Key Points
The genome of iPSCs has been edited to encode antigenically-distinct human platelet alloantigens.
The iPSC-derived megakaryocyte progenitor cells express the designed alloantigens for diagnostic, investigative, and future therapeutic use.
Abstract
Human platelet alloantigens (HPAs) reside on functionally important platelet membrane glycoproteins and are caused by single nucleotide polymorphisms in the genes that encode them. Antibodies that form against HPAs are responsible for several clinically important alloimmune bleeding disorders, including fetal and neonatal alloimmune thrombocytopenia and posttransfusion purpura. The HPA-1a/HPA-1b alloantigen system, also known as the PlA1/PlA2 polymorphism, is the most frequently implicated HPA among whites, and a single Leu33Pro amino acid polymorphism within the integrin β3 subunit is responsible for generating the HPA-1a/HPA-1b alloantigenic epitopes. HPA-1b/b platelets, like those bearing other low-frequency platelet-specific alloantigens, are relatively rare in the population and difficult to obtain for purposes of transfusion therapy and diagnostic testing. We used CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR associated protein 9) gene-editing technology to transform Leu33+ megakaryocytelike DAMI cells and induced pluripotent stem cells (iPSCs) to the Pro33 allotype. CD41+ megakaryocyte progenitors derived from these cells expressed the HPA-1b (PlA2) alloantigenic epitope, as reported by diagnostic NciI restriction enzyme digestion, DNA sequencing, and western blot analysis using HPA-1b–specific human maternal alloantisera. Application of CRISPR/Cas9 technology to genetically edit this and other clinically-important HPAs holds great potential for production of designer platelets for diagnostic, investigative, and, ultimately, therapeutic use.
Introduction
In addition to their well-described roles in platelet adhesion and thrombus formation, each of the major human platelet membrane glycoproteins is encoded in the human gene pool in multiple allelic isoforms, most of which differ from the predominant wild-type allele by only a single amino acid. A subset of these polymorphic isoforms is immunogenic in man—that is, the 3-D structures encompassing the polymorphic amino acid—are capable of eliciting an alloimmune response in appropriately mismatched individuals. The resulting alloantibodies bind to exposed target epitopes on the platelet surface, resulting in rapid clearance from circulation of the opsonized platelets by liver and splenic macrophages.1
Alloantibodies to platelet-specific antigens are responsible for 2 clinically important bleeding disorders: posttransfusion purpura (PTP) and neonatal alloimmune thrombocytopenia (NAIT, variously referred to in the literature as NATP, FNIT, and FNAIT).2 PTP is a rare syndrome in which a multiparous woman, after receiving a blood transfusion, enigmatically clears not only the transfused platelets, but her own as well, leading to severe thrombocytopenia, bruising, and petechiae. Unlike PTP, NAIT is a fairly common disorder, complicating 1 in 350 pregnancies,3 leading to mild to severe fetal and/or neonatal thrombocytopenia in approximately 1 in 1000 births.3,4 Although many infants recover uneventfully, NAIT is the leading cause of severe thrombocytopenia in the fetus and neonate, often producing bleeding serious enough to require transfusion with “antigen-negative” platelets. The most destructive consequences of NAIT, however, are intracranial hemorrhage and intrauterine death as early as 20 to 24 weeks of gestation.5 Despite advances in treatment, NAIT remains the leading cause of intracranial hemorrhage in term infants,6-10 often leading to lifelong disability.
The first human platelet alloantigen system was identified serologically more than 50 years ago and termed Zw11 or Platelet Antigen 1 (PlA1),12 respectively. The PlA epitope is controlled by a single Leu33Pro amino acid polymorphism within the PSI domain of platelet membrane glycoprotein (GP)IIIa (= the integrin β3 subunit),13 and work performed in many laboratories since that time has led to the identification of 37 distinct human platelet–specific alloantigen (HPA) systems on 6 different glycoproteins.14 PlA1 (HPA-1a), however, remains the alloantigen that most commonly provokes PTP and NAIT, being responsible for ∼80% of the cases in which an alloantibody can be detected.
