Key Points
Activating mutations outside exon 10 of MPL were identified in 10% (7 of 69) of triple-negative cases of ET and PMF.
JAK2-V625F and JAK2-F556V were identified in 2 triple-negative cases of ET and were shown to activate JAK-STAT5 signaling.
Abstract
Essential thrombocythemia (ET) and primary myelofibrosis (PMF) are chronic diseases characterized by clonal hematopoiesis and hyperproliferation of terminally differentiated myeloid cells. The disease is driven by somatic mutations in exon 9 of CALR or exon 10 of MPL or JAK2-V617F in >90% of the cases, whereas the remaining cases are termed “triple negative.” We aimed to identify the disease-causing mutations in the triple-negative cases of ET and PMF by applying whole-exome sequencing (WES) on paired tumor and control samples from 8 patients. We found evidence of clonal hematopoiesis in 5 of 8 studied cases based on clonality analysis and presence of somatic genetic aberrations. WES identified somatic mutations in 3 of 8 cases. We did not detect any novel recurrent somatic mutations. In 3 patients with clonal hematopoiesis analyzed by WES, we identified a somatic MPL-S204P, a germline MPL-V285E mutation, and a germline JAK2-G571S variant. We performed Sanger sequencing of the entire coding region of MPL in 62, and of JAK2 in 49 additional triple-negative cases of ET or PMF. New somatic (T119I, S204F, E230G, Y591D) and 1 germline (R321W) MPL mutation were detected. All of the identified MPL mutations were gain-of-function when analyzed in functional assays. JAK2 variants were identified in 5 of 57 triple-negative cases analyzed by WES and Sanger sequencing combined. We could demonstrate that JAK2-V625F and JAK2-F556V are gain-of-function mutations. Our results suggest that triple-negative cases of ET and PMF do not represent a homogenous disease entity. Cases with polyclonal hematopoiesis might represent hereditary disorders.
Introduction
Essential thrombocythemia (ET), primary myelofibrosis (PMF), and polycythemia vera (PV) are the classical BCR-ABL–negative myeloproliferative neoplasms (MPNs). They are chronic diseases, characterized by clonal expansion of differentiated myeloid cells driven by somatic mutations. Most of the cases are sporadic, however, familial clustering has also been observed. In 95% of PV cases and 50% to 60% of ET and PMF cases the disease is driven by an acquired mutation in the JAK2 gene V617F.1-4 JAK2-V617F leads to the constitutive activation of the Janus kinase 2 (JAK2) and subsequently the JAK-signal transducer and activator of transcription (STAT) signaling pathway. The remaining cases of PV carry mutations in exon 12 of JAK2. Although JAK2 mutations in MPNs are acquired, in recent years several families with hereditary thrombocytosis (HT) have been described to have germline JAK2 mutations.5-7 The second most commonly mutated gene in ET and PMF is CALR encoding calreticulin. Mutations in exon 9 of CALR have been described in 25% and 35% of patients with ET and PMF, respectively.8,9 There are over 40 different CALR mutation types reported in the literature and they all result in a frameshift into alternative reading frame 1 of the last exon of the CALR gene and hence the acquisition of a novel peptide at the C terminus of the mutant protein.8,9 The functional consequence of CALR mutations is not yet well understood, but the available data suggest that similar to JAK2 mutations, CALR mutations also lead to the activation of the JAK-STAT signaling pathway.8 Besides JAK2 and CALR, a small proportion (5%-10%) of ET and PMF patients carry somatic mutations in exon 10 of MPL, encoding the thrombopoietin receptor (MPL).10,11 The most common mutation is W515L, however, W515K, W515A, W515S, and others have been reported.11,12 The mutations cluster in the intracellular juxtamembrane domain of the receptor, which prevents its activation in the absence of the ligand.13 Germline mutations in MPL have been associated with cases of HT. MPL-S505N in exon 10 was found in 1 family, although it was also reported as somatic in sporadic cases of ET.12,14 Mutations in JAK2, CALR, and MPL are usually mutually exclusive and account for >90% of ET and PMF cases.8 However, in 12% of ET and 5% of PMF cases the disease driver is currently unknown and this group of patients is referred to as the triple negative. In clinical practice, the diagnostic workup of these patients is limited to analyzing exon 14 of JAK2, exon 10 of MPL, and exon 9 of CALR for the presence of mutations. Mutations in other genes such as TET2, ASXL1, and CBL have been described in all MPN patients, however, they co-occur with JAK2, MPL, and CALR mutations and are found across a variety of myeloid malignancies.15-18 Most of these mutations were shown to be involved in establishing clonal hematopoiesis or disease progression.
In this study, we aimed to identify the disease-causing mutation in triple-negative cases of ET and PMF by applying whole-exome sequencing (WES) primarily for the purpose of identifying somatic mutations. In cases where somatic mutations could not be identified, we looked for germline mutations that might be relevant for the phenotype. Finally, we aimed to assess the functional consequence of identified mutations.
