Key Points
Targeted NGS of relapsed/refractory CLL reveals a high incidence of concurrent mutations that mostly affect the TP53, ATM, and SF3B1 genes.
Concurrent mutations of the TP53, ATM, and/or SF3B1 genes confer short survival in patients with relapsed/refractory CLL.
Abstract
Although TP53, NOTCH1, and SF3B1 mutations may impair prognosis of patients with chronic lymphocytic leukemia (CLL) receiving frontline therapy, the impact of these mutations or any other, alone or in combination, remains unclear at relapse. The genome of 114 relapsed/refractory patients included in prospective trials was screened using targeted next-generation sequencing of the TP53, SF3B1, ATM, NOTCH1, XPO1, SAMHD1, MED12, BIRC3, and MYD88 genes. We performed clustering according to both number and combinations of recurrent gene mutations. The number of genes affected by mutation was ≥2, 1, and 0 in 43 (38%), 49 (43%), and 22 (19%) respectively. Recurrent combinations of ≥2 mutations of TP53, SF3B1, and ATM were found in 22 (19%) patients. This multiple-hit profile was associated with a median progression-free survival of 12 months compared with 22.5 months in the remaining patients (P = .003). Concurrent gene mutations are frequent in patients with relapsed/refractory CLL and are associated with worse outcome.
Introduction
Chronic lymphocytic leukemia (CLL) is characterized by its unique immunophenotype of CD19+CD5+CD23+sIgdim expressing clonal mature B cells and also by a highly variable clinical course. With the emergence of new classes of drugs such as the inhibitor of phosphatidylinositol 3-kinase, idelalisib,1 and the irreversible inhibitor of Bruton tyrosine kinase, ibrutinib,2 available treatment options have increased significantly and allow us to begin to develop models of treatment stratification. Over the last 3 years, the CLL genome has been thoroughly characterized by next-generation sequencing (NGS).3 Pioneer reports using this approach unraveled somatic mutations recurrently targeting multiple genes, among which TP53, SF3B1, NOTCH1, MYD88, and ATM were the most frequent.4-9 Large retrospective studies in untreated patients from historical cohorts have recently shown the adverse prognostic impact of the TP53, NOTCH1, and SF3B1 mutations on time to treatment and overall survival (OS).10-12 In addition, mutations in these genes may also be associated with poor progression-free survival (PFS) in frontline patients treated in prospective trials.13,14 Conversely, the pejorative impact of TP53, SF3B1, and NOTCH1 mutations on the clinical outcome of patients with relapsed/refractory (R/R) CLL is controversial.15-17 Compared with untreated CLL, relapse, as advanced disease, may be associated with a high level of genomic diversification.9,18 This process might result in accumulation and co-occurrence of these or other genomic events leading to interactions that could be of prognostic relevance. Here, we therefore investigated relapsed/refractory CLL genomes within a clinical trial setting using a comprehensive NGS gene panel.
Methods
Patients and treatment
The present study reports on 114 patients with R/R CLL treated by conventional immunochemotherapy in the setting of prospective clinical trials (flow chart in supplemental Figure 1 available on the Blood Web site). Fifty-five patients were enrolled into the ICLL01 trial by the French CLL Intergroup Groupe Coopératif Français d'étude de la Leucémie Lymphoïde Chronique et de la Maladie de Waldenström–Groupe Ouest-Est des Leucémies Aiguës et Autres Maladies du Sang prior to a planned interim analysis (phase 2 salvage treatment with bendamustine, ofatumumab, and methylprednisolone in relapsed CLL).19 The remaining patients were treated in 2 phase 2 trials from the UK National Cancer Research Network and selected on the basis of the availability of DNA samples; 37 were enrolled in the CLL201 trial (a randomized phase 2 trial of fludarabine, cyclophosphamide, and mitoxantrone with or without rituximab in previously treated CLL)20 and 26 in the CLL202 trial (a phase 2 study of subcutaneous alemtuzumab plus fludarabine in patients with fludarabine refractory CLL).21 Studies were approved by all relevant institutional ethical committees and regulatory review bodies and were conducted in accordance with the Declaration of Helsinki. Patient characteristics are shown in supplemental Table 1. Fludarabine-refractoriness was defined by a response less than a partial remission to a fludarabine-based regimen or a remission lasting <6 months on discontinuation of treatment with a fludarabine-based regimen.22 Median follow-up was 23.6 months. The study has been approved by the institutional review board (CPP Sud-Est 6) of Clermont-Ferrand University hospital in France and the French CLL Intergroup committee (samples from ICLL01 trial) and the UK National Cancer Research Institute CLL Trials biobanks (samples from CLL201 and CCLL202 trials).