Despite the availability of numerous DNA-based platforms for the rapid genotyping of each of the HPAs,15-19 identification of a platelet alloantigen-specific antibody in the maternal sera is still required to make a positive diagnosis of NAIT,10 and, less commonly, for post-transfusion refractoriness.20 Determination of antibody specificity, and in some cases titer, is also critical for guiding prenatal treatment to reduce the likelihood of prenatal bleeding and intracranial hemorrhage in utero, facilitating postnatal management, and managing future pregnancies.10,21,22 Platelets bearing low-frequency platelet alloantigens, however, are often difficult or impossible to obtain, and their lack of availability represents a significant barrier for diagnosing and developing effective therapies for NAIT. The purpose of the present investigation was to combine recent advances in gene editing and platelet production technologies to generate antigenically distinct, alloantigen-specific megakaryocyte progenitors for diagnostic and investigative use.
Materials and methods
Guide RNA plasmid constructs
gRNAs were designed using the clustered regularly interspaced short palindromic repeats (CRISPR) Design Tool (http://crispr.mit.edu/) to minimize off-target effects and selected to precede a 5′-NGG protospacer-adjacent motif (PAM). gRNAs used in this study were: gRNA1: 5′-AAGTCCAGCAATCAGAGCTA-3′ and gRNA2: 5′-TGTCTTACAGGCCCTGCCTC-3′. Oligos were annealed and cloned into the BbsI site of the Cas9n expression plasmids px461 or px462 (Addgene, Cambridge, MA).
Single-stranded homology-directed repair template
A single-stranded oligo-deoxynucleotide (ssODN), 181 nucleotides in length, having the sequence 5′-ACTCGGGCCTCACTCACTGGGAACTCGATGGATTCTGGGGCACAGTTATCCTTCAGCAGATTCTCCTTCAGGTCACAGCGAGGTGAGCCGGGTGGCAGGGCCTGTAAGACAGGAGCCCAAAGAGAAGTCCAGCAATCAGAGCTATGCCGACTCTCTACCTCCTGCAGGCCCTACCACTTCC-3′, was synthesized by Integrated DNA Technologies (IDT, Coralville, IA). This oligo corresponds to the antisense strand and, in addition to containing the CTG→CCG PlA1 to PlA2 substitution, contains silent mutations within the recognition sequence and the PAM sequence, of gRNA2 to avoid repetitive digestions by Cas9n.
Cell lines and transfection
2 × 106 DAMI cells were cultured at 37°C in 5% CO2 in Iscove’s Modified Dulbecco’s Medium with 10% horse serum, penicillin (100 U/mL), and streptomycin (100 μg/mL), and transfected with 1 μg of each guide plasmid and 40 pmol of the ssODN single-stranded homology-directed repair (HDR) template using the Amaxa Cell Line Nucleofector Kit C (Lonza, Allendale, NJ) and Nucleofector Program X-005. Transfection efficiency was assessed by visualizing GFP expression using fluorescence microscopy.
Human iPS.K3 cells23 (gift of Dr Steven Duncan, Medical College of Wisconsin) were grown on StemAdhere Defined Matrix-coated plates (Stemcell Technologies, Vancouver, BC, Canada) in mTeSR1/MEF-conditioned medium (50:50) containing 4 ng/mL bFGF (Thermo Fisher Scientific, Grand Island, NY) at 37°C in 4% O2/5% CO2. After incubation with 10 µM ROCK inhibitor Y27632 (StemRD Inc., Burlingame, CA), 2 × 105 cells were transfected with 0.5 μg of each guide plasmid and 40 pmol of the HDR oligonucleotide using the Amaxa P3 Primary Cell 4D Nucleofector Kit (Lonza) and Nucleofector Program CB-150. The cells were then plated on DR4 MEF feeder cells supplemented with 10 μM Y27632. Twenty-four–hour posttransfection puromycin was applied at a concentration of 1 μg/mL for 24 hours. Single clones were harvested at 12 to 14 days post–puromycin selection and replated on StemAdhere-coated plates. Karyotyping of the induced pluripotent stem cell (iPSC) lines was performed by Dynacare Laboratories (Milwaukee, WI) after genotyping to identify the correct lines and every 15 passages routinely during culture.