Methods
Patient samples
The study included 79 patients diagnosed with ET (N = 67) or PMF (N = 12), followed at the Department of Hematology Oncology, Fondazione Istituto di Ricovero e Cura a Carattere Scientifico (IRCCS) Policlinico San Matteo (Pavia, Italy) and the Medical University of Vienna (Vienna, Austria). All patients provided written informed consent in accordance with the Declaration of Helsinki and the study was approved by local ethics committees. Diagnostic criteria were applied as previously reported and later revised.3,19,20 Genomic DNA was isolated according to standard procedures from peripheral blood granulocytes, whole-blood samples, and immunomagnetically purified circulating T cells. JAK2, MPL, and CALR mutation analyses were performed as previously described, and presence of these mutations was excluded in all patients included in the study.8
Microarray analysis
DNA samples were processed according to the manufacturer’s instructions and hybridized to Genome-Wide Human SNP 6.0 arrays (Affymetrix). The data were analyzed using Genotyping Console version 3.0.2 software (Affymetrix), by applying previously reported criteria for annotation.21
WES and Sanger sequencing
DNA libraries were generated from genomic DNA isolated from granulocytes and CD3+ T lymphocytes using the TruSeq DNA Sample Prep kit (version 2; Illumina) with the TruSeq Exome Enrichment kit (Illumina) and the Nextera Rapid Capture Exome kit (Illumina) according to the manufacturer’s instructions. Sequencing was performed using the Illumina HiSeq 2000 platform, the 51-bp or 100-bp paired-end configuration, and Illumina version 3 reagents. Data analysis is described in supplemental Methods (see supplemental Data available at the Blood Web site).
Sanger sequencing was performed as described previously.8 Details and primer sequences are listed in supplemental Methods.
X-inactivation–based clonality assay
Clonal analysis of hematopoietic cells was performed through the study of methylation pattern and allelic expression of HUMARA, PGK, and IDS genes on peripheral blood granulocytes, whereas T lymphocytes were used as a control tissue, as previously described.22
Functional analysis of identified mutations
Methods used for generation of DNA constructs, virus production and transduction, cell proliferation and viability assays performed in Ba/F3 cells, and western blotting are described in supplemental Methods. Details of the luciferase assay performed in γ2A cells are also described in supplemental Methods.
Results
WES identifies novel mutations in triple-negative PMF and ET
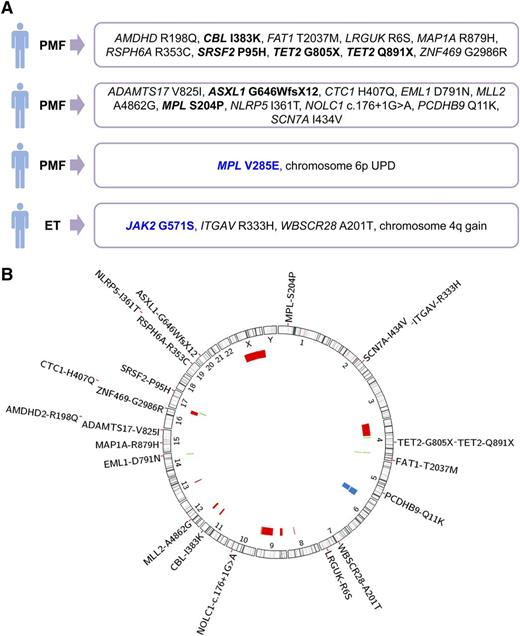
To identify disease-driving mutations in triple-negative MPN, we selected 4 cases of ET and 4 cases of PMF in which mutations in MPL exon 10, CALR exon 9, and JAK2-V617F were excluded, as previously reported.8 WES was performed, in all 8 cases, on the DNA samples from granulocytes and T cells, which were considered as the tumor and the control tissue, respectively. The median number of 104 713 153 unique reads per sample was obtained, which resulted in median coverage of 110× across all target regions. The variants identified in the tumor tissue were annotated for functional consequence and several filtering criteria were applied, including subtraction of variants present in the control tissue, in order to detect potential somatic mutations (supplemental Methods). Candidate somatic mutations were validated using Sanger sequencing on samples from both tissues, to exclude false positive and insufficiently covered germline variants. Using this approach, we identified somatic mutations in 3 of 8 patients. Two patients diagnosed with PMF carried 10 somatic mutations each, whereas 2 somatic mutations were found in 1 patient with ET (Figure 1A). In both PMF patients, we identified mutations in genes known to be relevant for myeloid malignancies, CBL, TET2, ASXL1, SRSF2, including a somatic mutation affecting the thrombopoietin receptor MPL-S204P. We did not detect any recurrent novel somatic mutations in the 8 studied patients.
Overview of genetic aberrations identified in 28 patients with triple-negative ET or PMF. (A) Genetic findings from 4 of 8 studied patients, in which somatic genetic aberrations were identified using WES or microarray analysis. Somatic mutations are marked in black and germline mutations in blue. (B) Genomic overview of somatic mutations identified in 8 patients analyzed by WES and chromosomal aberrations found in 27 patients, detected with Affymetrix Genome-Wide Human SNP 6.0 microarrays. The position and size of the colored bars indicate the chromosomal aberrations. Red bars depict deletions, blue bars depict UPDs, and green bars depict gains.
