. The similarity between IDH2 R140Q clone 11 (7 or 28 days treatment) and clone 17 (8 days treatment) is demonstrated by their significant level of DMP overlap (supplemental Figure 2B). Therefore, the observed hypermethylation is specific to IDH2 R140Q expression, and AGI-6780 can induce demethylation of genes hypermethylated by the mutant protein.
Methylation changes strongly enriched in leukemia- and lymphoma-related gene signatures
Analysis of the probes demethylated on treatment with AGI-6780 against the MSigDB/chemical and genetic perturbation data set18 showed that many of the gene signatures modulated by methylation are associated with leukemia and melanoma, as well as differentiation and development (supplemental Table 1). Because the effect of methylation on gene expression varies depending on the methylation site (ie, promoter region or gene body),24-26 we focused the analysis on probes found in promoter regions. We identified significant gene signatures in which 4 of the top 5 signatures were associated with leukemia and lymphoma, indicating that the methylation changes may be targeting pathways affecting leukemic tumorigenesis (Table 1). Although the demethylation associated with AGI-6780 treatment reverses only a subset of the hypermethylation induced by expression of IDH2 R140Q, the compound does affect the methylation status of genes that are highly relevant to leukemic pathogenesis.
Table of gene signatures enriched with genes that are significantly demethylated at promoter regions after 28 d treatment with AGI-6780 (1 μM)
| Overlapping genes . | Signature genes . | Percentage overlapping . | Gene signature name in MSigDB . | Q-value . |
|---|---|---|---|---|
| 17 | 186 | 9% | TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_HSC_DN* | 1.56E−13 |
| 24 | 414 | 6% | MULLIGHAN_MLL_SIGNATURE_2_UP* | 1.81E−13 |
| 23 | 378 | 6% | MULLIGHAN_MLL_SIGNATURE_1_UP* | 1.86E−13 |
| 17 | 223 | 8% | THEILGAARD_NEUTROPHIL_AT_SKIN_WOUND_DN | 3.45E−12 |
| 13 | 131 | 10% | PICCALUGA_ANGIOIMMUNOBLASTIC_LYMPHOMA_DN* | 1.96E−11 |
| Overlapping genes . | Signature genes . | Percentage overlapping . | Gene signature name in MSigDB . | Q-value . |
|---|---|---|---|---|
| 17 | 186 | 9% | TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_HSC_DN* | 1.56E−13 |
| 24 | 414 | 6% | MULLIGHAN_MLL_SIGNATURE_2_UP* | 1.81E−13 |
| 23 | 378 | 6% | MULLIGHAN_MLL_SIGNATURE_1_UP* | 1.86E−13 |
| 17 | 223 | 8% | THEILGAARD_NEUTROPHIL_AT_SKIN_WOUND_DN | 3.45E−12 |
| 13 | 131 | 10% | PICCALUGA_ANGIOIMMUNOBLASTIC_LYMPHOMA_DN* | 1.96E−11 |
Overlapping genes, overlap between gene signature genes and sample genes; signature genes, number of genes in signature; percentage overlapping, percentage of genes in both sample and gene signature; gene signature, gene signature name in MSigDB; Q-value, significance score with multiple hypothesis testing correction applied.
Signatures related to leukemia or lymphoma.
