Key Points
CD37 positivity predicts significantly better survival for DLBCL, and is superior to other prognostic factors in GCB-DLBCL.
CD37 loss is an important risk factor for R-CHOP resistance in both GCB- and ABC-DLBCL.
Abstract
CD37 (tetraspanin TSPAN26) is a B-cell surface antigen widely expressed on mature B cells. CD37 is involved in immune regulation and tumor suppression but its function has not been fully elucidated. We assessed CD37 expression in de novo diffuse large B-cell lymphoma (DLBCL), and investigated its clinical and biologic significance in 773 patients treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) and 231 patients treated with CHOP. We found that CD37 loss (CD37−) in ∼60% of DLBCL patients showed significantly decreased survival after R-CHOP treatment, independent of the International Prognostic Index (IPI), germinal center B-cell–like (GCB)/activated B-cell–like (ABC) cell of origin, nodal/extranodal primary origin, and the prognostic factors associated with CD37−, including TP53 mutation, NF-κBhigh, Mychigh, phosphorylated STAT3high, survivinhigh, p63−, and BCL6 translocation. CD37 positivity predicted superior survival, abolishing the prognostic impact of high IPI and above biomarkers in GCB-DLBCL but not in ABC-DLBCL. Combining risk scores for CD37− status and ABC cell of origin with the IPI, defined as molecularly adjusted IPI for R-CHOP (M-IPI-R), or IPI plus immunohistochemistry (IHC; IPI+IHC) for CD37, Myc, and Bcl-2, significantly improved risk prediction over IPI alone. Gene expression profiling suggested that decreased CD20 and increased PD-1 levels in CD37− DLBCL, ICOSLG upregulation in CD37+ GCB-DLBCL, and CD37 functions during R-CHOP treatment underlie the pivotal role of CD37 status in clinical outcomes. In conclusion, CD37 is a critical determinant of R-CHOP outcome in DLBCL especially in GCB-DLBCL, representing its importance for optimal rituximab action and sustained immune responses. The combined molecular and clinical prognostic indices, M-IPI-R and IPI+IHC, have remarkable predictive values in R-CHOP–treated DLBCL.
Introduction
The leukocyte surface antigen CD37 (TSPAN26), a member of the tetraspanin superfamily, is widely expressed on normal and malignant mature B cells and downregulated in plasma cells.1-4 Most B-cell malignancies express CD37, including B-cell non-Hodgkin lymphoma (NHL) and B-cell chronic lymphocytic leukemia (B-CLL).5 CD37 was detected at variable levels in 60% of Burkitt lymphoma cell lines.6 Although CD37 expression in neoplastic B cells correlated with the maturation stage of their corresponding B-cell counterparts, B-CLL has lower CD37 levels than do normal mature circulating B lymphocytes.3
Tetraspanins are considered as “molecular facilitators” of signaling transduction, involved in a wide range of biological processes including cell growth, survival, adhesion, trafficking, intercellular communication via exosomes, metastasis, and immune responses.1,4,7-9 CD37 forms complexes with other tetraspanins and major histocompatibility complex (MHC) class II on B cells. CD37 is important for T-cell–B-cell interaction, immunoglobulin G (IgG)/IgA production, and a balance between immune responses and tolerance,1,2,4,10-13 although its role in adaptive immunity is controversal.1,13-15 Using a Cd37−/− mouse model and a confirmative cohort of patients with diffuse large B-cell lymphoma (DLBCL), our study group recently showed that loss of CD37 and interaction between CD37 and suppressor of cytokine signaling 3 (SOCS3) leads to constitutive activation of the interleukin 6 (IL6)-AKT-STAT3 pathway, spontaneous development of germinal center–derived lymphoma, and poorer clinical outcomes.16
CD37 could be targeted by monoclonal antibodies in patients with CLL and NHL expressing high levels of CD37. Although anti-CD37 antibody development predates rituximab (a chimeric monoclonal anti-CD20 IgG1 antibody) by nearly a decade, anti-CD37 antibodies (with otlertuzumab/TRU-016 most common) are in the spotlight only recently1,5,17-20 and have shown promise in phase 1/2 clinical trials for CLL and NHL.1 Upon cross-ligation with anti-CD37 antibodies, CD37 transduces both death signals (from the N-terminal domain associated with Src homology region 2 domain-containing phosphatase-1 [SHP1], LYN, and phosphatidylinositol 3-kinase γ [PI3Kγ]) and opposing survival signals (from the C-terminal domain recruiting p85 and PI3Kδ).10
DLBCL is the most common and heterogeneous NHL. Although the addition of rituximab to cyclophosphamide, doxorubicin, vincristine, prednisone (CHOP) significantly improves clinical outcomes, approximately one-third of DLBCL patients still have refractory disease or relapse.21-23 Currently, DLBCL risk stratification relies mainly on the International Prognostic Index (IPI), which is based on patients’ clinical features. However, the IPI cannot identify high-risk subgroups in the rituximab era24,25 because it was originally developed for CHOP outcome prediction from multivariate survival analyses in CHOP-treated patients.26 Unfortunately, robust and reproducible biomarkers in DLBCL are also lacking.27,28 Gene expression profiling (GEP) subdivides DLBCL into 2 major molecular subtypes, germinal center B-cell–like (GCB) DLBCL and activated B-cell–like (ABC) DLBCL, and patients with ABC-DLBCL have poorer survival.29 In ABC-DLBCL, B-cell receptor (BCR) signaling is chronically active with constitutive activation of antiapoptotic NF-κB; comparably, GCB-DLBCL has tonic BCR signaling with PI3K pathway activation (either proapoptotic or antiapoptotic).30,31
In this study, we assessed CD37 status and its prognostic effects in large cohorts of patients with DLBCL, and correlated CD37 status with tumor biology at both the protein and messenger RNA (mRNA) levels to determine the underlying mechanisms.
Methods
Patients
A total of 1037 patients with de novo DLBCL were studied as a part of the International DLBCL R-CHOP Consortium Program, including 806 rituximab plus CHOP (R-CHOP)-treated patients (discovery cohort, n = 560; validation cohort, n = 246) and 231 CHOP-treated patients. The study was conducted in accordance with the Helsinki Declaration and was approved as being of minimal or no risk or as exempt by the institutional review boards of all participating medical centers.