Overview of genetic aberrations identified in 28 patients with triple-negative ET or PMF. (A) Genetic findings from 4 of 8 studied patients, in which somatic genetic aberrations were identified using WES or microarray analysis. Somatic mutations are marked in black and germline mutations in blue. (B) Genomic overview of somatic mutations identified in 8 patients analyzed by WES and chromosomal aberrations found in 27 patients, detected with Affymetrix Genome-Wide Human SNP 6.0 microarrays. The position and size of the colored bars indicate the chromosomal aberrations. Red bars depict deletions, blue bars depict UPDs, and green bars depict gains.
As in 5 of 8 patients we did not detect a single somatic mutation by WES; we analyzed the WES data for the presence of germline mutations in genes involved in MPN pathogenesis by applying the same filtering criteria as described for analysis of somatic mutations, except for the subtraction of variants present in the control tissue (supplemental Methods). In 1 PMF patient, we identified a germline mutation in MPL-V285E and validated it by Sanger sequencing. In the ET patient with 2 identified somatic mutations, we detected and validated a germline variant in JAK2-G571S (Figure 1A). The germline mutations in MPL and JAK2 were not annotated as common variants in any of the databases we used for the analysis of the WES data.
Granulocyte DNA samples of the 8 selected patients were, in addition to WES, analyzed for acquired chromosomal copy number alterations and uniparental disomies (UPDs) using genome-wide single-nucleotide polymorphism microarrays. Due to DNA quality, the data were obtained for 7 of 8 cases. We identified a somatic UPD of chromosome 6p in 1 patient with PMF (supplemental Figure 1) and a gain on chromosome 4q in a patient with ET. The patient with 6p UPD carried a germline MPL-V285E mutation and no detectable somatic mutations, whereas the ET patient with 4q gain carried the 2 validated somatic mutations in ITGAV and WBSCR28 (Figure 1A).
Chromosomal aberrations often target genomic regions that carry the genes relevant for the disease. We integrated the microarray data from an additional 20 triple-negative MPN patients, with the microarray and WES data from the 8 patients described previously; however, we did not identify any somatic mutations in the regions affected with chromosomal aberrations (Figure 1B). Chromosomal aberrations were detected in 7 of 27 cases, suggesting that the disease is clonal in at least 25.9% of the cases. UPD of chromosome 6p was observed in 2 cases, however, the target gene remains unknown.
As 3 of 8 patients studied by WES and microarrays were female, we performed X-inactivation–based clonality analysis. Clonal hematopoiesis was observed in 1 PMF case and polyclonal pattern in 1 ET case, whereas in the second case of ET the patient was not informative at the analyzed loci. Taking the results of WES, microarray analysis, and clonality assay in consideration, we have found evidence of clonal hematopoiesis in 5 of 8 triple-negative MPN patients (4 of 4 with PMF and 1 of 4 with ET) included in the study, suggesting that the disease might be polyclonal in the other 3 cases. Two triple-negative MPN patients carried mutations affecting the extracellular domain of the thrombopoietin receptor, in contrast to the previously reported mutations, which cluster in the juxtamembrane domain encoded by exon 10 of the MPL gene.
Targeted sequencing of all coding exons of MPL and JAK2 genes
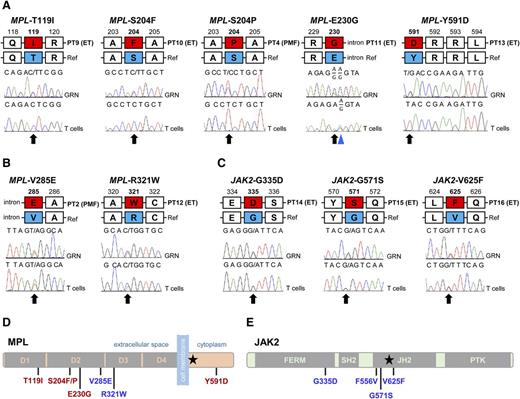
To determine the frequency of the MPL mutations outside of exon 10, in triple-negative MPN patients, we performed Sanger sequencing of all coding exons of MPL on samples from 61 triple-negative MPN cases (53 patients with ET and 8 patients with PMF), in addition to the 8 cases who were subject to WES. Variants outside exon 10 of MPL were identified in 5 of 61 cases. The control tissue was available for 4 patients and we could determine the origin of identified mutations. MPL-T119I, MPL-S204F, and MPL-E230G were somatic mutations (Figure 2A), whereas MPL-R321W was germline (Figure 2B). We identified a second triple-negative case with the MPL-S204P mutation and diagnosis of ET, however, the control tissue was not available. The same mutation was confirmed to be somatic in the PMF case subjected to WES (Figure 2A). We also detected a variant R321Q in 1 case of triple-negative ET, however, this variant was reported with a variant frequency of 3% in an East Asian population in the 1000 Genomes Project, and therefore was not considered further for functional analysis.