RUNX1T1 shows methylation and 2HG-dependent expression changes
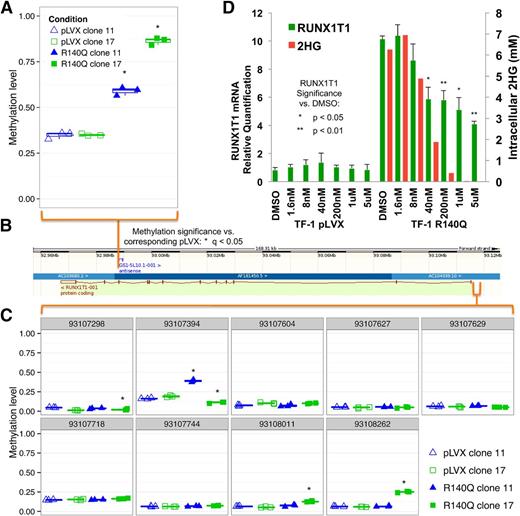
Further analysis of the global methylation changes induced by expression of IDH2 R140Q in TF-1 cells showed both gene body and promoter hypermethylation. Methylation and hydroxymethylation of gene bodies, defined as the area downstream of transcription start sites (intragenic methylation), has been correlated with an increase, rather than a decrease, in gene expression.24-27 Analysis of genes highly methylated in TF-1 R140Q cells showed that the runt-related transcription factor, translocated to 1 (RUNX1T1), had increased methylation within its gene body in both clones (Figure 3A-B). However, no probes with significant hypermethylation in both clones were observed in the promoter region of the gene (Figure 3B-C). Interestingly, when highly expressed, RUNX1T1 has been shown to block hematopoietic differentiation.28 In TF-1 R140Q cells, increased RUNX1T1 gene body methylation was accompanied by an increase in mRNA transcript expression, as measured by RT-PCR, and the increase could be reversed by treatment with doses of AGI-6780 that lower 2HG (Figure 3D). To assess whether downregulation of RUNX1T1 expression might relieve the block in differentiation in TF-1 R140Q cells, we performed a knockdown of RUNX1T1 in these cells and assessed several markers of differentiation, but did not find them to be increased, indicating that RUNX1T1 downregulation alone is not sufficient to drive differentiation (supplemental Figure 7).
RUNX1T1 methylation, expression, and 2HG level correlation. (A) Methylation of RUNX1T1 is enriched in the intragenic region in TF-1 R140Q cells (clone 11 and clone 17) compared with TF-1 pLVX (n = 12). (B) Gene map: black and white bar (top), chromosomal position; light green bar (bottom), gene structure, with exons as maroon vertical bars. (C) Methylation in the promoter region in TF-1 R140Q cells (clone 11 and clone 17) compared with TF-1 pLVX (n = 3; gray bar, chromosomal position). (D) Increase in RUNX1T1 mRNA expression is observed in TF-1 R140Q cells, which can be reversed (as measured by real-time PCR) after 7 days of treatment with AGI-6780. The decrease in expression also correlates with a dose-dependent decrease in the production of 2HG on compound treatment. Error bars = standard deviation.
RUNX1T1 methylation, expression, and 2HG level correlation. (A) Methylation of RUNX1T1 is enriched in the intragenic region in TF-1 R140Q cells (clone 11 and clone 17) compared with TF-1 pLVX (n = 12). (B) Gene map: black and white bar (top), chromosomal position; light green bar (bottom), gene structure, with exons as maroon vertical bars. (C) Methylation in the promoter region in TF-1 R140Q cells (clone 11 and clone 17) compared with TF-1 pLVX (n = 3; gray bar, chromosomal position). (D) Increase in RUNX1T1 mRNA expression is observed in TF-1 R140Q cells, which can be reversed (as measured by real-time PCR) after 7 days of treatment with AGI-6780. The decrease in expression also correlates with a dose-dependent decrease in the production of 2HG on compound treatment. Error bars = standard deviation.