Differentiation of iPS.K3 cells
Wild-type iPS.K3 cells and CRISPR-edited PlA2 iPS.K3 cells were differentiated to HPCs as previously described.24,25 Briefly, cells were cultured in feeder-free conditions before plating on Matrigel for differentiation. The medium and cytokine changes were followed as described with the following modification. The GSK-3β inhibitor, CHIR99021 (Cayman, Ann Arbor, MI) (0.5-1 μM) was used instead of Wnt3a. Cells were cultured at 37°C, 5% CO2, 5% O2, and 90% N2 for 7 to 9 days, and loosely adherent HPCs were collected by carefully removing the supernatant. Cells were analyzed by flow cytometry for the surface expression of CD41a and CD235a.
Flow cytometry
Twenty-four hours posttransfection, DAMI cells were washed and resuspended in growth medium containing 25 mM HEPES buffer and filtered through 100 μm MACS SmartStrainers (Miltenyi Biotec, San Diego, CA). GFP+ cells were analyzed with a BD Biosciences (San Jose, CA) ARIA-IIIu Cell Sorter. Nontransfected cells were used as negative control. GFP+ cells were sorted as single cells into individual wells of 96-well plates. Analysis of iPSC-derived HPCs was performed using a FACSCanto II (BD Biosciences). The antibodies used were anti-CD235-APC and CD41a-PE (BD Biosciences). Flow cytometry data were analyzed using FlowJo software (Tree Star Inc., Ashland, OR).
Detection of introduced mutations in genomic DNA
Cells were harvested 72 hours after transfection, and DNA was extracted using a QIAamp DNA mini kit (Qiagen, Germantown, MD) according to the manufacturer’s protocol. The genomic region flanking the PlA1 site was amplified using polymerase chain reaction (PCR) primer GPIIIa fw2: 5′-CGTGGAATTCGCTGGTCTACCAGGCATCTT-3′ and GPIIIa rev2: 5′-CCGAAGCTTACCTTGTGCTCTATGCCCAC-3′. PCR products were purified using QiaQuick Spin Column (Qiagen). Purified PCR products (400 ng) were mixed with 1× Taq DNA polymerase PCR buffer, denatured at 95°C, and reannealed to form DNA heteroduplexes. The reannealed PCR products were treated with Surveyor nuclease (IDT) following the manufacturer’s protocol and analyzed on a 2% agarose gel. Quantification was based on relative band intensities. The percentage of DNA mismatches was determined by the equation 100 × {1-[1-(b+c)/(a+b+c)]1/2}, where a is the integrated intensity of the undigested PCR product and b and c are the integrated intensities of each cleavage product.
Genotyping
Genomic DNA was extracted from each clone of DAMI and iPS.K3 cells using the QuickExtract DNA Extraction Solution (Epicenter, Madison, WI) following the manufacturer’s protocol. The region surrounding the PlA1/PlA2 polymorphism was amplified using GPIIIa fw1: 5′-CGTGGAATTCGGCATCTTACTGTACAGGCT-3′ and GPIIIa rev1: 5′- GGCAAGCTTAAGACTTCCTCCTCAGACCT-3′. PCR products were purified using QiaQuick Spin Column, digested with NciI (New England Biolabs Inc., Ipswich, MA) and analyzed on 2% agarose gels.
Immunoprecipitation and western blot analysis
2 × 107 DAMI cells or 3 × 106 iPSC-derived HPCs were lysed in 20 mM Tris (pH7.4), 150 mM NaCl, 1% Triton X-100, 1 mM ethylenediaminetetraacetic acid, 10 mM N-ethylmaleimide, and protease inhibitor cocktail (Thermo Fisher Scientific). Lysates were centrifuged at 17 000 × g for 15 minutes at 4°C. Supernatants were collected, precleared with protein G sepharose, and then incubated with the anti-GPIIIa monoclonal antibody (mAb) AP3 overnight at 4°C. Immune complexes were collected on protein G sepharose beads, eluted with nonreducing SDS sample buffer, and loaded onto 4% to 20% polyacrylamide gels. After electrophoresis, the samples were electrotransferred onto polyvinylidene fluoride membrane (EMD Millipore, Billerica, MA) and immunoblotted with human anti-PlA2 antisera, the PlA1-selective murine mAb, SZ21 (Beckman Coulter, Brea, CA), AP3, or a mouse mAb specific for β-actin (Sigma, St. Louis, MO). Bound antibodies were visualized using species-specific peroxidase-conjugated donkey anti-human IgG (H+L) or goat anti-mouse IgG (H+L) secondary antibodies from Jackson ImmunoResearch Laboratories (West Grove, PA).