Mutations outside exon 10 of MPL and exons 12 and 14 of JAK2 identified in triple-negative ET and PMF. (A) Somatic mutations in MPL identified in 4 cases of ET and 1 case of PMF. A common polymorphism is marked with the blue triangle. (B) Germline mutations in MPL identified in single cases of ET and PMF. (C) Germline JAK2 mutations found in 3 cases of triple-negative ET. Red boxes in panels A-C represent the amino acid change; black arrows indicate mutated bases. GRN, granulocyte; PT, patient; Ref, reference. (D) Schematic representation of the thrombopoietin receptor and the positions of mutations identified in triple-negative ET and PMF. Somatic mutations are indicated in red. (E) Schematic representation of the JAK2 kinase and the mutations found in ET. Black stars (★) in panels D and E indicate positions of MPL-W515K and JAK2-V617F, respectively.
Mutations outside exon 10 of MPL and exons 12 and 14 of JAK2 identified in triple-negative ET and PMF. (A) Somatic mutations in MPL identified in 4 cases of ET and 1 case of PMF. A common polymorphism is marked with the blue triangle. (B) Germline mutations in MPL identified in single cases of ET and PMF. (C) Germline JAK2 mutations found in 3 cases of triple-negative ET. Red boxes in panels A-C represent the amino acid change; black arrows indicate mutated bases. GRN, granulocyte; PT, patient; Ref, reference. (D) Schematic representation of the thrombopoietin receptor and the positions of mutations identified in triple-negative ET and PMF. Somatic mutations are indicated in red. (E) Schematic representation of the JAK2 kinase and the mutations found in ET. Black stars (★) in panels D and E indicate positions of MPL-W515K and JAK2-V617F, respectively.
Mutations outside exon 10 of MPL were identified in 7 of 69 cases of triple-negative MPNs analyzed by WES (N = 8) and Sanger sequencing (N = 61) (Figure 2A-B, supplemental Table 3). All mutations affect the extracellular domain of the thrombopoietin receptor (Figure 2D).
In addition to these 69 cases, we performed targeted sequencing of all coding exons of MPL in a case of ET, which was initially diagnosed as JAK2 and MPL-negative, but developed a JAK2-V617F positivity 5.5 years following the diagnosis. We tested the diagnostic sample for CALR mutations, however, it was negative. When we sequenced the entire coding region of MPL, we identified a somatic MPL-Y591D mutation in the diagnostic sample (Figure 2A), which remained throughout the 5.5 years follow-up. Because the clinical data indicate that the MPL-Y591D mutation was sufficient to cause the phenotype at diagnosis, we included this mutation for the functional validations.
As WES revealed a germline JAK2-G571S mutation in 1 of 8 triple-negative MPN patients, we performed Sanger sequencing for all coding exons of JAK2 on DNA samples from additional 49 triple-negative MPN patients (41 patients with ET and 8 with PMF) which were also analyzed for mutations in MPL (supplemental Table 3). We identified variants in JAK2 in 4 of 49 analyzed cases. All patients with JAK2 variants were diagnosed with ET. JAK2-G335D and JAK2-V625F were confirmed as germline mutations, whereas the control tissue was not available from patients with JAK2-F556V and a second case of JAK2-G571S (Figure 2C,E). Mutations affecting exons 8, 13, and 15 of the JAK2 gene were identified in a total of 5 of 57 triple-negative MPN cases.
Clonality analysis was performed for 23 of 39 female patients included for targeted sequencing of both MPL and JAK2. Clonal hematopoiesis was revealed in 3 cases, whereas 10 cases were polyclonal and 10 cases were either noninformative or showed an ambiguous result of the analysis. In regard to patients with germline mutations, the patient carrying JAK2-V625F was not informative, whereas the patient with MPL-R321W had clonal hematopoiesis. Other patients with germline mutations (JAK2-G335D, JAK2-G571S, and MPL-V285E) were male. All the patients with germline MPL and JAK2 mutations where considered as sporadic as the family history was either negative for the presence of MPN phenotype in family members or unavailable. Overview of molecular data and results of clonality analyses performed for all the patients included in the study, is listed in supplemental Table 3.
Functional analysis of identified MPL and JAK2 mutations
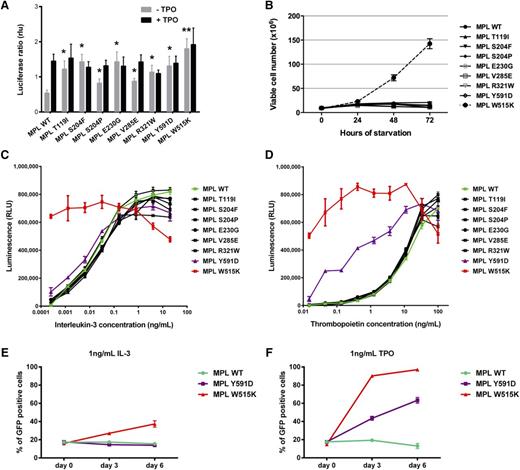
We next performed functional assays to determine whether the identified MPL mutants were able to induce ligand-independent JAK-STAT signaling, similar to other pathogenic mutants such as S505N and W515.23 We used a luciferase reporter assay in JAK2-deficient γ2A cells where we transiently expressed the complementary DNAs for wild-type or mutant MPL, JAK2, STAT5, the STAT5 reporter Spi-Luc, and pRL-TK for transfection control.23 As shown in Figure 3A, expression of MPL mutants supported cytokine-independent JAK2-STAT5 signaling. Of interest, detection of activity required longer times (48 hours) than for the usual pathogenic mutants W515K or S505N, which exhibit constitutive activity at 24 hours posttransfection.23 These data suggest that the identified mutations have a milder effect compared with the common W515K mutant.