Genetic characterization
Cytogenetic aberrations and IGHV status were, respectively, analyzed by fluorescence in situ hybridization (FISH) for 17p and 11q regions and Sanger sequencing.23 For prognostic analyses, patients were distributed in 3 groups according to their cytogenetic features by FISH in a hierarchical model: (1) 17p deletion, (2) 11q deletion without 17p deletion, and (3) no 17p or 11q deletion. Copy number variations were generated for 59 patients using HumanOmni1-Quad BeadChip (Illumina) and following the manufacturer’s instructions.
Somatic gene mutational status was determined by targeted NGS. A TruSeq Custom amplicon panel of 9 recurrently mutated genes in CLL (ie, TP53, SF3B1, ATM, NOTCH1, XPO1, SAMHD1, MED12, BIRC3, and MYD88) was designed using Design Studio (http://designstudio.illumina.com/; supplemental Methods). Sequencing libraries were prepared according to the manufacturer’s instructions and run on the Illumina MiSeq instrument (Illumina). According to Illumina RTA (v.1.18.42), a mean of 14.1 M reads were obtained per run, of which 96.8% were identified reflecting an acceptable signal-to-noise ratio. The yield was 4.1 Gb, and 95.9% of reads were above Q30 across 6 MiSeq runs. The data were analyzed using in-house bioinformatics pipeline consisting of a combination of 2 analyses. After demultiplexing and generation of FASTQ files, the data were processed in MiSeq Reporter (v.2.3.32.0) using a banded Smith-Waterman algorithm to align the sequences to the reference genome and Illumina Somatic Variant caller (variant caller suitable for the detection of somatic variants in cancer samples). A second alignment followed by variant calling using the packages Stampy (v. 1.0.22)24 and Platypus (v.0.5.1)25 was used to specifically identify additional insertions and deletions (InDels) not detected with MiSeq Reporter. The Variant Call Format files were annotated using Annovar26 and filtered according to depth and variant allele frequency (VAF), position, and function of the variant in the coding sequence (hg19 refGene), common polymorphism databases (dbSNP v.132 and 137, 1000 genomes Apr2012), the cancer-specific database Catalogue of Somatic Mutations in Cancer (v.67), our in-house CLL specific database, and the prediction score Sorting Intolerant From Tolerant (from ljb2). All alignments and annotations were performed using the hg19 reference genome (supplemental Figure 2). Median coverage of the genes was 2266×; 97.1% and 92.1% of patients had a mean coverage across all genes >100× and 500×, respectively (supplemental Figure 3). Only NOTCH1 had a less uniform coverage across its targets. However, the amplicon covering the hotspot 7541_7542delCT performed as well as the other genes, with 95.6% of patients with ≥100× and 87.9% with ≥500×.
Subclonal distribution
We inferred the likely subclonal distribution using the VAF from the sequencing data and corrected for copy number with information obtained from 17p and 11q FISH data and from the B allele frequency distributions derived from the whole genome array where available. We then accounted for some degree of germ-line contamination and assessed the tumor purity with an error rate of ±5% for the VAFs. If the VAF was <55%, we supposed it was a heterozygous mutation rather than a subclonal homozygous mutation. If 2 mutations in the same patient showed a VAF difference of <5%, we took the average to calculate the percentage of cell carrying the mutations assuming they occurred in the same clone; if the VAF difference was >5%, we presumed that the 2 mutations were independent (the mutation with the highest VAF occurred prior the second lower VAF mutation).
Statistical analyses
Overall response rate (ORR) and complete remission rate (CRR) were defined using the International Workshop on CLL Criteria.27 Categorical factors between groups of patients were compared using Pearson χ2 or Fisher’s exact test. Logistic regression analysis was used to examine the influence of various factors on ORR. Survival time data were calculated using the Kaplan-Meier method for OS and PFS and were compared by log-rank testing. Proportional hazards models (Cox regression) were fitted to investigate effects of prognostic factors for PFS and OS. All variables with P < .1 were included. Multivariate Cox models were established after backward selection. Interactions were tested and the proportional hazard hypothesis verified by visual display of the Schönfeld residuals.28 The models were validated by a bootstrapping process. In each step, 1000 bootstrap samples with replacements were created from the training set.29 The bootstrap estimates of each covariate coefficient and standard error were averaged from those replicates. The analysis was performed with SPSS software v22 (IBM).
Results
Frequency of gene mutations and correlation to other CLL features
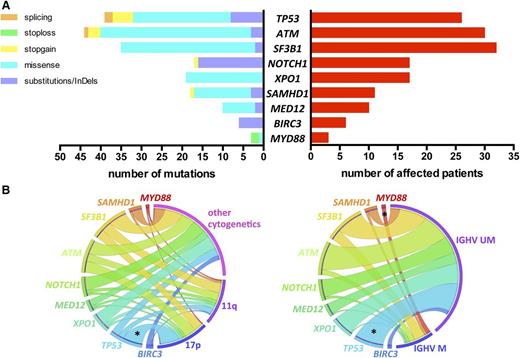
We identified a total of 191 mutations (average of 1.68 per sample) in 92 (81%) patients: 135 missense mutations, 40 substitutions/InDels, 12 nonsense, and 4 splicing mutations, distributed as shown in Figure 1A (list of all mutations in supplemental Table 2).