Genetic and immunohistochemical analysis
Tissue microarrays prepared from the diagnostic formalin-fixed, paraffin-embedded (FFPE) blocks were stained with a CD37 monoclonal antibody (clone 2B8; Thermo Fisher Scientific).16 FFPE tissue sections were also stained for IgA, IgG, IgM, p53, MDM2, p63, NF-κB subunits, phosphorylated STAT3 (p-STAT3), Myc, Bcl-2, Bcl-6, CD10, GCET1, FOXP1, MUM1/IRF4, BLIMP-1, Ki-67, CD5, CD30, CXCR4, PI3K, p-AKT, and survivin, and assessed for TP53 mutations and MYC/BCL2/BCL6 translocations, as described in supplemental Methods (available on the Blood Web site). GEP was performed on Affymetrix GeneChips Plus version 2.0 using total RNAs extracted from FFPE tissues.32
Statistical analysis
Correlations between CD37 and the clinical factors and biomarkers were analyzed using the χ2 test, the Fisher exact test, the unpaired Student t test (2-tailed), and Spearman rank correlation. Survival analysis was performed using the Kaplan-Meier method and the log-rank (Cox-Mantel) test with GraphPad Prism 6 software. Overall survival (OS) was calculated from the date of diagnosis to the date of death from any cause or last follow-up for censored patients. Progression-free survival (PFS) was calculated from the date of diagnosis to the date of disease progression, recurrence, or death from any cause. Multivariate analysis was conducted using Cox proportional hazard regression models with SPSS software, version 19.0 (IBM, Armonk, NY). P values ≤.05 were considered statistically significant.
Results
DLBCL patients with CD37 surface expression have significantly better clinical outcomes
CD37 surface expression was scored in 5% increments and CD37 staining was evaluable for 527 cases in the discovery cohort. Figure 1A-B shows representative CD37− and CD37+ immunohistochemistry results, respectively. Staining for CD37 was positive (≥5%) at variable expression levels in 40% (212 of 527) of the discovery cohort (Figure 1C). The ABC-DLBCL subgroup had a higher frequency of CD37+ patients than the GCB-DLBCL subgroup (46.8% vs 33.6%; Table 1), and a higher mean level of CD37 protein (P = .002; Figure 1D) and CD37 mRNA (P < .0001; supplemental Figure 1A). Cell of origin according to the B-cell–associated gene signature classification33 did not show significant differences between the CD37+ and CD37− groups, although in the GCB subtype, CD37+ DLBCL had a nonsignificantly higher frequency of centrocyte cell of origin (supplemental Figure 1B).
Expression and prognostic effect of CD37 antigen in patients with DLBCL. (A-B) Representative CD37− and CD37+ (red) IHC results (×60). Cell nuclei were counterstained with hematoxylin (blue). Images were obtained with an Olympus AX70 microscope with a DP71 camera. (C) Histogram of CD37 IHC scores in the DLBCL discovery cohort. (D) A scatter plot for CD37 expression in DLBCL and comparison between GCB and ABC cell of origin. (E) Patients with CD37− DLBCL had significantly worse OS and PFS compared with patients with CD37+ DLBCL, with a HR of 2.80 and 95% CI of 1.95 to 3.38 for OS, and a HR of 2.89 and 95% CI of 2.07 to 3.45 for PFS. (F-H) The adverse prognostic effect of CD37 loss was independent of GCB and ABC cell of origin, high and low IPI scores, and primary nodal (NL) and primary extranodal (ENL) origin.
Expression and prognostic effect of CD37 antigen in patients with DLBCL. (A-B) Representative CD37− and CD37+ (red) IHC results (×60). Cell nuclei were counterstained with hematoxylin (blue). Images were obtained with an Olympus AX70 microscope with a DP71 camera. (C) Histogram of CD37 IHC scores in the DLBCL discovery cohort. (D) A scatter plot for CD37 expression in DLBCL and comparison between GCB and ABC cell of origin. (E) Patients with CD37− DLBCL had significantly worse OS and PFS compared with patients with CD37+ DLBCL, with a HR of 2.80 and 95% CI of 1.95 to 3.38 for OS, and a HR of 2.89 and 95% CI of 2.07 to 3.45 for PFS. (F-H) The adverse prognostic effect of CD37 loss was independent of GCB and ABC cell of origin, high and low IPI scores, and primary nodal (NL) and primary extranodal (ENL) origin.
Comparisons of clinicopathologic characteristics between CD37+ patients and CD37− patients with DLBCL, GCB-DLBCL, or ABC-DLBCL
| Variable . | CD37+ . | CD37− . | P . | CD37+ GCB . | CD37− GCB . | P . | CD37+ ABC . | CD37− ABC . | P . |
|---|---|---|---|---|---|---|---|---|---|
| n (%) . | n (%) . | n (%) . | |||||||
| Patients | 212 (100) | 315 (100) | 90 (100) | 178 (100) | 118 (100) | 134 (100) | .0023 | ||
| Age, y | |||||||||