Functional analysis of MPL mutants. (A) STAT-dependent transcriptional activity induced by wild-type MPL or mutants of MPL. γ2A cells which are JAK2-deficient were transiently transfected with wild-type MPL (or mutants), JAK2, STAT5, and with Firefly STAT5 luciferase reporter spi-Luc and pRL-TK vector coding for Renilla luciferase. Luminescence was measured 48 hours after transfection. After transfection, medium was changed after 5 hours and replaced either with culture medium or medium supplemented with TPO (10 ng/mL). Shown are average units + SEM of 1 representative experiment performed in triplicate out of 3. **P < .001; *P < .05. (B) Number of viable Ba/F3 cells expressing wild-type MPL or different MPL mutants, in the absence of IL-3, was measured every 24 hours for 72 hours. The data are shown as the average of 2 biological replicates performed each in triplicate ± SEM. (C-D) The viability of Ba/F3 cells expressing wild-type MPL or different mutants was assessed after 72 hours in the presence of increasing concentrations of IL-3 (C) and TPO (D). Error bars represent SEM. (E-F) Ba/F3 cells expressing wild-type MPL (MPL wt-puro) but not GFP were mixed with Ba/F3 cells expressing either wild-type MPL (MPL wt-GFP), MPL Y591D (MPL Y591D-GFP), or MPL W515K (MPL W515K-GFP) and GFP in a 4:1 ratio and cultured in the presence of 1 ng/mL IL-3 (E) or 1 ng/mL TPO (F). The GFP positivity of the mixed culture was assessed every 72 hours by flow cytometric analysis. The experiment was performed in triplicate, with error bars representing SEM. SEM, standard error of the mean.
Functional analysis of MPL mutants. (A) STAT-dependent transcriptional activity induced by wild-type MPL or mutants of MPL. γ2A cells which are JAK2-deficient were transiently transfected with wild-type MPL (or mutants), JAK2, STAT5, and with Firefly STAT5 luciferase reporter spi-Luc and pRL-TK vector coding for Renilla luciferase. Luminescence was measured 48 hours after transfection. After transfection, medium was changed after 5 hours and replaced either with culture medium or medium supplemented with TPO (10 ng/mL). Shown are average units + SEM of 1 representative experiment performed in triplicate out of 3. **P < .001; *P < .05. (B) Number of viable Ba/F3 cells expressing wild-type MPL or different MPL mutants, in the absence of IL-3, was measured every 24 hours for 72 hours. The data are shown as the average of 2 biological replicates performed each in triplicate ± SEM. (C-D) The viability of Ba/F3 cells expressing wild-type MPL or different mutants was assessed after 72 hours in the presence of increasing concentrations of IL-3 (C) and TPO (D). Error bars represent SEM. (E-F) Ba/F3 cells expressing wild-type MPL (MPL wt-puro) but not GFP were mixed with Ba/F3 cells expressing either wild-type MPL (MPL wt-GFP), MPL Y591D (MPL Y591D-GFP), or MPL W515K (MPL W515K-GFP) and GFP in a 4:1 ratio and cultured in the presence of 1 ng/mL IL-3 (E) or 1 ng/mL TPO (F). The GFP positivity of the mixed culture was assessed every 72 hours by flow cytometric analysis. The experiment was performed in triplicate, with error bars representing SEM. SEM, standard error of the mean.
To further study the functional consequence of MPL mutations, we expressed the MPL wild-type or mutants in the interleukin-3 (IL-3)-dependent murine cell line Ba/F3. To test how the expression of MPL mutants affects proliferation of Ba/F3 cells, we performed a starvation assay for 72 hours (Figure 3B). None of the mutants conferred cytokine-independent growth of Ba/F3 cells or hypersensitivity to IL-3, except for the W515K which was used as a positive control (Figure 3B-C; supplemental Figure 2A). Because most of the mutations clustered in the extracellular domain of the thrombopoietin receptor, we tested the cell viability in the dilution series of thrombopoietin (TPO) to determine whether the mutations confer hypersensitivity to TPO (Figure 3D; supplemental Figure 2B). We did not observe a difference between the cells expressing the mutants affecting the extracellular domain of MPL compared with the wild type. However, Ba/F3 cells expressing MPL-Y591D showed marked hypersensitivity to TPO. To confirm that the MPL-Y591D mutation gives proliferative advantage to the Ba/F3 cells, we performed a competition assay in which the cells expressing MPL wild type (pMSCV-HA-MPLwt-puro) were mixed in 4:1 ratio with cells expressing MPL-Y591D and green fluorescent protein (GFP) (pMx-IRES-HA-MPLY591D-GFP). We assessed the GFP positivity by flow cytometry analysis at days 0, 3, and 6. In 2 independent biological replicates we could show that the GFP positivity of mixed culture reaches 60% in the presence of 1 ng/mL TPO in 6 days, meaning that expression of MPL-Y591D confers proliferative advantage in vitro (Figure 3F; supplemental Figure 2D). The effect on proliferation was lower than the one observed for MPL-W515K, and also absent when the cells were cultured in the presence of IL-3 (Figure 3E; supplemental Figure 2C).