Results
CRISPR-mediated conversion of PlA1 homozygous DAMI cells to PlA2
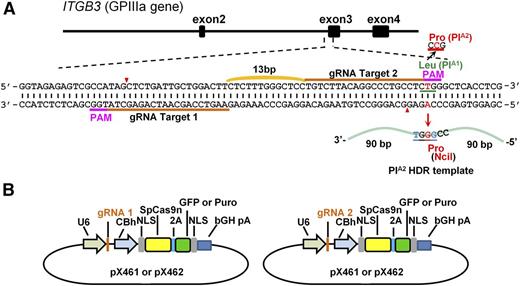
Because iPSCs do not express the GPIIb-IIIa (CD41/CD61) complex unless they are subjected to a rather lengthy differentiation process, conditions for CRISPR-mediated genome editing, including selection of guide RNAs (gRNAs) and homology-directed repair (HDR) oligonucleotides, were first optimized using DAMI cells, a human polyploid megakaryocytic cell line that constitutively expresses the common PlA1 allelic isoform of GPIIIa.26
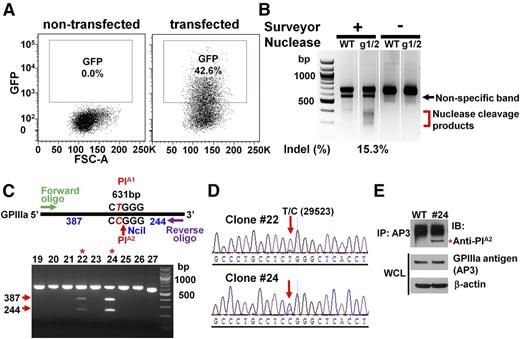
To convert the PlA1 allelic form of GPIIIa, which differs from PlA2 by a single T29523C nucleotide substitution in the ITGB3 gene, to PlA2, we designed 2 gRNAs targeting opposite strands of the ITGB3 gene (Figure 1A) and introduced them into px461, which encodes the single-strand nickase Cas9n and green fluorescent protein (GFP) (Figure 1B). GFP-encoding px461 plasmids harboring each gRNA sequence were transfected into DAMI cells together with a 181 nucleotide PlA2 HDR template (supplemental Figure 1, available on the Blood Web site), and the resulting GFP+ cells were sorted by flow cytometry to enrich for transfected cells (Figure 2A). After cell expansion, surveyor nuclease digestion of a genomic DNA hybridized/rehybridized PCR amplicon spanning the Cas9n cleavage site revealed partial cleavage products of 270-371 bp (Figure 2B), indicating efficient gRNA-directed double nicking by Cas9n. PCR amplicon of genomic DNA from 27 GFP+ single-cell clones was digested with NciI, revealing 2 clones (#22 and #24) that carried the PlA2 polymorphism (Figure 2C). DNA sequence analysis (Figure 2D) confirmed heterozygous replacement of the PlA2 HDR template in these cells. Based on the band intensity of the NciI cleavage products, it appears that approximately half of the PlA1 alleles in clone #24 were CRISPR-converted to PlA2, whereas only one-fourth were converted in clone #22—expected because of the polyploid nature of the DAMI cell population. Finally, immunoprecipitation/western blot analysis using a well-characterized human anti-PlA2 maternal alloantiserum demonstrated that at least a subpopulation of GPIIIa molecules from clone #24 now expresses the Pro33, PlA2 alloantigenic epitope (Figure 2E).