| <60 | 79 (37.3) | 143 (45.4) | .064 | 42 (46.7) | 89 (50.0) | .90 | 34 (28.8) | 52 (38.8) | .095 |
| ≥60 | 133 (62.7) | 172 (54.6) | 48 (53.3) | 89 (50.0) | 84 (71.2) | 82 (61.2) | |||
| Sex | |||||||||
| Female | 82 (38.7) | 135 (42.9) | .34 | 37 (41.1) | 74 (41.6) | .94 | 43 (36.4) | 60 (44.8) | .18 |
| Male | 130 (61.3) | 180 (57.1) | 53 (58.9) | 104 (58.4) | 75 (63.6) | 74 (55.2) | |||
| Stage | |||||||||
| I/II | 105 (51.5) | 138 (45.0) | .15 | 53 (61.6) | 88 (50.9) | .10 | 49 (43.0) | 48 (36.6) | .31 |
| III/IV | 99 (48.5) | 169 (55.0) | 33 (38.4) | 85 (49.1) | 65 (57.0) | 83 (63.4) | |||
| B symptoms | |||||||||
| No | 132 (66.7) | 193 (63.3) | .44 | 61 (73.5) | 114 (66.7) | .27 | 68 (61.3) | 76 (58.0) | .61 |
| Yes | 66 (33.3) | 112 (36.7) | 22 (26.5) | 57 (33.3) | 43 (38.7) | 55 (42.0) | |||
| LDH level | |||||||||
| Normal | 75 (39.1) | 107 (36.8) | .61 | 34 (43.6) | 65 (39.6) | .56 | 40 (36.4) | 42 (33.9) | .69 |
| Elevated | 117 (60.9) | 184 (63.2) | 44 (56.4) | 99 (60.4) | 70 (63.6) | 82 (66.1) | |||
| No. of extranodal sites | |||||||||
| 0-1 | 161 (78.2) | 228 (75.7) | .81 | 69 (80.2) | 132 (78.1) | .69 | 89 (76.7) | 93 (72.1) | .41 |
| ≥2 | 45 (21.8) | 73 (24.3) | 17 (19.8) | 37 (21.9) | 27 (23.3) | 36 (27.9) | |||
| ECOG performance status | |||||||||
| 0-1 | 175 (89.3) | 220 (79.4) | .0044 | 74 (92.5) | 126 (81.8) | .028 | 97 (86.6) | 91 (75.8) | .037 |
| ≥2 | 21 (10.7) | 57 (20.6) | 6 (7.5) | 28 (18.2) | 15 (13.4) | 29 (24.2) | |||
| Size of largest tumor | |||||||||
| <5cm | 98 (60.9) | 131 (56.7) | .41 | 45 (67.2) | 73 (55.7) | .12 | 50 (54.9) | 58 (59.2) | .56 |
| ≥5cm | 63 (39.1) | 100 (43.3) | 22 (32.8) | 58 (44.3) | 41 (45.1) | 40 (40.8) | |||
| IPI score | |||||||||
| 0-2 | 132 (67.3) | 173 (61.3) | .21 | 64 (78.0) | 106 (66.7) | .07 | 64 (58.2) | 64 (53.3) | .46 |
| 3-5 | 64 (32.7) | 109 (38.7) | 18 (22.0) | 53 (33.3) | 46 (41.8) | 56 (46.7) | |||
| Therapy response* | |||||||||
| CR | 189 (89.2) | 215 (68.3) | <.0001 | 82 (91.1) | 124 (69.7) | <.0001 | 103 (87.3) | 89 (66.4) | .0001 |
| PR | 18 (8.5) | 52 (16.5) | 6 (6.7) | 25 (14) | 12 (10.2) | 27 (20.1) | |||
| SD | 0 (0) | 23 (7.3) | 0 (0) | 14 (7.9) | 0 (0) | 9 (6.7) | |||
| PD | 5 (2.3) | 25 (7.9) | 2 (2.2) | 15 (8.4) | 3 (2.5) | 9 (6.7) | |||
| Primary disease | |||||||||
| Extranodal | 76 (36.9) | 114 (36.4) | .91 | 33 (37.5) | 62 (35.2) | .72 | 40 (35.1) | 51 (38.1) | .63 |
| Nodal | 130 (63.1) | 199 (63.6) | 55 (62.5) | 114 (64.8) | 74 (64.9) | 83 (61.9) | |||
| IgA IHC, % | |||||||||
| 0 | 210 (99.1) | 303 (96.2) | .045 | 89 (98.9) | 171 (96.1) | .20 | 117 (99.2) | 129 (96.3) | .13 |
| 100 | 2 (0.9) | 12 (3.8) | 1 (1.1) | 7 (3.9) | 1 (0.8) | 5 (3.7) | |||
| IgG IHC, % | |||||||||
| 0 | 189 (89.2) | 283 (89.8) | .80 | 82 (91.1) | 159 (89.3) | .65 | 103 (87.3) | 121 (90.3) | .45 |
| 100 | 23 (10.8) | 32 (10.2) | 8 (8.9) | 19 (10.7) | 15 (12.7) | 13 (9.7) | |||
| IgM IHC | |||||||||
| Negative | 142 (67.0) | 239 (75.9) | .025 | 76 (84.4) | 154 (86.5) | .65 | 63 (52.9) | 82 (61.2) | .19 |
| Positive | 70 (33.0) | 76 (24.1) | 14 (15.6) | 24 (13.5) | 56 (47.1) | 52 (38.8) | |||
| Variable . | CD37+ . | CD37− . | P . | CD37+ GCB . | CD37− GCB . | P . | CD37+ ABC . | CD37− ABC . | P . |
|---|---|---|---|---|---|---|---|---|---|
| n (%) . | n (%) . | n (%) . | |||||||
| Patients | 212 (100) | 315 (100) | 90 (100) | 178 (100) | 118 (100) | 134 (100) | .0023 | ||
| Age, y | |||||||||
| <60 | 79 (37.3) | 143 (45.4) | .064 | 42 (46.7) | 89 (50.0) | .90 | 34 (28.8) | 52 (38.8) | .095 |
| ≥60 | 133 (62.7) | 172 (54.6) | 48 (53.3) | 89 (50.0) | 84 (71.2) | 82 (61.2) | |||
| Sex | |||||||||
| Female | 82 (38.7) | 135 (42.9) | .34 | 37 (41.1) | 74 (41.6) | .94 | 43 (36.4) | 60 (44.8) | .18 |
| Male | 130 (61.3) | 180 (57.1) | 53 (58.9) | 104 (58.4) | 75 (63.6) | 74 (55.2) | |||
| Stage | |||||||||
| I/II | 105 (51.5) | 138 (45.0) | .15 | 53 (61.6) | 88 (50.9) | .10 | 49 (43.0) | 48 (36.6) | .31 |
| III/IV | 99 (48.5) | 169 (55.0) | 33 (38.4) | 85 (49.1) | 65 (57.0) | 83 (63.4) | |||
| B symptoms | |||||||||
| No | 132 (66.7) | 193 (63.3) | .44 | 61 (73.5) | 114 (66.7) | .27 | 68 (61.3) | 76 (58.0) | .61 |
| Yes | 66 (33.3) | 112 (36.7) | 22 (26.5) | 57 (33.3) | 43 (38.7) | 55 (42.0) | |||
| LDH level | |||||||||
| Normal | 75 (39.1) | 107 (36.8) | .61 | 34 (43.6) | 65 (39.6) | .56 | 40 (36.4) | 42 (33.9) | .69 |
| Elevated | 117 (60.9) | 184 (63.2) | 44 (56.4) | 99 (60.4) | 70 (63.6) | 82 (66.1) | |||
| No. of extranodal sites | |||||||||
| 0-1 | 161 (78.2) | 228 (75.7) | .81 | 69 (80.2) | 132 (78.1) | .69 | 89 (76.7) | 93 (72.1) | .41 |