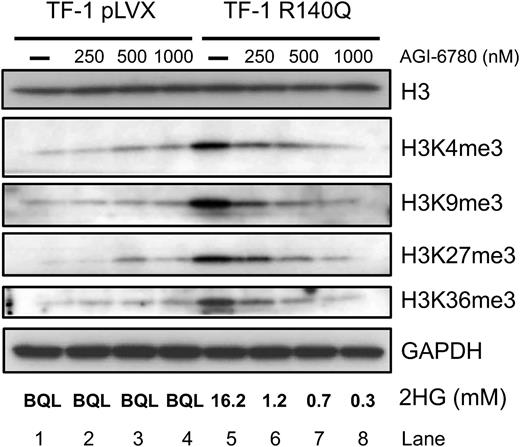
Histone methylation increases with IDH2 mutation and is reversed by AGI-6780
High levels of 2HG have also been shown to competitively inhibit αKG-dependent histone demethylases,11 allowing mutant IDH to induce hypermethylation at H3K4, H3K9, H3K27, and H3K36.4 Consistent with these studies, expression of IDH2 R140Q in TF-1 cells led to a marked increase in histone methylation at all 4 histone marks (Figure 4, lanes 1 and 5). Treatment with AGI-6780 (250-1000 nM) for 7 days induced a dose-dependent decrease in histone methylation (Figure 4, lanes 5-8) that was not seen in TF-1 pLVX control cells (Figure 4, lanes 1-4) and that correlated with a decrease in intracellular 2HG. Taken together, these data demonstrate that AGI-6780 can reverse IDH2 R140Q-induced changes in DNA and histone methylation and suggest that inhibitor treatment could have an effect on the biology of these cells, at least in part, through epigenetic rewiring.
AGI-6780 reverses IDH2 R140Q-induced histone hypermethylation expression in a dose-dependent manner. Expression of R140Q increases H3K4me3, H3K9me3, H3K27me3, and H3K36me3 methylation. Seven-day treatment with AGI-6780 reverses the hypermethylation in a dose-dependent manner at all 4 marks.
AGI-6780 reverses IDH2 R140Q-induced histone hypermethylation expression in a dose-dependent manner. Expression of R140Q increases H3K4me3, H3K9me3, H3K27me3, and H3K36me3 methylation. Seven-day treatment with AGI-6780 reverses the hypermethylation in a dose-dependent manner at all 4 marks.
IDH2 R140Q inhibition reverses HBG promoter DNA and histone methylation and releases differentiation block
To probe histone methylation at a finer resolution, we investigated the interplay between hemoglobin γ (HBG), a marker of early, fetal differentiation states, and EPO, an erythroid differentiation inducer, with histone and DNA methylation marks. Using methylation-dependent DNA restriction digestion to probe the methylation status of the HBG promoter, we found that in the absence of EPO, the promoter was hypermethylated (∼98%) in both the TF-1 R140Q and TF-1 pLVX cells (Figure 5A). EPO treatment lowered the methylation of the HBG promoter to 60% to 70% in TF-1 pLVX cells but had no effect in TF-1 R140Q cells, indicating that R140Q expression prevents EPO-induced demethylation. After 7 days of AGI-6780 treatment, EPO-treated cells were able to increase HBG expression in TF-1 R140Q cells (Figure 5B), and DNA methylation of the HBG promoter decreased to the level found in TF-1 pLVX (Figure 5A). This suggests that the inhibition of mutant IDH2 after treatment with AGI-6780 can induce HBG expression and promote differentiation in TF-1 R140Q cells.
AGI-6780 releases TF-1 R140Q differentiation block by allowing EPO-induced demethylation; reduced histone mark in the γ-globin (HBG) promoter. (A) Pretreatment of TF-1 R140Q cells with AGI-6780 allows EPO-induced demethylation of the HBG promoter. (IDH2, EPO, and AGI-6780 conditions apply to all subfigures; error bars show standard deviation). (B) RT-qPCR for HBG expression shows that treatment of IDH2 R140Q cells with AGI-6780, which decreased DNA hypermethylation at this promoter, can restore expression of HBG. GAPDH expression used for control. (C) AGI-6780 induced dissociation of H3K9me3 from the HBG promoter (as measured by ChIP, using an anti-H3K9me3 antibody) and restored differentiation and expression of HBG, as measured by RT-PCR and the red color change of the cells in the microtubes. Fold enrichment of the HBG promoter with H3K9me3 compared with input DNA was determined by qPCR. Nonspecific enrichment was measured by immunoprecipitation with α-IgG and lack of association of H3K9me3, with the GAPDH gene promoter serving as controls. (D) Picture of the cell pellets and 2HG levels (mM) at cell harvest.