Incidence of gene mutations and correlation with genomic features. (A) Number and type of mutation (left) and number of affected patients (right) of each sequenced gene. (B) Circos diagrams illustrating pairwise co-occurrence of gene mutations with cytogenetics (left, n = 110 patients) and IGHV status (right, n = 96 patients). The length of the arc depicts the frequency of the marker and the width of the ribbon corresponds to the proportion of co-occurrence with the second marker. Co-occurrences among genes mutations or among cytogenetics are not shown. 17p, 17p deletion; 11q, 11q deletion; IGHV M, mutated IGHV; IGHV UM, unmutated IGHV; *P < .05.
Incidence of gene mutations and correlation with genomic features. (A) Number and type of mutation (left) and number of affected patients (right) of each sequenced gene. (B) Circos diagrams illustrating pairwise co-occurrence of gene mutations with cytogenetics (left, n = 110 patients) and IGHV status (right, n = 96 patients). The length of the arc depicts the frequency of the marker and the width of the ribbon corresponds to the proportion of co-occurrence with the second marker. Co-occurrences among genes mutations or among cytogenetics are not shown. 17p, 17p deletion; 11q, 11q deletion; IGHV M, mutated IGHV; IGHV UM, unmutated IGHV; *P < .05.
TP53, NOTCH1, and SF3B1 mutations occurred in 26 (22.8%), 17 (14.9%), and 32 (28.1%) patients, respectively (Figure 1A). The usual distribution of mutations in these 3 genes was found. TP53 mutations were mostly missense (62%) and located in the DNA-binding domain. NOTCH1 mutations were virtually confined (88%) to the 2-bp deletion (c.7541_7542delCT) in exon 34. SF3B1 mutations were all missense, occurring most frequently as p.K700E/c.A2098G (46%). Thirty (26.3%) patients harbored ATM mutations. Consistent with previous observations, ATM mutations were distributed across the whole gene; 9 mutations clustered in the 3′ terminal region of the gene encoding FAT and PI3kinase functional domains. Mutations in the other genes sequenced were distributed as follows: XPO1 mutations in 17 (14.9%), SAMHD1 mutations in 11 (9.6%), MED12 mutations in 10 (8.8%), BIRC3 mutations in 6 (5.3%), and MYD88 mutations in 3 (2.6%) patients (Figure 1A).
A correlation analysis of gene mutations with other CLL features revealed few significant relations (Figure 1B). As expected, significant overrepresentation of mut-TP53 was noted in the 17p deletion subset (65.4% vs 9.4%; P < .001) and in IGHV unmutated cases (26.9% vs 0%; P = .01). Of note, 55% of 11q-deleted patients had either mut-SF3B1 (n = 9) and/or mut-ATM (n = 7) without reaching statistical significance. All patients with mut-MYD88 had mutated IGHV (n = 3), and all mut-BIRC3 occurred in IGHV unmutated cases (n = 6).
Distribution of gene mutations
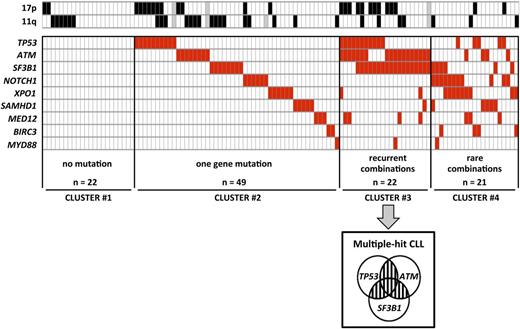
To analyze the relative distribution of gene mutations, we performed clustering according to the number of mutated genes (0 in cluster 1, 1 in cluster 2, ≥2 in clusters 3 and 4) and the recurrence of combinations of 2 gene mutations (≥5% of cases in cluster 3 vs <5% of cases in cluster 4) (Figure 2). Twenty-two (19.3%) patients did not have any mutations (cluster 1) and 49 (43%) patients had 1 gene mutated only (cluster 2). The remaining 43 (37.7%) patients had ≥2 genes mutated (clusters 3 and 4). Recurrent combinations of mutations (affecting >5% of patients) were found in a group of 22 (19.3%) patients. These combinations of mutations comprised ≥2 of the following genes: TP53, SF3B1, and ATM (cluster 3, so called multiple-hit [MH] profile). Remarkably, these 3 gene mutations were found more frequently associated with each other than alone.