| ≥2 | 45 (21.8) | 73 (24.3) | 17 (19.8) | 37 (21.9) | 27 (23.3) | 36 (27.9) | |||
| ECOG performance status | |||||||||
| 0-1 | 175 (89.3) | 220 (79.4) | .0044 | 74 (92.5) | 126 (81.8) | .028 | 97 (86.6) | 91 (75.8) | .037 |
| ≥2 | 21 (10.7) | 57 (20.6) | 6 (7.5) | 28 (18.2) | 15 (13.4) | 29 (24.2) | |||
| Size of largest tumor | |||||||||
| <5cm | 98 (60.9) | 131 (56.7) | .41 | 45 (67.2) | 73 (55.7) | .12 | 50 (54.9) | 58 (59.2) | .56 |
| ≥5cm | 63 (39.1) | 100 (43.3) | 22 (32.8) | 58 (44.3) | 41 (45.1) | 40 (40.8) | |||
| IPI score | |||||||||
| 0-2 | 132 (67.3) | 173 (61.3) | .21 | 64 (78.0) | 106 (66.7) | .07 | 64 (58.2) | 64 (53.3) | .46 |
| 3-5 | 64 (32.7) | 109 (38.7) | 18 (22.0) | 53 (33.3) | 46 (41.8) | 56 (46.7) | |||
| Therapy response* | |||||||||
| CR | 189 (89.2) | 215 (68.3) | <.0001 | 82 (91.1) | 124 (69.7) | <.0001 | 103 (87.3) | 89 (66.4) | .0001 |
| PR | 18 (8.5) | 52 (16.5) | 6 (6.7) | 25 (14) | 12 (10.2) | 27 (20.1) | |||
| SD | 0 (0) | 23 (7.3) | 0 (0) | 14 (7.9) | 0 (0) | 9 (6.7) | |||
| PD | 5 (2.3) | 25 (7.9) | 2 (2.2) | 15 (8.4) | 3 (2.5) | 9 (6.7) | |||
| Primary disease | |||||||||
| Extranodal | 76 (36.9) | 114 (36.4) | .91 | 33 (37.5) | 62 (35.2) | .72 | 40 (35.1) | 51 (38.1) | .63 |
| Nodal | 130 (63.1) | 199 (63.6) | 55 (62.5) | 114 (64.8) | 74 (64.9) | 83 (61.9) | |||
| IgA IHC, % | |||||||||
| 0 | 210 (99.1) | 303 (96.2) | .045 | 89 (98.9) | 171 (96.1) | .20 | 117 (99.2) | 129 (96.3) | .13 |
| 100 | 2 (0.9) | 12 (3.8) | 1 (1.1) | 7 (3.9) | 1 (0.8) | 5 (3.7) | |||
| IgG IHC, % | |||||||||
| 0 | 189 (89.2) | 283 (89.8) | .80 | 82 (91.1) | 159 (89.3) | .65 | 103 (87.3) | 121 (90.3) | .45 |
| 100 | 23 (10.8) | 32 (10.2) | 8 (8.9) | 19 (10.7) | 15 (12.7) | 13 (9.7) | |||
| IgM IHC | |||||||||
| Negative | 142 (67.0) | 239 (75.9) | .025 | 76 (84.4) | 154 (86.5) | .65 | 63 (52.9) | 82 (61.2) | .19 |
| Positive | 70 (33.0) | 76 (24.1) | 14 (15.6) | 24 (13.5) | 56 (47.1) | 52 (38.8) | |||
P values designating statistical significance are highlighted in bold.
CR, complete remission; LDH, lactate dehydrogenase; PD, progressive disease; PR, partial response; SD, stable disease.
We calculated P values as CR vs other responses. Clinicopathologic data were not available for some cases due to clinical data unavailability and tissue exhaustion.
At clinical presentation, patients with CD37− DLBCL had a higher frequency of Eastern Cooperative Oncology Group (ECOG) performance status >1 than patients with CD37+ DLBCL (20.6% vs 10.7%; Table 1). Although the CD37+ DLBCL group had a trend toward more elderly (≥60 years) patients (P = .064), these patients had a significantly higher complete response rate and significantly increased OS and PFS rates compared with the CD37− DLBCL patients (P < .0001; Figure 1E). The favorable effect of CD37 expression was independent of GCB/ABC subtype, high/low IPI, and nodal/extranodal primary origin (Figure 1F-H). Among the CD37+ patients, CD37 high/low levels did not seem to be prognostic, although in some CD37+ ABC-DLBCL patients with high CD37 levels, the favorable effect of CD37 expression was decreased (supplemental Figure 1C). The significant impact of CD37+/− status was confirmed in an independent validation cohort (n = 246) (P = .0003 for OS and P = .0065 for PFS; supplemental Figure 1D).
CD37 loss is associated with lower levels of CD37 and CD20 mRNA expression
CD37 mRNA levels were obtained from the GEP data and correlated with CD37 protein levels. The CD37+ group had a significantly higher mean level of CD37 mRNA than the CD37− group (Figure 2A; supplemental Figure 1A). Using the Spearman rank correlation method, CD37 protein and CD37 mRNA levels were significantly correlated (r = 0.25, P = 7.57e-8). In addition, CD37 negativity was associated with downregulation of many BCR signaling–related genes (Table 2). CD20/MS4A1 mRNA levels significantly correlated with both CD37 protein (r = 0.209, P = 8.16e-6) and CD37 mRNA levels (r = 0.406, P = 3.40e-21), likely reflecting that both CD37 and CD20 are expressed in mature B cells with BCR and downregulated in plasma cells. Moreover, tested in 13 DLBCL cell lines, CD20 and CD37 protein levels measured by flow cytometry also showed significant correlation (Spearman rank correlation: r = 0.771, P = .002). In 6 cell lines (CD20−/CD37−: Oci-Ly19, SU-DHL6, WSU-NHL; CD20+/CD37+: Oci-Ly8, SU-DHL5, and SU-DHL2), CD20 and CD37 levels showed strong correlation, confirmed by 2 different CD37 monoclonal antibodies (WR17, HH1) (linear regression, R2 = 0.9737; Figure 2B). As control, CD19 expression was also measured and detected on all DLBCL cell lines.