AGI-6780 releases TF-1 R140Q differentiation block by allowing EPO-induced demethylation; reduced histone mark in the γ-globin (HBG) promoter. (A) Pretreatment of TF-1 R140Q cells with AGI-6780 allows EPO-induced demethylation of the HBG promoter. (IDH2, EPO, and AGI-6780 conditions apply to all subfigures; error bars show standard deviation). (B) RT-qPCR for HBG expression shows that treatment of IDH2 R140Q cells with AGI-6780, which decreased DNA hypermethylation at this promoter, can restore expression of HBG. GAPDH expression used for control. (C) AGI-6780 induced dissociation of H3K9me3 from the HBG promoter (as measured by ChIP, using an anti-H3K9me3 antibody) and restored differentiation and expression of HBG, as measured by RT-PCR and the red color change of the cells in the microtubes. Fold enrichment of the HBG promoter with H3K9me3 compared with input DNA was determined by qPCR. Nonspecific enrichment was measured by immunoprecipitation with α-IgG and lack of association of H3K9me3, with the GAPDH gene promoter serving as controls. (D) Picture of the cell pellets and 2HG levels (mM) at cell harvest.
We also assessed the histone methylation changes at the HBG promoter via ChIP, using an H3K9me3 antibody. H3K9me3 is a histone mark often linked to silenced genes and is associated with the HBG promoter in both TF-1 R140Q and TF-1 pLVX cells in the absence of EPO (Figure 5C). Interestingly, IDH2 R140Q expression increased the association of H3K9me3 histones with this promoter, and AGI-6780 treatment reduced the level back to that seen in TF-1 pLVX cells; however, this level is still not sufficient to induce differentiation in the absence of EPO (Figure 5D). Treatment with EPO lowered the association of H3K9me3 histones with the HBG promoter in TF-1 pLVX cells, increased HBG mRNA expression (Figure 5B), and induced a color change associated with erythroid differentiation (Figure 5D). In contrast, TF-1 R140Q cells retained a high level of H3K9me3 at the HBG promoter and failed to express HBG mRNA or change color. However, when TF-1 R140Q cells were treated with AGI-6780, EPO induced a reduction in H3K9me3 levels at the HBG promoter, increased HBG mRNA expression, and produced the color change associated with erythroid differentiation (Figure 5B-D). Importantly, neither EPO nor AGI-6780 had any effect on H3K9me3 associated with GAPDH or when ChIP was performed using an IgG control antibody. Taken together, these data suggest that AGI-6780 is able to reverse both histone and DNA hypermethylation and rescue a differentiation block in TF-1 R140Q cells.
Discussion
We present data showing that IDH2 R140Q mutant expression leads to DNA and histone hypermethylation that can be reversed on treatment with AGI-6780, a potent IDH2 R140Q inhibitor. Previous studies have shown that IDH1 and IDH2 mutations functionally block cellular differentiation and that this block is dependent on elevated levels of 2HG.9,10 The results reported here provide mechanistic insight into the roles IDH mutations and elevated 2HG play in blocking cellular differentiation through epigenetic reprograming.
To more clearly understand the mechanism of this epigenetic rewiring, we performed ChIP in the TF-1 R140Q system and found that on compound treatment, when 2HG levels dropped below 1 mM, association of H3K9me3 at the HBG promoter was decreased. H3K9me3 is a histone mark associated with JMJD2A, a histone demethylase that plays a pivotal role in regulating heterochromatin structure and cellular differentiation through transcriptional repression.6,7,29,30 Interestingly, it has been reported that JMJD2A is one of the most sensitive enzymes to inhibition by 2HG.11 Similarly, targeted inhibition of IDH2 R140Q also reduced the level of DNA methylation and correlated with transcriptional activation of genes involved in cellular differentiation. Because IDH mutations and high levels of 2HG have been shown to disrupt TET2 activity and promote DNA hypermethylation, it will be important to understand their role in tumorigenesis.5 On the basis of this data, to epigenetically rewire TF-1 R140Q cells to differentiate, it may be necessary to reduce the levels of 2HG low enough to at least restore the activity of JMJD2A and TET2. As there are many histone and DNA demethylases that are affected by elevated levels of 2HG,11,31 it remains to be seen whether restoring the activity of these 2 enzymes alone is sufficient to inhibit tumor growth associated with IDH2 R140Q mutations; further experiments will be done to test this hypothesis.