Strategy used to convert the PlA1 allelic form of GPIIIa to PlA2. (A) Schematic illustration of the ITGB3 locus, showing the location of the 2 gRNA binding sites (orange bars) and the protospacer adjacent motif (PAM) sequences (magenta) necessary to guide Cas9n to its cleavage site (red arrowheads). A 181 bp PlA2 HDR template was designed to introduce the Leu→Pro amino acid polymorphism. The T>C mutation responsible for the PlA1/PlA2 polymorphism (highlighted in red) is flanked by 90 nucleotide homology arms and creates an NciI site at the target locus that can be used for genotyping.13 The HDR template also contains 2 silent mutations (highlighted in blue) to prevent recleavage by Cas9n (see “Materials and methods”). (B) The gRNAs were cloned into the BbsI site of the CRISPR vectors px461 or px462, which encode GFP or a puromycin-resistance gene, respectively, as well as Cas9n. The use of 2 different guides to direct the Cas9n nickase to nearby sites at this locus significantly reduces the incidence of off-target mutations relative to that incurred using a single-guide RNA and the double-strand nuclease Cas9.27,28
Strategy used to convert the PlA1 allelic form of GPIIIa to PlA2. (A) Schematic illustration of the ITGB3 locus, showing the location of the 2 gRNA binding sites (orange bars) and the protospacer adjacent motif (PAM) sequences (magenta) necessary to guide Cas9n to its cleavage site (red arrowheads). A 181 bp PlA2 HDR template was designed to introduce the Leu→Pro amino acid polymorphism. The T>C mutation responsible for the PlA1/PlA2 polymorphism (highlighted in red) is flanked by 90 nucleotide homology arms and creates an NciI site at the target locus that can be used for genotyping.13 The HDR template also contains 2 silent mutations (highlighted in blue) to prevent recleavage by Cas9n (see “Materials and methods”). (B) The gRNAs were cloned into the BbsI site of the CRISPR vectors px461 or px462, which encode GFP or a puromycin-resistance gene, respectively, as well as Cas9n. The use of 2 different guides to direct the Cas9n nickase to nearby sites at this locus significantly reduces the incidence of off-target mutations relative to that incurred using a single-guide RNA and the double-strand nuclease Cas9.27,28
Conversion of PlA1-homozygous DAMI cells to PlA2 using CRISPR/Cas9n-directed gene editing. (A) GFP+ (∼40%) DAMI cells, transfected with px461-gRNA1, px461-gRNA2, and PlA2-encoding HDR template, were enriched with fluorescence-activated cell sorting and expanded. (B) Surveyor assay detected the insertions/deletions (indels), indicative of Cas9n-mediated cleavage at the PlA locus, in GFP+ CRISPR-edited cells. The red bracket indicates the range of expected fragment sizes. (C) Genomic DNA from single-cell GFP+ DAMI clones was PCR-amplified and digested with NciI to identify those clones encoding the PlA2 allelic isoform of GPIIIa. Red arrows indicate the expected NciI digestion products. Red asterisks indicate PlA2+ clones #22 and #24. (D) DNA sequence analysis showed the presence of the HDR-introduced T>C 29523 point mutation in clones #22 and #24. The red arrow highlights the heterozygous partial allelic substitution expected in the multiploid DAMI cell line. (E) Cell lysates from wild-type and clone #24 DAMI cells were immunoprecipitated using the GPIIIa-specific mAb, AP3, followed by immunoblotting with human maternal anti-PlA2 antiserum. The relative equivalence of antigen loading was determined by immunoblotting whole-cell lysates (WCL) with AP3 and anti-β-actin antibodies. Note that clone #24, but not wild-type DAMI cells, has a PlA2-reactive band (red asterisk).
Conversion of PlA1-homozygous DAMI cells to PlA2 using CRISPR/Cas9n-directed gene editing. (A) GFP+ (∼40%) DAMI cells, transfected with px461-gRNA1, px461-gRNA2, and PlA2-encoding HDR template, were enriched with fluorescence-activated cell sorting and expanded. (B) Surveyor assay detected the insertions/deletions (indels), indicative of Cas9n-mediated cleavage at the PlA locus, in GFP+ CRISPR-edited cells. The red bracket indicates the range of expected fragment sizes. (C) Genomic DNA from single-cell GFP+ DAMI clones was PCR-amplified and digested with NciI to identify those clones encoding the PlA2 allelic isoform of GPIIIa. Red arrows indicate the expected NciI digestion products. Red asterisks indicate PlA2+ clones #22 and #24. (D) DNA sequence analysis showed the presence of the HDR-introduced T>C 29523 point mutation in clones #22 and #24. The red arrow highlights the heterozygous partial allelic substitution expected in the multiploid DAMI cell line. (E) Cell lysates from wild-type and clone #24 DAMI cells were immunoprecipitated using the GPIIIa-specific mAb, AP3, followed by immunoblotting with human maternal anti-PlA2 antiserum. The relative equivalence of antigen loading was determined by immunoblotting whole-cell lysates (WCL) with AP3 and anti-β-actin antibodies. Note that clone #24, but not wild-type DAMI cells, has a PlA2-reactive band (red asterisk).