CD37 and CD20 expression show correlations but the prognostic significance of CD37 is independent of CD20 expression in DLBCL. (A) In both GCB- and ABC-DLBCL, the CD37− group had significantly lower mean levels of CD37 and CD20 mRNA expression compared with the CD37+ group. (B) Geometric mean fluorescence intensities (gMFIs) of CD37 and CD20 protein expression in 3 different CD37− and 3 different CD37+ DLBCL cell lines measured by flow cytometry. CD20 expression on the plasma membrane of DLBCL cells was detected by a nontherapeutic CD20 antibody (2H7; BioLegend) (upper) and by therapeutic Rituxan (Roche) (lower). Results are representative for 2 independent experiments. Coefficient for linear regression: R2 = 0.9737 (left) and R2 = 0.8043 (right). Dotted lines show 95% CI. (C) Low CD20 mRNA expression (less than mean) correlated with significantly worse OS and PFS, especially in ABC-DLBCL. (D) However, CD37+ patients with lower CD20 mRNA levels had significantly better OS and PFS than CD37− patients with higher CD20 mRNA levels. (E) In GCB-DLBCL, CD37 antigen status but not CD20 mRNA levels predicted survival. In ABC-DLBCL, CD37 status and CD20 mRNA levels showed prognostic impact independent of each other, but CD37 status showed stronger prognostic impact. (F) In an independent CHOP-treated DLBCL cohort, overall CD37+ patients had significantly better PFS than CD37− patients. However, the favorable impact was limited in patients with low (≤50%) CD37 levels and patients with high (>50%) CD37 expression showed similar PFS with CD37− patients.
CD37 and CD20 expression show correlations but the prognostic significance of CD37 is independent of CD20 expression in DLBCL. (A) In both GCB- and ABC-DLBCL, the CD37− group had significantly lower mean levels of CD37 and CD20 mRNA expression compared with the CD37+ group. (B) Geometric mean fluorescence intensities (gMFIs) of CD37 and CD20 protein expression in 3 different CD37− and 3 different CD37+ DLBCL cell lines measured by flow cytometry. CD20 expression on the plasma membrane of DLBCL cells was detected by a nontherapeutic CD20 antibody (2H7; BioLegend) (upper) and by therapeutic Rituxan (Roche) (lower). Results are representative for 2 independent experiments. Coefficient for linear regression: R2 = 0.9737 (left) and R2 = 0.8043 (right). Dotted lines show 95% CI. (C) Low CD20 mRNA expression (less than mean) correlated with significantly worse OS and PFS, especially in ABC-DLBCL. (D) However, CD37+ patients with lower CD20 mRNA levels had significantly better OS and PFS than CD37− patients with higher CD20 mRNA levels. (E) In GCB-DLBCL, CD37 antigen status but not CD20 mRNA levels predicted survival. In ABC-DLBCL, CD37 status and CD20 mRNA levels showed prognostic impact independent of each other, but CD37 status showed stronger prognostic impact. (F) In an independent CHOP-treated DLBCL cohort, overall CD37+ patients had significantly better PFS than CD37− patients. However, the favorable impact was limited in patients with low (≤50%) CD37 levels and patients with high (>50%) CD37 expression showed similar PFS with CD37− patients.
Differential expression of tetraspanins and BCR signaling-related genes between CD37− and CD37+ patients with GCB- or ABC-DLBCL
| CD37− vs CD37+ in GCB/ABC DLBCL . | |||
|---|---|---|---|
| Genes . | Downregulation in CD37− . | Genes . | Upregulation in CD37− . |
| CD37 | ↓ in both GCB (P = .036) and ABC (P < .0001) | CD63 | ↑ in ABC (P < .0001) |
| CD20 | ↓ in both GCB (P = .0008) and ABC (P = .0009) | MS4A4A (CD20L1) | ↑ in GCB (P = .0073) and ABC (P = .059) |
| CD79A | ↓ in both GCB (P = .002) and ABC (P = .009) | MS4A6A (CD20L3) | Trend of ↑ in ABC (P = .057) |
| CD79B | ↓ in ABC (P = .032) | AKT1 | ↑ in GCB (P = .019) |
| CD22 | ↓ in both GCB (P = .016) and ABC (P = .011) | ||
| CD23 | ↓ in GCB (P = .016) | ||
| STAP1 | ↓ in GCB (P = .0012) | ||
| SWAP70 | ↓ in GCB (P = .0036) | ||
| SYK | ↓ in ABC (P = .045) | ||
| CARD11 | ↓ in ABC (P = .05) | ||
| BCL10 | Trend of ↓ in ABC (P = .07) | ||
| MALT1 | ↓ in GCB (P = .021) | ||
| CD40 | ↓ in GCB (P = .016) | ||
| BLNK | Trend of ↓ in ABC (P = .064) | ||
| MYC | Trend of ↓ in ABC (P = .08) | ||
| REL | ↓ in GCB (P = .0031) | ||
| PIK3C2B | ↓ in GCB (P = .046) and ABC (P = .07) | ||
| A20/TNFAIP3 | ↓ in GCB (P = .018) | A20/TNFAIP3 | ↑ in ABC (P = .036) |
| CD37− vs CD37+ in GCB/ABC DLBCL . | |||
|---|---|---|---|
| Genes . | Downregulation in CD37− . | Genes . | Upregulation in CD37− . |
| CD37 | ↓ in both GCB (P = .036) and ABC (P < .0001) | CD63 | ↑ in ABC (P < .0001) |
| CD20 | ↓ in both GCB (P = .0008) and ABC (P = .0009) | MS4A4A (CD20L1) | ↑ in GCB (P = .0073) and ABC (P = .059) |
| CD79A | ↓ in both GCB (P = .002) and ABC (P = .009) | MS4A6A (CD20L3) | Trend of ↑ in ABC (P = .057) |
| CD79B | ↓ in ABC (P = .032) | AKT1 | ↑ in GCB (P = .019) |
| CD22 | ↓ in both GCB (P = .016) and ABC (P = .011) | ||
| CD23 | ↓ in GCB (P = .016) | ||
| STAP1 | ↓ in GCB (P = .0012) | ||
| SWAP70 | ↓ in GCB (P = .0036) | ||
| SYK | ↓ in ABC (P = .045) | ||
| CARD11 | ↓ in ABC (P = .05) | ||
| BCL10 | Trend of ↓ in ABC (P = .07) | ||
| MALT1 | ↓ in GCB (P = .021) | ||
| CD40 | ↓ in GCB (P = .016) | ||
| BLNK | Trend of ↓ in ABC (P = .064) | ||
| MYC | Trend of ↓ in ABC (P = .08) | ||
| REL | ↓ in GCB (P = .0031) | ||
| PIK3C2B | ↓ in GCB (P = .046) and ABC (P = .07) | ||
| A20/TNFAIP3 | ↓ in GCB (P = .018) | A20/TNFAIP3 | ↑ in ABC (P = .036) |
The P values were obtained by the unpaired Student t test (2-tailed).