The functional data suggest that all identified MPL mutations are gain-of-function mutations, leading to the activation of JAK2-STAT5 signaling. MPL-Y591D is the only mutation which gives an obvious but mild proliferative advantage to the cells in vitro.
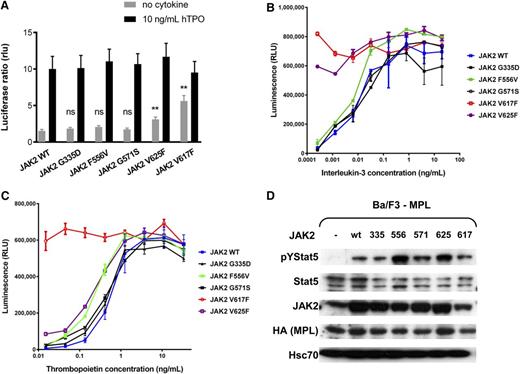
We used the luciferase reporter assay in JAK2-deficient γ2A cells to assess the ligand-independent JAK-STAT5 signaling activation of identified JAK2 mutants in the presence of MPL (Figure 4A) and erythropoietin receptor (supplemental Figure 3A).23 Only the expression of JAK2-V625F supported cytokine-independent JAK2-STAT5 signaling in the presence of MPL, whereas no difference was observed for other mutants compared with the wild type. Upon expressing all of the JAK2 mutants or the wild type in Ba/F3-MPL cells, we tested the cell viability in the serial dilution of IL-3 and TPO concentrations. The cells expressing JAK2-V625F showed hypersensitivity to both IL-3 and TPO (Figure 4B-C; supplemental Figure 3B-C), whereas for JAK2-F556V–expressing cells we could also observe hypersensitivity to TPO. In addition, we examined the phosphorylation of STAT5 in the absence of cytokine in the transfected cell lines. We detected increased phosphorylation of STAT5 in the absence of cytokines in the cells expressing JAK2-V625F and JAK2-F556V (Figure 4D). Thus, increased activation of JAK-STAT signaling is likely responsible for the hypersensitivity to TPO observed for the 2 mutants.
Functional analysis of JAK2 mutants. (A) STAT-dependent transcriptional activity induced by wild-type JAK2 or mutants of JAK2. γ2A cells which are JAK2-deficient were transiently transfected with wild-type JAK2 (or wild-type JAK2 and mutant JAK2 at a 1:1 ratio to represent heterozygous condition), MPL, STAT5, and with Firefly STAT5 luciferase reporter spi-Luc and pRL-TK vector coding for Renilla luciferase. Luminescence was measured 24 hours after transfection. After transfection, medium was changed after 5 hours and replaced either with culture medium or medium supplemented with TPO (10 ng/mL). Shown are means ± SEM of 3 independent experiments done in triplicate. **P < .001; *P < .05. (B-C) The viability of Ba/F3 cells expressing wild-type MPL together with wild-type JAK2 or different mutants was assessed after 72 hours in the presence of increasing concentrations of IL-3 (B) and TPO (C). Error bars represent SEM. (D) The activation of STAT5 in starving condition. Ba/F3 cells expressing the wild-type MPL only, or together with wild-type JAK2 or JAK2 mutants were cultured for 48 hours on TPO (1 ng/mL) and then starved for 4 hours in serum-free medium without TPO. Western blot was performed on the cell lysates with antibodies against pYSTAT5, STAT5, hemagglutinin-tag (HA), and JAK2. An antibody against Hsc70 was used as loading control.
Functional analysis of JAK2 mutants. (A) STAT-dependent transcriptional activity induced by wild-type JAK2 or mutants of JAK2. γ2A cells which are JAK2-deficient were transiently transfected with wild-type JAK2 (or wild-type JAK2 and mutant JAK2 at a 1:1 ratio to represent heterozygous condition), MPL, STAT5, and with Firefly STAT5 luciferase reporter spi-Luc and pRL-TK vector coding for Renilla luciferase. Luminescence was measured 24 hours after transfection. After transfection, medium was changed after 5 hours and replaced either with culture medium or medium supplemented with TPO (10 ng/mL). Shown are means ± SEM of 3 independent experiments done in triplicate. **P < .001; *P < .05. (B-C) The viability of Ba/F3 cells expressing wild-type MPL together with wild-type JAK2 or different mutants was assessed after 72 hours in the presence of increasing concentrations of IL-3 (B) and TPO (C). Error bars represent SEM. (D) The activation of STAT5 in starving condition. Ba/F3 cells expressing the wild-type MPL only, or together with wild-type JAK2 or JAK2 mutants were cultured for 48 hours on TPO (1 ng/mL) and then starved for 4 hours in serum-free medium without TPO. Western blot was performed on the cell lysates with antibodies against pYSTAT5, STAT5, hemagglutinin-tag (HA), and JAK2. An antibody against Hsc70 was used as loading control.