Clustering diagram of gene mutations distribution. Rows correspond to cytogenetics (11q or 17p deletion) or sequenced gene and columns represent individual patients. Patients are clustered according to (i) number of genes mutations (0 in cluster 1, 1 in cluster 2, ≥2 in clusters 3 and 4) and (ii) recurrence of combinations (≥5% of cases in cluster 3 or <5% of cases in cluster 4). Color coding is based on the marker status (white, no 11q deletion, no 17p deletion or no gene mutation; black, 17p deletion or 11q deletion; red, gene mutated; gray, missing data).
Clustering diagram of gene mutations distribution. Rows correspond to cytogenetics (11q or 17p deletion) or sequenced gene and columns represent individual patients. Patients are clustered according to (i) number of genes mutations (0 in cluster 1, 1 in cluster 2, ≥2 in clusters 3 and 4) and (ii) recurrence of combinations (≥5% of cases in cluster 3 or <5% of cases in cluster 4). Color coding is based on the marker status (white, no 11q deletion, no 17p deletion or no gene mutation; black, 17p deletion or 11q deletion; red, gene mutated; gray, missing data).
Relative frequencies of each cluster were significantly affected by neither clinical features nor IGHV status and were well balanced among trials (supplemental Table 3). Conversely, in line with the well-known link between TP53 mutation and 17p deletion, MH patients were more frequent in the 17p deleted subset (42.3%) compared with the 11q deleted subset (8.3%) and other cytogenetics (13.3%; P = .002; Figure 2).
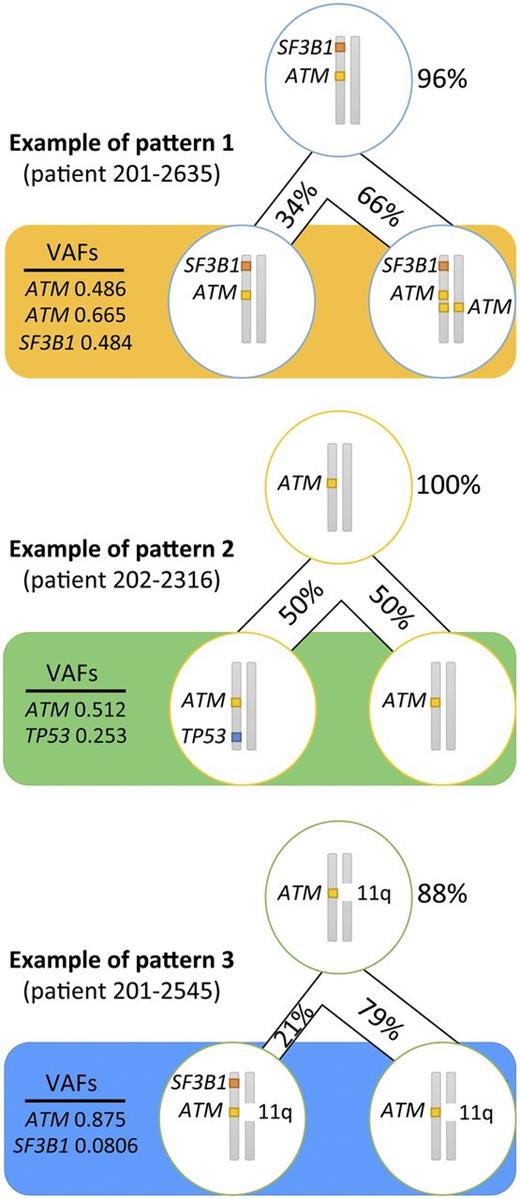
In an attempt to infer the likely subclonal distribution of the mutations carried by the patients with an MH profile from a combination of their respective VAFs and the results from FISH and copy number variations analyses (supplemental Table 2), we established that in ≥8 patients (80%) with a TP53 mutation, this mutation occurred as a second hit. ATM mutations, although generally accepted as a secondary event in CLL leukemogenesis, arose before TP53 or SF3B1 mutations in 15 patients (83%), whereas SF3B1 came first in just over half of patients with this mutation (n = 11; 65%). We discovered 3 recurrent patterns of clonal evolution: ATM and SF3B1 mutations arose in the same clone (pattern 1 in 5 patients); ATM was mutated first and then TP53 was mutated second (pattern 2 in 5 patients); and ATM mutation came first and then SF3B1 mutation originated later (pattern 3 in 4 patients) (Figure 3).
Recurrent patterns of clonal evolution in the multiple-hit profile cluster. Only the loci for TP53, ATM, and SF3B1 are represented. The figures in percentage (%) represent the proportion of cells carrying the mutations. The VAFs represent the frequency of alleles affected by the mutation. The top cell represents the closest common ancestor. The bottom cells represent the cell population when the sample was collected (pretreatment).