We further treated these 6 cell lines with rituximab (Rituxan and MabThera) and compared the cytotoxic effect. As expected, CD20−/CD37−/CD19+ cell lines had less Rituxan binding and were resistant to rituximab treatment in contrast to CD20+/CD37+/CD19+ cell lines (Figure 2B; supplemental Figure 2A). Likewise, in the discovery cohort, patients with lower (less than mean) CD20 mRNA levels had significantly poorer OS and PFS in the overall DLBCL and ABC-DLBCL cohorts, and poorer OS in the GCB-DLBCL cohort (Figure 2C).
In order to investigate whether CD37’s prognostic significance was actually due to its association with CD20 mRNA levels, first we incorporated both CD20 mRNA and CD37 factors into the survival analysis (Figure 2D-E; supplemental Figure 2B). We found CD37+ status significantly predicted favorable OS and PFS regardless of high/low CD20 mRNA levels; furthermore, CD37+ DLBCL patients with low CD20 mRNA levels still had significantly better OS and PFS than CD37− DLBCL patients with high CD20 mRNA levels, especially in cases of GCB-DLBCL. On the other hand, CD20 mRNA levels only showed significant impact on OS and PFS in CD37− DLBCL overall (but not in the GCB/ABC subset), and on PFS in the CD37+ ABC subgroup.
Second, to eliminate the confounding prognostic effects conferred by CD20, we studied CD37 expression in a CHOP-treated cohort (n = 231). We found CD37 positivity still correlated with significantly better survival, yet with less predictive power (P = .022 for OS and P = .013 for PFS), more significantly in GCB-DLBCL than in ABC-DLBCL (supplemental Figure 3A). However, this favorable impact was limited in patients with relatively low CD37 levels (≤50%) whereas it was lost in CD37high (>50%) patients (Figure 3F). Comparing between CHOP-treated and R-CHOP–treated patients, CD37high patients especially had improved survival with R-CHOP treatment regardless of CD20 mRNA levels (supplemental Figure 3B-C).
Correlation analysis and the robust prognostic effect of CD37 expression in patients with DLBCL. (A) A distribution plot showing that CD37− GCB-DLBCL (denoted by the yellow bar) more frequently had TP53 mutations (highlighted in red) or high levels of nuclear p50 (yellow), Myc (green), p-STAT3 (orange), and p65 (lighter red) expression compared with CD37+ GCB-DLBCL (denoted by the blue bar). (B) A distribution plot showing that CD37− ABC-DLBCL more frequently had high nuclear p50 (yellow) and survivin (pink) expression and BCL6 translocation (green), whereas CD37+ ABC-DLBCL more frequently had PI3K (blue) and CXCR4 (purple) overexpression. Each column in panels A and B represents 1 patient; cases without indicated abnormalities detected are shown in light blue (negative) or white (unknown). (C) In GCB-DLBCL, CD37 positivity predicted significantly improved survival, regardless of presence of TP53 mutations, p50high, Mychigh, p-STAT3high, GCET1high, and to a lesser extent, MYC translocations. Conversely, the adverse effect of CD37 negativity was independent of all these biomarkers. Particularly, CD37− patients without TP53 mutations and p50/Myc overexpression remained to have significantly worse survival than patients with CD37+ GCB-DLBCL. (D) In GCB-DLBCL, CD37 positivity predicted significantly better survival even when the patients had high IPI scores. (E) In ABC-DLBCL, the adverse prognostic effect of CD37 negativity was independent of p50, survivin, p63, PI3K, and CXCR4 expression and BCL6 translocations. In particular, CD37− patients without p50 and survivin overexpression remained to have significantly worse survival than patients with CD37+ ABC-DLBCL. (F) In ABC-DLBCL, CD37 and IPI had independent prognostic impact. The cutoffs for high/positive expression as indicated by p50+, Myc+, p-STAT3+, GCET1+, survivin+, p63+, PI3K+, and CXCR4+ in the figures were ≥20%, ≥70%, ≥50%, ≥50%, >25%, >5%, ≥70%, and ≥20%, respectively.
Correlation analysis and the robust prognostic effect of CD37 expression in patients with DLBCL. (A) A distribution plot showing that CD37− GCB-DLBCL (denoted by the yellow bar) more frequently had TP53 mutations (highlighted in red) or high levels of nuclear p50 (yellow), Myc (green), p-STAT3 (orange), and p65 (lighter red) expression compared with CD37+ GCB-DLBCL (denoted by the blue bar). (B) A distribution plot showing that CD37− ABC-DLBCL more frequently had high nuclear p50 (yellow) and survivin (pink) expression and BCL6 translocation (green), whereas CD37+ ABC-DLBCL more frequently had PI3K (blue) and CXCR4 (purple) overexpression. Each column in panels A and B represents 1 patient; cases without indicated abnormalities detected are shown in light blue (negative) or white (unknown). (C) In GCB-DLBCL, CD37 positivity predicted significantly improved survival, regardless of presence of TP53 mutations, p50high, Mychigh, p-STAT3high, GCET1high, and to a lesser extent, MYC translocations. Conversely, the adverse effect of CD37 negativity was independent of all these biomarkers. Particularly, CD37− patients without TP53 mutations and p50/Myc overexpression remained to have significantly worse survival than patients with CD37+ GCB-DLBCL. (D) In GCB-DLBCL, CD37 positivity predicted significantly better survival even when the patients had high IPI scores. (E) In ABC-DLBCL, the adverse prognostic effect of CD37 negativity was independent of p50, survivin, p63, PI3K, and CXCR4 expression and BCL6 translocations. In particular, CD37− patients without p50 and survivin overexpression remained to have significantly worse survival than patients with CD37+ ABC-DLBCL. (F) In ABC-DLBCL, CD37 and IPI had independent prognostic impact. The cutoffs for high/positive expression as indicated by p50+, Myc+, p-STAT3+, GCET1+, survivin+, p63+, PI3K+, and CXCR4+ in the figures were ≥20%, ≥70%, ≥50%, ≥50%, >25%, >5%, ≥70%, and ≥20%, respectively.