PlA1 to PlA2 conversion of human iPSCs
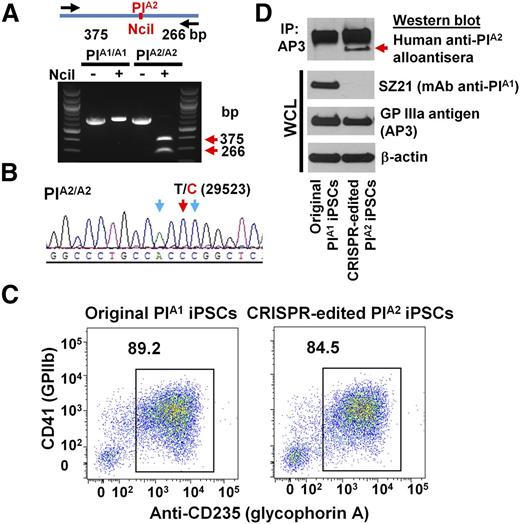
Having optimized the conditions for editing the ITGB3 locus in DAMI cells, we applied a similar protocol to edit iPS.K3 cells—a footprint-free cell line that was reprogrammed from human foreskin fibroblasts with nonepisomal plasmids.23 DNA sequencing (not shown) of genomic DNA of iPS.K3 cells in and around the PlA polymorphism showed them to be homozygous for the PlA1 allele. gRNAs 1 and 2 were cloned into the CRISPR/Cas9 vector, px462, which expresses a puromycin resistance gene (Figure 1B) and cotransfected with the PlA2 HDR template into iPS.K3 cells using Nucleofection. Clones from puromycin-resistant colonies were manually picked and expanded 2 weeks postplating and subjected to diagnostic NciI restriction enzyme digestion to identify clones in which biallelic conversion of PlA1 to PlA2 had taken place. Figure 3A shows the NciI digestion pattern of one such homozygous PlA2 clone, the T>C 29523 genotype of which was verified by DNA sequencing (Figure 3B).
Conversion of iPSCs from PlA1 to PlA2. (A) Genomic DNA, isolated from iPSCs that had been transfected with px462-gRNA1, px462-gRNA2, and PlA2-ssODN, was PCR amplified and digested with NciI, which differentiates the PlA1 allelic isoform from PlA2. Red arrows indicate the expected fragment sizes of a typical clone that had been converted to PlA2. (B) Sequencing data confirmed the T>C 29523 point mutation in CRISPR-edited PlA2 iPSCs. The red arrow indicates the target T>C mutation. Blue arrows indicate silent mutations that were intentionally introduced into the repair oligo to prevent digestion of the final edited genome by Cas9n. (C) Allele-specific expression of GPIIb-IIIa (CD41) on both native and CRISPR-edited iPSC-derived day 8 HPCs. Nonadherent HPCs express abundant levels of the CD41/CD61 complex (integrin αIIb-β3) as well as CD235 (glycophorin A). Note that both cell lines were similarly double-positive. (D) Cell lysates from wild-type, PlA1, and CRISPR-edited PlA2 iPSC–derived HPCs were immunoprecipitated with AP3, followed by immunoblotting with human maternal anti-PlA2 antiserum. Note that the anti-PlA2 antiserum is positive for GPIIIa expressed in the gene-edited, but not native, iPSC line (red arrow), whereas the PlA1-selective mAb, SZ21, binds GPIIIa from native, but not gene-edited, iPSCs.