Recurrent patterns of clonal evolution in the multiple-hit profile cluster. Only the loci for TP53, ATM, and SF3B1 are represented. The figures in percentage (%) represent the proportion of cells carrying the mutations. The VAFs represent the frequency of alleles affected by the mutation. The top cell represents the closest common ancestor. The bottom cells represent the cell population when the sample was collected (pretreatment).
Predictive markers of response to salvage therapy
To investigate the clinical significance of gene mutations and their distribution, we evaluated their impact on the prospectively recorded therapeutic response. Univariate analysis on response is reported in supplemental Table 4. In the entire cohort, ORR and CR rates were 73.9% and 20.7%, respectively. Poor ORR was strongly predicted only by fludarabine-refractoriness (55.9% vs 81.8%, P = .004) and TP53 disruption (57.1% vs 81.3%, P = .007). If considered individually, no other gene mutation influenced ORR. Conversely, the MH profile was associated with poorer ORR (42.9%) compared with other genomic clusters (86.4%, 83.3%, and 71.4% in clusters 1, 2, and 4 respectively; P = .002). Logistic regression analysis revealed that ORR was significantly and independently influenced by MH profile (odds ratio [OR] = 5.16; 95% confidence interval [CI] = 1.62-16.45; P = .006) and fludarabine-refractoriness (OR = 4.89; 95% CI = 1.77-13.55; P = .002) but not by TP53 disruption (OR = 2.52; 95%CI = 0.89-7.11; P = .081). In terms of CRR, a significantly lower response rate was noted in the TP53 disrupted (8.6% vs 26.7%, P = .03) and ATM mutated (3.4% vs 26.8%; P = .008) patients. Although unexpected, NOTCH1 mutations conferred a better CRR (41.2% vs 17%; P = .045). Furthermore, none of the patients with an MH profile achieved complete remission compared with 24% for the remaining patients (P = .006).
Survival analyses
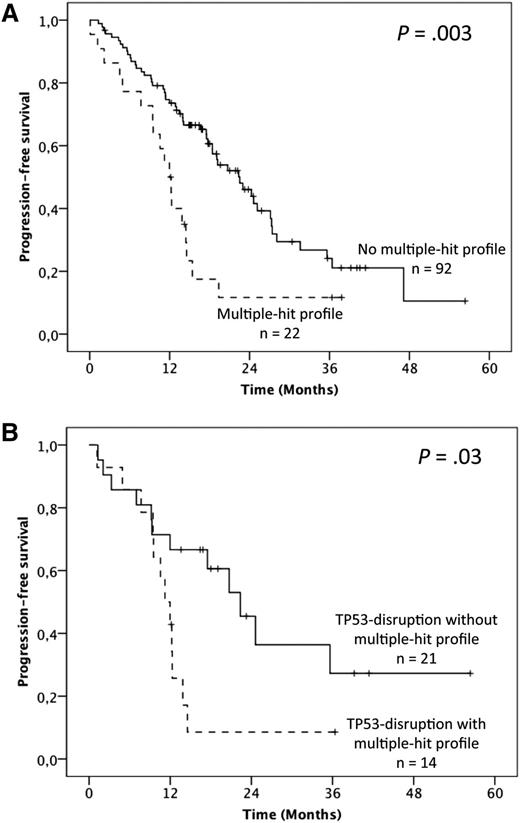
To address the question of the prognostic relevance of gene mutations, we first performed univariate analysis on PFS (supplemental Table 4). Overall, median PFS was 19.1 months but was shorter in fludarabine-refractory cases (12.3 vs 22.5 months; P = .001). No significant influence of either the 17p or 11q deletion was found. Furthermore, when considered individually, TP53 mutations were the only gene mutations that adversely impacted PFS (12 vs 20.7 months; P = .004). Patients harboring the MH profile displayed a significant shorter median PFS of 12 months compared with 22.5 months in the other patients (P = .003; 19.1 months in cluster 1, 23 months in cluster 2, and 17.5 months in cluster 4; Figure 4A; supplemental Table 4). Multivariate analysis for PFS confirmed the independent and adverse prognostic value of the MH profile (hazard ratio [HR] = 2.24; 95% CI = 1.24-4.03; P = .007), as well as those of fludarabine-refractoriness (HR = 3.36; 95% CI = 1.61-7.058; P = .001), whereas the TP53 mutation was not significant anymore after the bootstrapping process (Table 1). Interestingly, among the TP53-disrupted patients, the MH profile retained its prognostic impact with a median PFS of 11.2 vs 22.4 months for the TP53-disrupted patients with wild-type (WT)-SF3B1 and WT-ATM (P = .03; Figure 4B).
Impact of the multiple-hit profile on PFS in univariate analysis. (A) Kaplan-Meier curves of PFS according to the presence of the multiple-hit profile (combinations of mutations comprising ≥2 of the following genes: TP53, SF3B1, and ATM) (A) in the whole population and (B) in the TP53 disruption subgroup.