Together, these results suggest that although CD20 levels may partially contribute to the prognostic effects of CD37 status in CD37− DLBCL and CD37+ ABC-DLBCL patients treated with R-CHOP, CD37 positivity predicts better survival (more remarkable in GCB-DLBCL), independent of CD20 expression, but to certain extent, dependent on the use of rituximab.
CD37 loss in DLBCL is associated with adverse prognostic factors, including TP53 mutations, NF-κB activation, and MYC translocation
To get a better understanding of the underlying molecular mechanisms, we first correlated CD37 status with a spectrum of genetic/phenotypical biomarkers. Compared with the CD37+ group, the CD37− group more frequently had GCB cell of origin, TP53 mutation, MYC rearrangement, increased p50, RelB, and p65 (NF-κB) nuclear expression, and the IgA+ immunophenotype. Conversely, the CD37+ group more frequently had ABC cell of origin, p63+, PI3Khigh, CXCR4high, GCET1high, FOXP1high, MUM1/IRF4high, and the IgM+ immunophenotype.
To eliminate the confounding effect of the GCB predominance in CD37− DLBCL on the molecular differences, we further compared the CD37− and CD37+ groups in GCB-DLBCL and ABC-DLBCL separately. In GCB-DLBCL, CD37− was associated with TP53 mutations and nuclear expression of p50, p65, Myc, and p-STAT3. In ABC-DLBCL, CD37− was associated with BCL6 translocations and nuclear expression of p50, RelB, and survivin. In contrast, CD37+ GCB-DLBCL was associated with GCET1 and BCL-6 expression, and CD37+ ABC-DLBCL was associated with PI3K, CXCR4, GCET1, MUM1/IRF4, and FOXP1 expression (Figure 3A-B).
Predictive value of CD37 expression is robust, especially in GCB-DLBCL
To examine whether the adverse effect of CD37 loss depends on its associated molecular abnormalities and to identify prognostic determinants, we incorporate both CD37 and associated biomarkers into the survival analyses. As shown in Figure 3C and supplemental Figure 4A, in GCB-DLBCL, CD37 loss correlated with significantly decreased PFS/OS rates with and without TP53 mutation, p50high, p65high, Mychigh, p-STAT3high, GCET1low, BCL-6low, and MYC rearrangement, although TP53 mutation, MYC rearrangement, and Mychigh had additive adverse effects to CD37 loss. In particular, patients with CD37− GCB-DLBCL without TP53 mutation, p50high, and Mychigh expression (the 3 adverse factors most strongly associated with CD37− in GCB-DLBCL) remained to have significantly worse survival than patients with CD37+ GCB-DLBCL (OS, P = .0015; PFS, P = .0011). Conversely, CD37 positivity robustly predicted significantly better survival in GCB-DLBCL. In fact, this predictive value completely abolished the prognostic significance of TP53 mutation, p50high, Mychigh, p-STAT3high, and GCET1high expression in CD37+ GCB-DLBCL, although not that of MYC rearrangement. Even more strikingly, the IPI lost prognostic significance in patients with CD37+ GCB-DLBCL (Figure 3D).
Similarly, in ABC-DLBCL, CD37 loss predicted significantly worse survival with and without p50high, survivinhigh, RelB+, p63−, CXCR4high, PI3Khigh, FOXP1high, MUM1high, and BCL6 translocations. In particular, patients with CD37− ABC-DLBCL without p50high and survivinhigh expression (the 2 prognostic factors most strongly associated with CD37− in ABC-DLBCL) still had significantly poorer survival than patients with CD37+ ABC-DLBCL (OS, P = .0017; PFS, P = .0009). However, ABC vs GCB subtype of CD37+ DLBCL patients had significantly worse survival (Figure 1F), and among CD37+ ABC-DLBCL patients, high IPI, TP53 mutation, and Mychigh had significant adverse impact, in contrast to their lack of apparent effect in patients with CD37+ GCB-DLBCL (Figure 3E-F; supplemental Figure 4B).
Multivariate survival analysis shows remarkable predictive values of CD37 status in GCB-DLBCL
We further performed multivariate survival analysis for CD37−/+ status using Cox regression models. After adjustment of clinical variables and CD37 status-associated prognostic factors, CD37 status remained to be a significant prognostic factor in overall DLBCL, GCB-DLBCL, and ABC-DLBCL. CD37− status had the most significant risk prediction power among all the molecular biomarkers in the Cox model for DLBCL overall (for OS: hazard ratio [HR], 3.43; 95% confidence interval [CI], 1.96-6.02; P < .001) (for PFS: HR, 3.91; 95% CI, 2.28-6.71; P < .001), followed by ABC cell of origin (Table 3).