Conversion of iPSCs from PlA1 to PlA2. (A) Genomic DNA, isolated from iPSCs that had been transfected with px462-gRNA1, px462-gRNA2, and PlA2-ssODN, was PCR amplified and digested with NciI, which differentiates the PlA1 allelic isoform from PlA2. Red arrows indicate the expected fragment sizes of a typical clone that had been converted to PlA2. (B) Sequencing data confirmed the T>C 29523 point mutation in CRISPR-edited PlA2 iPSCs. The red arrow indicates the target T>C mutation. Blue arrows indicate silent mutations that were intentionally introduced into the repair oligo to prevent digestion of the final edited genome by Cas9n. (C) Allele-specific expression of GPIIb-IIIa (CD41) on both native and CRISPR-edited iPSC-derived day 8 HPCs. Nonadherent HPCs express abundant levels of the CD41/CD61 complex (integrin αIIb-β3) as well as CD235 (glycophorin A). Note that both cell lines were similarly double-positive. (D) Cell lysates from wild-type, PlA1, and CRISPR-edited PlA2 iPSC–derived HPCs were immunoprecipitated with AP3, followed by immunoblotting with human maternal anti-PlA2 antiserum. Note that the anti-PlA2 antiserum is positive for GPIIIa expressed in the gene-edited, but not native, iPSC line (red arrow), whereas the PlA1-selective mAb, SZ21, binds GPIIIa from native, but not gene-edited, iPSCs.
Wild-type PlA1 homozygous iPS.K3 cells and their CRISPR-edited progeny were then differentiated into hematopoietic progenitor cells (HPCs) using a previously described serum-free, feeder-free, adherent differentiation system.24,25 The HPCs generated with this method possess erythroid, megakaryocyte, and myeloid multilineage potential, and coexpress the CD41/CD61 GPIIb-IIIa complex, as well as CD235 (glycophorin A). As shown in Figure 3C, HPCs from both iPSC lines express similar levels of CD41+ and CD235+ on their surface, demonstrating importantly that the CRISPR-modified cells retained the full ability to differentiate. Finally, GPIIIa from the PlA2, but not wild-type, iPSC line expressed the PlA2 allelic isoform, as shown by its specific reactivity with a human anti-PlA2 alloantiserum, and its concomitant loss of SZ21 binding (Figure 3D). Taken together, these data demonstrate successful CRISPR-mediated homozygous conversion of PlA1 to PlA2 human iPSCs and their subsequent differentiation into GPIIb-IIIa–expressing HPCs.
An unintended consequence of CRISPR/Cas9 technology is the occasional introduction of off-target mutations elsewhere in the genome that may affect cell growth and differentiation. This problem can be mitigated in part by using a single-strand Cas9 nickase in combination with 2 different gRNAs that target opposite strands surrounding the sequence to be edited (Figure 1A). To evaluate putative off-target effects of the pair of the guide sequences used in this study, we PCR-amplified the top five off-target sites predicted (http://crispr.mit.edu/) for each of our guide sequences (supplemental Table 1) in our PlA2 iPS.K3 cell line, but found no mutations at these loci (supplemental Figure 2).
Discussion
Despite the availability of genotyping for platelet-specific alloantigens, platelet immuno-diagnostics continues to be hampered by the technical complexities of HPA antibody detection—still the gold standard in making a clinical diagnosis of NAIT. Though the majority of human platelet alloantigenic determinants have now been characterized, platelets expressing them are often unavailable, and their detection is additionally hampered by instability or loss of the epitopes after detergent solubilization and storage.29 Finally, serological typing is complicated by the fact that ∼25% of multiparous women produce antibodies against Class I human leukocyte antigens (HLAs)30 that mask detection of platelet-specific alloantigenic epitopes. Taken together, laboratories charged with resolving difficult cases of NAIT have struggled to translate basic scientific discoveries into improved clinical care of families afflicted by this serious condition. The goal of the present investigation, therefore, was to exploit the convergence of CRISPR/Cas9 gene editing and iPSC → platelet technologies to create human platelet progenitors expressing low-frequency platelet alloantigens for diagnostic, investigative, and, perhaps future, therapeutic use.