Impact of the multiple-hit profile on PFS in univariate analysis. (A) Kaplan-Meier curves of PFS according to the presence of the multiple-hit profile (combinations of mutations comprising ≥2 of the following genes: TP53, SF3B1, and ATM) (A) in the whole population and (B) in the TP53 disruption subgroup.
Multivariate analysis for PFS (treatment adjusted)
| Prognostic factors . | HR . | 95% CI . | P . | P* . |
|---|---|---|---|---|
| Fludarabine-refractoriness | 3.36 | 1.65-6.86 | .001 | .004 |
| TP53 mutation | 1.81 | 1.02-3.21 | .043 | .077 |
| Multiple-hit profile | 2.24 | 1.24-4.03 | .007 | .015 |
| Prognostic factors . | HR . | 95% CI . | P . | P* . |
|---|---|---|---|---|
| Fludarabine-refractoriness | 3.36 | 1.65-6.86 | .001 | .004 |
| TP53 mutation | 1.81 | 1.02-3.21 | .043 | .077 |
| Multiple-hit profile | 2.24 | 1.24-4.03 | .007 | .015 |
Including variables with P < .1 in univariate analysis (fludarabine refractoriness, TP53 mutation, ATM mutation, multiple-hit profile and trial).
After bootstrapping process (1000 samples).
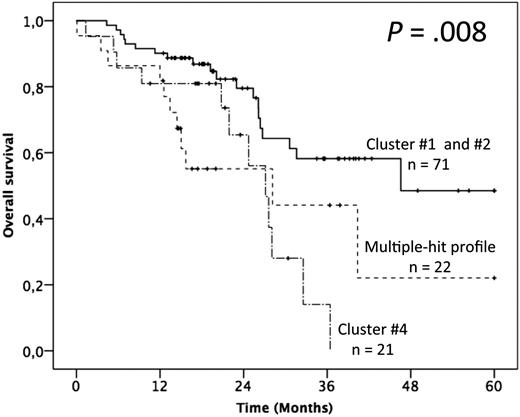
In terms of OS, advanced Binet staging and fludarabine-refractoriness adversely impacted the outcome, whereas no gene mutation alone appeared as discriminant (supplemental Table 4). The patients with >1 gene mutation (ie, clusters 3 and 4) had, however, a significantly poorer outcome than others (ie, clusters 1 and 2; median OS of 28.2 and 27.1 months vs not reached and 46.6 months respectively; P = .019; Figure 5; supplemental Table 4). Multivariate analysis confirmed the independent adverse impact of either >1 gene mutated (model A: HR = 2.91; 95% CI = 1.52-5.60; P = .001) or the MH profile only (model B: HR = 2.85; 95% CI = 1.37-5.94; P = .005; Table 2). Interestingly, besides the retained impact of fludarabine-refractoriness, multivariate analysis uncovered an adverse and independent impact of the BIRC3 mutation, although it did not retain significance after bootstrapping.
Impact of the multiple-hit profile on OS in univariate analysis. (A) Kaplan-Meier curves of OS according to the presence of the multiple-hit profile (combinations of mutations comprising ≥2 of the following genes: TP53, SF3B1, and ATM), cluster 4, and clusters 1 and 2.
Impact of the multiple-hit profile on OS in univariate analysis. (A) Kaplan-Meier curves of OS according to the presence of the multiple-hit profile (combinations of mutations comprising ≥2 of the following genes: TP53, SF3B1, and ATM), cluster 4, and clusters 1 and 2.
Multivariate analyses for OS (treatment adjusted)
| Multivariate models . | Prognostic factors . | HR . | 95% CI . | P . | P* . |
|---|---|---|---|---|---|
| Model A | Fludarabine-refractoriness | 4.1 | 1.75-9.64 | .001 | .005 |
| BIRC3 mutation | 3.41 | 1.15-10.12 | .027 | .835 | |
| Cluster #3 & #4 | 2.91 | 1.52-5.60 | .001 | .003 | |
| Model B | Fludarabine-refractoriness | 3.53 | 1.51-8.22 | .003 | .014 |
| BIRC3 mutation | 4.79 | 1.57-14.61 | .006 | .786 | |
| Multiple-hit profile | 2.85 | 1.37-5.94 | .005 | .025 |
| Multivariate models . | Prognostic factors . | HR . | 95% CI . | P . | P* . |
|---|---|---|---|---|---|
| Model A | Fludarabine-refractoriness | 4.1 | 1.75-9.64 | .001 | .005 |
| BIRC3 mutation | 3.41 | 1.15-10.12 | .027 | .835 | |
| Cluster #3 & #4 | 2.91 | 1.52-5.60 | .001 | .003 | |
| Model B | Fludarabine-refractoriness | 3.53 | 1.51-8.22 | .003 | .014 |
| BIRC3 mutation | 4.79 | 1.57-14.61 | .006 | .786 | |
| Multiple-hit profile | 2.85 | 1.37-5.94 | .005 | .025 |
Both models include variables with P < .1 in univariate analysis (Binet/RAI staging, fludarabine-refractoriness, BIRC3 mutation, trial). In addition, model 1 includes clusters 3 and 4 vs 1 and 2, and model 2 includes cluster 3 (multiple-hit profile) vs 1, 2, and 4.