Discussion
We identified somatic and germline mutations outside exon 10 of MPL in 10.1% of cases (7 of 69) of triple-negative ET and PMF. Somatic mutations of MPL at positions T119I, S204F, S204P, and E230G were detected in exons 3, 4, and 5, respectively, whereas germline mutations V285E and R321W were affecting exon 6 of MPL. In 1 additional triple-negative case of ET, we identified a somatic mutation in exon 12 of MPL, MPL-Y591D, at diagnosis, however, the patient became JAK2-V617F positive in a 5.5-year follow-up. Germline mutations of the JAK2 at positions G335D, G571S, and V625F, and mutation of unknown origin F556V in exons 8, 13, and 15 of JAK2 were found in 8.8% (5 of 57) cases of triple-negative ET and PMF. All mutations were heterozygous. Taken together, noncanonical mutations (outside of exons usually examined for diagnostic purposes) of MPL and JAK2 are present in about 18.9% of triple-negative MPN cases. We could demonstrate that all identified MPL mutations lead to constitutive activation of JAK-STAT5 signaling, and that MPL-Y591D provides proliferative advantage to Ba/F3 cells. We could demonstrate that JAK2-V625F is a gain-of-function mutation, whereas for F556V we found evidence of constitutive activation of JAK-STAT5 signaling by Western immunodetection. However, the other JAK2 mutants do not significantly influence the JAK-STAT5 signaling in the presence of erythropoietin receptor or MPL in vitro.
Mutations outside exon 10 of MPL have previously been reported in the literature, but their origin was not determined, mainly due to lack of control tissue availability. MPL-S204F mutation associated with chromosome 1p UPD and heterozygous MPL-S204P mutation have previously been identified in 2 JAK2-V617F–negative PMF cases,24,25 whereas MPL-Y591D was reported in a JAK2-V617F–positive PV patient with chromosome 1p UPD.24 In all 3 cases, the control tissue was not available, and the functional relevance of the mutations was not shown. We could demonstrate that somatic MPL-Y591D mutation was present at diagnosis in a triple-negative ET patient who, during follow-up, acquired JAK2-V617F. Hitchcock et al have shown that MPL-Y591 is a part of the YRRL motif (Figure 2A), which, through interaction with AP2, regulates surface expression and degradation of MPL upon stimulation.26 In the same study, the authors showed that cells expressing MPL-Y591F have a proliferation advantage and an increased JAK2, STAT5, and AKT activation upon stimulation.26 It is unclear why MPL-Y591D is found together with JAK2-V617F. Further studies will be necessary to determine whether combination of these mutations has a specific effect on the patient’s phenotype.
Germline MPL mutations have been reported in cases of HT. Clinical features of ET and HT are similar and that is why some of the HT cases could be diagnosed as sporadic ET, especially if the family history is unknown or absent. The differences in the biology of the 2 diseases are mainly that HT exhibit polyclonal hematopoiesis, lack somatic mutations, and have generally better prognosis without disease progression.27 The first germline MPL mutation associated with HT was S505N in exon 10, which was also reported as somatic in sporadic ET.12,14 MPL-Y252H mutation was reported in a pediatric case of ET with clonal hematopoiesis without evidence of the mutation origin,28 whereas P106L was reported as homozygous in several Arabic families with HT.29 In our cohort, 2 activating germline mutations were identified in cases initially diagnosed as PMF (V285E) and ET (R321W). We cannot exclude the possibility that these mutations could have been acquired in common progenitors, as the only available germline control was DNA isolated from T cells. Both patients had clonal hematopoiesis, as detected by presence of acquired UPD of chromosome 6 in a patient with PMF (supplemental Figure 1) and the X-inactivation–based clonality assay in the patient with ET. We can only speculate that MPL-V285E and MPL-R321W are either not sufficient to cause the observed phenotypes and that an additional event is necessary, likely the target of 6p aberration in the case with PMF, as we did not detect any other somatic mutations in this patient, or that the 6p aberration and presence of clonal hematopoiesis are associated with disease progression.
The 4 somatic mutations we identified were affecting the extracellular ligand binding domain of the MPL,30,31 whereas the 2 germline mutations affected the membrane proximal extracellular domain which appears to play a role in negative regulation of receptor activation. We initially hypothesized that the mutations might affect the affinity to TPO, however, the luciferase assay we performed in γ2A cells showed that all identified mutants activated STAT5-induced transcription in the absence of stimulation with TPO.