Gene signature over-representation analysis of the genes that undergo demethylation on treatment with AGI-6780 (of which almost all are hypermethylated by IDH2 R140Q expression) identified the RUNX1-RUNX1T1 gene signature as the top-scoring hit. The RUNX1-RUNX1T1 (AML-ETO) fusion protein has been shown to repress transcription of developmentally significant transcription factors such as CEBPA, GATA1, and RARA, as well as repress genes involved in granulocytic and erythroid development.32 It is interesting to note that although TF-1 cells do not carry the RUNX1-RUNX1T1 fusion, IDH2 R140Q expression induced transcriptional activation of RUNX1T1 and its expression was reversed by AGI-6780. It will be important to understand which histone and/or DNA demethylase activities are altered by mutant IDH and whether inhibition of the mutant enzyme can restore normal epigenetic patterning and translate to clinical efficacy.
The reversal of DNA methylation by IDH2 inhibition also suggests that a potential route to a synergistic effect may be achieved by combining IDH inhibition with DNA methyltransferase inhibitors such as azacitidine or decitabine, as low doses of these compounds lead to sustained decreases in promoter DNA methylation and altered gene expression at critical regulatory pathways.33 In earlier-stage development, current efforts to inhibit the histone methyltransferase activity of EZH2 have shown a dose-dependent decrease of H3K27me3 specific to SMARCB1 mutant background, very much like the H3K27me3 seen specifically in IDH2 mutant background with AGI-6780.34 Therefore, it may be interesting to test combination therapies of AGI-6780 with DNA and/or histone methylation inhibitors for an improved effect at gene loci that affect cell cycle, differentiation, or metabolism. Although further investigation of the specific histone and DNA methylation activities is warranted, the identification of a rapid reversal of IDH2 mutant-induced histone hypermethylation, followed by reversal of DNA hypermethylation at genes relevant to leukemic differentiation, adds an important link among IDH2 mutation, cellular differentiation, and tumorigenesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Ingo Mellinghoff and Lew Cantley for helpful discussions about this work, Dan Rohle (I. Mellinghoff laboratory) for methylation chip processing, Hilary Luderer for helpful comments on the manuscript, and the anonymous reviewers for their valuable comments and suggestions that improved the quality of the article.
Authorship
Contribution: A.K., F.W., K.E.Y., and W.L. designed the study. A.K., F.W., E.H., S.S., K.S., and C.G. performed experiments; J.T. provided AGI-6780; A.K. analyzed the data; A.K., K.E.Y., and W.L. wrote the paper; and H.Y., S.M., M.D., S.A., D.P.S., S.A.B., S.M.S., W.L., and K.E.Y. guided the research.
Conflict-of-interest disclosure: All authors except for S.S. are employees of and have ownership interest (including some with patents) in Agios Pharmaceuticals. S.S. is an employee of Blueprint Medicines and has ownership interest in Agios Pharmaceuticals and Blueprint Medicines.
The current affiliation for S.S. is Blueprint Medicines, Cambridge, MA.
Correspondence: Wei Liu, Agios Pharmaceuticals, 38 Sidney St, Suite 2, Cambridge, MA 02139; e-mail: wei.liu@agios.com; and Katharine Yen, Agios Pharmaceuticals, 38 Sidney St, Suite 2, Cambridge, MA 02139; e-mail: katharine.yen@agios.com.
References
Author notes
A.K. and F.W. contributed equally to this study.