After bootstrapping process (1000 samples).
Discussion
Here, we present the largest NGS study of patients with R/R CLL recruited prospectively into clinical trials to date. We reveal a high incidence of concurrent mutations that mostly affect the TP53, ATM, and SF3B1 genes. This MH profile is associated with a significantly poorer response to salvage conventional immunochemotherapy and with a significant and independent shorter survival than those of patients of similar clinical phenotype with single or no mutation.
Thus far, the incidence of gene mutations has largely been reported in NGS studies of heterogeneous and mostly treatment-naïve patients or focusing on the TP53, SF3B1, and NOTCH1 genes only using Sanger sequencing.10-17 Now that main CLL genomic drivers have been uncovered by whole exome sequencing or whole genome sequencing studies and targeted NGS has been demonstrated as an accurate and reproducible technique in CLL,30,31 it offers an affordable and more sensitive tool for mutational screening of patients with CLL in clinical practice. We focused on previously identified CLL drivers and used stringent quality filters and visual inspection to remove all possible false-positive calls. Next, we annotated all remaining variants with the latest versions of public and cancer-specific databases, in silico prediction scores, and the CLL literature to systematically identify all variants of potential pathogenicity. This approach is, however, limited by the quality of existing databases.
Our targeted NGS analysis confirms the increased incidence of the TP53, NOTCH1, and SF3B1 mutations in relapsing patients, matching the range of what was previously reported in Sanger sequencing studies including R/R CLLs.10,15,16 Indeed, the frequency of NOTCH1 mutation remains comparable at around 12% to 14.9% among both previous studies and our study. Statistically significant differences in the incidence of TP53 mutations were found between our study (22.8% of cases) and the European Research Initiative on CLL study (13%)10 and the CLL2H (39%)15 but not CLL3X trials (30%).16 The frequency of SF3B1 mutations was also statistically different between our study (28.1%) and the ERIC study (16%), but not the CLL2H (17.5%) or CLL3X (26%). These differences in the mutation incidence might be explained by different inclusion criteria such as definitions of relapse and refractoriness. In addition, although our study is not reporting on gene mutations occurring only in very minor subclones (ie, VAF < 8%), targeting NGS is more sensitive than Sanger Sequencing and is exon-wide as opposed to focused on mutation hotspots.
Furthermore, our study provides new insights in the spectrum of gene mutations of R/R CLL, which, overall, present with a high frequency of other gene mutations. Although poorly documented thus far, notably at relapse, the frequency of ATM mutations is markedly increased compared with series of patients in need of first treatment.32,33 We also show that SAMHD1 mutations are more common compared with untreated patients34 in addition to mutations in XPO1 (14.9% vs 3.4%).11 The incidence of BIRC3 mutations is twofold higher than in untreated patients.10,12 Finally, MED12 mutations are more frequent in our cohort than in those recently reported (5.2%).35 By contrast, MYD88 is rarely affected at relapse, which is consistent with its association with mutated IGHV status.
To investigate the prognostic significance of gene mutations at relapse, we chose to focus our analysis on 114 patients coming from prospective clinical trials to avoid potential bias in terms of response evaluation or survival indicators. Although the patients come from 3 trials, response and survival do not significantly differ between them in univariate analysis, and we included trial in the multivariate analysis to avoid a potential trend of treatment effect.
Other recent prognostic studies of R/R CLL focused on the TP53, SF3B1, and NOTCH1 genes but not ATM.15-17 We confirm that neither SF3B1 nor NOTCH1 mutation adversely influenced the outcome when evaluated at relapse. The presence of NOTCH1 mutation is even associated with a significantly better PFS, which is supported by the previous analysis of the CLL2H trial.15 The present study also shows that, if considered individually, no other gene mutations except for TP53 or BIRC3 impact on prognosis in the present analysis. In line with such observations, Rossi et al proposed a hierarchical model in which these 2 genomic events were considered as conferring the worst outcome in CLL.12
Importantly, our work challenges conventional single gene analyses aimed at establishing a specific prognosis for each driver. Indeed, we clearly demonstrate that contrary to previous studies,36 most R/R CLL cases have >1 recurrently mutated gene and that these are not mutually exclusive. Therefore, an accurate prognostic dissection of R/R CLL requires the integration of such mutational complexity.