In 2007, the Yamanaka31,32 and Thomson33 labs reported that adult human fibroblasts can be reprogrammed, using a limited number of transcription factors, into pluripotent stem cells. Building upon this discovery, several groups have developed efficient protocols for differentiating iPSCs to HPCs,34,35 which can be expanded to megakaryocytes,36,37 and platelets.38-40 Although still a long way off from producing a transfusable number of platelets, the ability to generate and cryopreserve iPSC-derived megakaryocyte progenitor cells leaves open the possibility of maintaining an inexhaustible source of platelets and their progenitors for diagnostic and investigative applications. We sought to exploit this capability to produce antigenically-distinct megakaryocytes and progenitor cells from genetically customized iPSCs in sufficient quantities for characterization of their platelet-specific alloantigen expression and function by flow cytometry and other diagnostic methods.
Originally discovered as an ancient form of adaptive immunity that functions by incorporating short pieces of DNA into a series of clustered, regularly interspaced short palindromic repeats within the genomes of bacteria and archaea to direct degradation of foreign DNA,41 the CRISPR system of RNA-guided nucleases has largely supplanted earlier zinc finger and TALEN protein–guided nucleases as the preferred gene-editing tool.42 By incorporating a carefully designed gRNA sequence into a plasmid or lentiviral vector encoding a Cas nuclease, one can engineer double-43 or single-strand44 breaks at precise endogenous loci within the genome of almost any cell that can be transfected or transduced, including iPSCs,45 embryonic stem cells, and zygotes.46
In the present investigation, we combined these technologies to generate iPSC-derived HPCs that express allele-specific forms of clinically important human platelet alloantigens. Because it is the most frequent cause of NAIT and PTP in the Western world, we performed proof-of-concept genetic manipulations on the PlA alloantigen system and were able to successfully generate sufficient quantities of PlA1- and PlA2-specific HPCs to perform flow cytometric detection of these human platelet alloantigens—an assay that requires <10 μL of human serum. Intact human cells are normally not used for alloantibody detection because maternal sera containing platelet antigen–specific alloantibodies also often contain antibodies specific for Class I HLA that are present on the platelet surface.47,48 For this reason, time-consuming and technically demanding antigen-capture enzyme-linked immunosorbent assays are necessary that require hundreds of microliters of maternal alloantisera. HLA detection can be circumvented by introducing a stop codon into the β2 microglobulin (β2M) gene that encodes the light chain of Class I HLA molecules, which is required for trafficking of all Class I heavy chains to the cell surface.49 This tactic has been achieved using both siRNA technology in CD34+ hematopoietic stem cells50 and TALEN technology in iPSCs39 to produce HLA Class I–deficient platelets, and we have recently used CRISPR technology to generate a β2M– founder iPSC line (not shown) into which we plan to introduce polymorphisms that define each of the major HPAs. The availability of a potentially replenishable source of alloantigen-specific megakaryocyte and platelet progenitors should go a long way toward improving the diagnosis, treatment, and care of newborns with this all-to-common cause of morbidity and mortality.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Steven Duncan (Medical College of Wisconsin) for supplying the human iPS.K3 cell line and protocols for their culture, Amy Ludwig-Kubinski for excellent technical assistance, and Drs Debra Newman and David Wilcox for helpful comments.
This work was supported by grants P01 HL44612 (P.J.N.), P01 HL64190 (M.P.), and U01 HL099656 (M.P., D.L.F.) from the National Heart, Lung and Blood Institute of the National Institutes of Health.
Authorship
Contribution: N.Z. and C.J. conducted experiments and analyzed data; B.R.C. provided human anti-PlA2 alloantisera; N.Z., H.Z., S.R., and P.J.N. designed the CRISPR-mediated gene editing experiments; M.P. and D.L.F. designed the iPSC differentiation experiments; and N.Z. and P.J.N. wrote the manuscript.
Conflict-of-interest disclosure: B.R.C. consulted for Prophylix Pharma about platelet antigen and antibody detection. The remaining authors declare no competing financial interests.
Correspondence: Peter J. Newman, Blood Research Institute, BloodCenter of Wisconsin, PO Box 2178, Milwaukee, WI 53201; e-mail: peter.newman@bcw.edu.
References
Author notes
D.L.F. and P.J.N. contributed equally to this study.