Triple-negative status of patients with PMF was shown to associate with inferior overall and leukemia-free survival,32 however, in 2 of 12 triple-negative PMF patients with novel mutations identified in our study, MPL-S204P and MPL-V285E, we did not observe disease progression or leukemic transformation in a 12-year and 6-year follow-up, respectively. Although the number of patients is too low to perform statistical analyses, the clinical data indicate that these patients might have a better clinical course, in accordance with our molecular and functional findings. Analysis of larger patient cohorts will be necessary to confirm the effect of noncanonical MPL mutations on patients’ prognosis.
Germline JAK2 mutations clustering in the pseudokinase or kinase domain have been reported in several families with HT.5-7 In all cases, they were weak gain-of-function mutations. Here, we reported 4 different JAK2 mutations in 5 triple-negative ET cases. For JAK2-V625F and JAK2-F556V, we could demonstrate that they are gain-of-function mutations leading to increased phosphorylation of STAT5 and hypersensitivity to TPO. JAK2-G571S was found in 2 patients. In 1 case, we had evidence for presence of somatic mutations identified by WES and a gain of chromosome 4q, indicating a clonal disease. JAK2-G571S was previously reported in a CALR-positive patient with ET, and the lack of any hematologic phenotype in family members carrying JAK2-G571S indicated that this mutation might be a polymorphism not relevant for the disease.33 We did not find evidence for G571S and G335D mutations altering the function of JAK2. These JAK2 variants either do not play a role in the disease pathogenesis or require additional genetic events to exert a phenotype.
We cannot exclude that some of the patients with wild-type MPL and JAK2, as determined by Sanger sequencing, might carry low burden mutations in these genes, as sensitivity of this method is estimated to ≥10%. Our data indicate that inclusion of all coding exons of JAK2 and MPL in the targeted sequencing panels designed for next-generation sequencing–based approaches for detection of mutations, which are likely to enter the clinical practice in near future, is recommended. Analysis of bigger patient cohorts using such panels will provide more accurate estimation of the frequency of the noncanonical MPL and JAK2 mutations in triple-negative MPN, and their relevance for diagnostic purposes.
In the rest of the cases studied by WES, we did not identify a causative mutation due to absence of recurrent mutations. Similar results were obtained by other groups,34,35 who reported clonal hematopoiesis in 10 of 17 cases, similar to our study (evidence of clonality in 5 of 8 patients). The cases with polyclonal hematopoiesis are likely to be hereditary MPN-like disorders. The lack of recurrency (and the fact that in 1 female PMF patient with clonal hematopoiesis, we did not identify any somatic mutations) can be attributed to the limitations of the WES approach, which can only detect mutations in the coding exons of genes. RNA sequencing for detection of fusion oncogenes and whole-genome sequencing for covering the regulatory sequences in the genome might provide an answer to which mutations drive the disease in the remaining triple-negative MPN cases and whether they represent a heterogeneous disease entity.
The data reported in this article have been deposited in the ArrayExpress database (accession number E-MTAB-3778).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the Biomedical Sequencing Facility at CeMM and Edith Bogner for their technical assistance.
The grants Sonderforschungsbereich (SFB) from the Austrian Science Fund (FWF): F2812-B20 and F4702-B20, are acknowledged for their generous support to R.K. and H.N. J.D.M.F. acknowledges support from L’ORÉAL Österreich, For Women in Science Fellowship. Studies performed at the Department of Hematology Oncology, Fondazione IRCCS Policlinico San Matteo and University of Pavia were supported by grants from Associazione Italiana per la Ricerca sul Cancro (AIRC) and Ministero dell’Istruzione, dell’Università e della Ricerca (MIUR; PRIN 2010-2011) to M.C., and from the Italian Ministry of Health (GR-2009-1580521) and AIRC (MFAG-2014-15672) to E.R. In particular, M.C. acknowledges funding from the AIRC Special Program “Molecular Clinical Oncology 5 per mille” (AIRC Gruppo Italiano Malattie Mieloproliferative [AGIMM], project #1005). S.N.C was supported by the Ludwig Institute for Cancer Research, FNRS (FRSM), Belgium, the Salus Sanguinis Foundation, the Action de Recherche Concertée projects MEXP31C1 and ARC10/15-027 of the University Catholique de Louvain, the Fondation Contre le Cancer, Brussels, the PAI Programs BCHM61B5 and Belgian Medical Genetics Initiative BeMGI, Belgium. E.L. and I.C. received fellowships from FRIA and Télévie, Belgium. B.K. was supported by MUNI/A/1180/2014 and MH CZ-DRO (FNBr, 65269705).
Authorship
Contribution: J.D.M.F. and R.K. conceived and designed the experiments and wrote the paper; J.D.M.F., H.N., E.L., I.C., K.B., B.K., D.P., C.M., R.J., T.B., and M. Schalling performed the experiments; J.D.M.F., H.N., E.L., E.R., I.C., D.P., C.M., D.C., M. Schuster, C.B., S.N.C., and R.K. analyzed and interpreted the data; H.G., E.R., B.G., C.B., S.N.C., M.C., and R.K. contributed reagents/materials/analysis tools; and all authors contributed to the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Robert Kralovics, CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences, Lazarettgasse 14, AKH BT25.3, 1090 Vienna, Austria; e-mail: robert.kralovics@cemm.oeaw.ac.at.