Complex karyotype is defined by multiple numerical/structural cytogenetic changes and also discriminates poor outcome in patients with CLL undergoing salvage treatment.37,38 The number of gene mutations did not seem to correlate with number of cytogenetic abnormalities revealed by metaphase cytogenetics in the 46 documented patients of our series (data not shown). However, there is evidence from previous whole genome array studies that there is a strong correlation between genomic complexity and the presence of TP53 disruption.39,40 Therefore, large patient cohorts will be required to unravel potential independence of these phenomena.
Evolutionary principles in CLL as in other cancers supposes that genetic diversification may fuel clonal evolution.3,9,18,39 Using our targeted NGS approach on bulk leukemia cells, the likely subclonal distribution of the MH profile could only be inferred. Longitudinal genome-wide studies including multiple time points or single cell approaches are required to precisely reconstruct the tumor phylogeny.18 However, within the limitations of the single time point-targeted approach presented here, our results are intriguing and suggest different patterns of clonal evolution in which the TP53 mutation appears as a late event, which is in line with recently reported mathematical modeling.41 Furthermore, our data are consistent with the idea that genomic complexity, intratumor heterogeneity, and cooperation between certain mutations may drive CLL evolution. Indeed, the OS of patients with >1 mutated gene was significantly shorter in our study. In line with our results, genomic intratumor heterogeneity by itself was recently linked to poor outcome because the presence of subclonal drivers were shown to adversely influence survival.9,18,42 Conversely, regarding response to therapy and PFS, it seems that the combinations of 2 of the TP53, ATM, and SF3B1 gene mutations are the most powerful combination to provide therapeutic resistance. Interestingly, these 3 genes are all known to be involved in DNA damage response.43 Therefore, multiple hits of the TP53, ATM, and SF3B1 genes might specifically cooperate to drive tumor resistance in addition to being a surrogate for genomic complexity.
Clearly, our results demonstrate that the heterogeneity in survival after cytotoxic therapy of patients with R/R CLL is at least in part due to a specific acquired mutational architecture. It remains to be seen whether this will continue to be the case in the BCR inhibitors era. Our results argue for the systematic evaluation of mutational profiles including ATM status of patients with R/R CLL to assess the accurate prognosis within future clinical trials to establish whether those with ultra-high-risk disease are patients who will benefit most from a novel non–chemotherapy-based targeted multimodality regimen such as novel CD20-targeting antibodies, B-cell CLL/lymphoma 2 protein, or B-cell receptor inhibitor combinations.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Roselyne Delepine for data management of the ICLL01 trial and the Centre de Ressources Biologiques Auvergne for banking ICLL01 patient samples. The authors also acknowledge the Liverpool Biobank and the Leeds Institute of Clinical Trials Research, University of Leeds, for the CLL201 trial data. The authors acknowledge the patients who contributed to the studies.
NGS was in part granted by the Comité du Cantal de La Ligue contre le Cancer. The ICLL01 trial was supported by GSK and Mundipharma.
This publication includes independent research supported by the Health Innovation Challenge Fund (HICF-1009-026, WT091989/Z/10/Z), a parallel funding partnership between the Department of Health and Wellcome Trust. P.R., S.J.L.K., and A.S. are supported also by the NIHR Biomedical Research Centre, Oxford. The work was supported also by funding from the Wellcome Trust Core award grant 090532/Z/09/Z.
The views expressed are those of the authors and not necessarily those of the Department of Health or Wellcome Trust.
Authorship
Contribution: R.G., P.R., O.T., and A.S. designed the research, analyzed the data, and wrote the paper; B.P. performed statistical analyses; F.N.-K. performed FISH analyses; F.D. performed IGHV genes sequencing; P.R., R.C., A.T., M.C., A.B., R.A., and A.H. designed, performed, and analyzed NGS experiments; J.M., S.J.L.K., and J.T. supervised NGS work; S.d.G. and O.T. designed the ICLL01 trial and enrolled patients; M.-S.D., L.Y., and V.L enrolled patients in the ICLL01 trial; P.H., A.P., L.V., P.C., H.M.-B., and M.L.G.-T. provided samples; and all the authors approved the final version of this manuscript.
Conflict-of-interest disclosure: V.L. has received honoraria, consultancy, and advisory board fees from Roche, Janssen, GSK, and Gilead. The remaining authors declare no competing financial interests.
Correspondence: Anna Schuh, Oxford Molecular Diagnostics Centre, Molecular Haematology, Level 4, John Radcliffe Hospital, Oxford OX3 9DU, UK; e-mail: anna.schuh@oncology.ox.ac.uk.
References
Author notes
R.G. and P.R. contributed equally to this work.
O.T. and A.S. contributed equally to this work.