Multivariate survival analysis for CD37 expression
| . | OS . | PFS . | ||||
|---|---|---|---|---|---|---|
| HR . | 95% CI . | P . | HR . | 95% CI . | P . | |
| In overall DLBCL | ||||||
| Female sex | 0.67 | 0.42-1.07 | .09 | 0.74 | 0.48-1.15 | .18 |
| B symptoms | 1.68 | 1.04-2.73 | .035 | 1.69 | 1.06-2.68 | .026 |
| Tumor size ≥5 cm | 1.32 | 0.83-2.09 | .24 | 1.27 | 0.82-1.96 | .28 |
| IPI >2 | 2.35 | 1.49-3.70 | <.001 | 2.43 | 1.57-3.75 | <.001 |
| ABC subtype | 2.12 | 1.24-3.60 | .006 | 1.79 | 1.08-2.98 | .025 |
| TP53 mutation | 1.76 | 1.05-2.96 | .033 | 1.68 | 1.02-2.75 | .040 |
| p63+ | 0.71 | 0.47-1.07 | .097 | 0.81 | 0.55-1.19 | .29 |
| p50high | 1.09 | 0.70-1.71 | .70 | 1.13 | 0.73-1.74 | .58 |
| CXCR4high | 1.81 | 1.06-3.10 | .03 | 2.15 | 1.28-3.63 | .004 |
| CD37+* | 0.29 | 0.17-0.51 | <.001 | 0.26 | 0.15-0.44 | <.001 |
| CD37– | 3.43 | 1.96-6.02 | <.001 | 3.91 | 2.28-6.71 | <.001 |
| In GCB-DLBCL | ||||||
| Female sex | 0.51 | 0.26-1.01 | .053 | 0.57 | 0.31-1.06 | .075 |
| B symptoms | 1.27 | 0.67-2.42 | .46 | 1.16 | 0.65-2.10 | .61 |
| Tumor size ≥5 cm | 1.65 | 0.88-3.08 | .12 | 1.61 | 0.91-2.86 | .10 |
| IPI >2 | 3.66 | 1.90-7.01 | <.001 | 3.39 | 1.90-6.06 | <.001 |
| TP53 mutation | 1.36 | 0.64-2.89 | .42 | 1.40 | 0.70-2.79 | .35 |
| p50high | 0.77 | 0.35-1.69 | .51 | 0.71 | 0.33-1.57 | .40 |
| p65high | 1.90 | 0.81-4.46 | .14 | 1.48 | 0.65-3.36 | .35 |
| p-STAT3high | 0.73 | 0.28-1.89 | .51 | 1.20 | 0.54-2.69 | .65 |
| MYC-R+ | 3.01 | 1.06-3.98 | .038 | 2.46 | 1.03-3.04 | .043 |
| Mychigh† | 2.18 | 1.14-4.16 | .018 | 1.67 | 0.90-3.08 | .10 |
| CD37+* | 0.21 | 0.078-0.54 | .001 | 0.18 | 0.073-0.43 | <.001 |
| CD37− | 4.86 | 1.84-12.82 | .001 | 5.64 | 2.32-13.74 | <.001 |
| In ABC-DLBCL | ||||||
| Female sex | 0.86 | 0.51-1.46 | .58 | 0.94 | 0.57-1.55 | .80 |
| B symptoms | 1.57 | 0.93-2.66 | .098 | 1.52 | 0.91-2.54 | .11 |
| Tumor size ≥5 cm | 1.31 | 0.77-2.24 | .12 | 1.18 | 0.71-1.94 | .53 |
| IPI >2 | 2.75 | 1.65-4.56 | <.001 | 2.76 | 1.58-4.82 | <.001 |
| p50high | 1.87 | 1.10-3.16 | .02 | 1.73 | 1.07-2.81 | .027 |
| Survivinhigh | 1.61 | 0.89-2.88 | .11 | 1.47 | 0.85-2.55 | .17 |
| CXCR4high | 2.17 | 1.30-3.61 | .003 | 1.90 | 1.11-3.24 | .019 |
| p63+ | 0.58 | 0.34-1.00 | .049 | 0.62 | 0.38-1.00 | .048 |
| CD37+* | 0.39 | 0.23-0.67 | .001 | 0.40 | 0.23-0.67 | .001 |
| CD37− | 2.57 | 1.50-4.40 | .001 | 2.53 | 1.50-4.44 | .001 |
| In CD37+GCB-DLBCL | ||||||
| Female sex | 0.25 | 0.016-4.08 | .33 | 0.17 | 0.014-2.07 | .17 |
| B symptoms | 2.13 | 0.17-27.3 | .56 | 1.59 | 0.14-17.9 | .71 |
| Tumor size ≥5 cm | 2.61 | 0.31-22.2 | .38 | 1.44 | 0.24-8.62 | .69 |
| IPI >2 | 2.08 | 0.14-30.5 | .60 | 2.01 | 0.14-28.0 | .60 |
| TP53 mutation | 1.37 | 0.095-19.8 | .82 | 3.17 | 0.35-28.8 | .31 |
| Mychigh | 20.0 | 1.87-213.6 | .013 | 14.1 | 1.39-143 | .025 |
| CXCR4high | 5.45 | 0.61-48.7 | .13 | 6.65 | 0.85-51.8 | .071 |
| . | OS . | PFS . | ||||
|---|---|---|---|---|---|---|
| HR . | 95% CI . | P . | HR . | 95% CI . | P . | |
| In overall DLBCL | ||||||
| Female sex | 0.67 | 0.42-1.07 | .09 | 0.74 | 0.48-1.15 | .18 |
| B symptoms | 1.68 | 1.04-2.73 | .035 | 1.69 | 1.06-2.68 | .026 |
| Tumor size ≥5 cm | 1.32 | 0.83-2.09 | .24 | 1.27 | 0.82-1.96 | .28 |
| IPI >2 | 2.35 | 1.49-3.70 | <.001 | 2.43 | 1.57-3.75 | <.001 |
| ABC subtype | 2.12 | 1.24-3.60 | .006 | 1.79 | 1.08-2.98 | .025 |
| TP53 mutation | 1.76 | 1.05-2.96 | .033 | 1.68 | 1.02-2.75 | .040 |
| p63+ | 0.71 | 0.47-1.07 | .097 | 0.81 | 0.55-1.19 | .29 |
| p50high | 1.09 | 0.70-1.71 | .70 | 1.13 | 0.73-1.74 | .58 |
| CXCR4high | 1.81 | 1.06-3.10 | .03 | 2.15 | 1.28-3.63 | .004 |
| CD37+* | 0.29 | 0.17-0.51 | <.001 | 0.26 | 0.15-0.44 | <.001 |
| CD37– | 3.43 | 1.96-6.02 | <.001 | 3.91 | 2.28-6.71 | <.001 |
| In GCB-DLBCL | ||||||
| Female sex | 0.51 | 0.26-1.01 | .053 | 0.57 | 0.31-1.06 | .075 |
| B symptoms | 1.27 | 0.67-2.42 | .46 | 1.16 | 0.65-2.10 | .61 |
| Tumor size ≥5 cm | 1.65 | 0.88-3.08 | .12 | 1.61 | 0.91-2.86 | .10 |
| IPI >2 | 3.66 | 1.90-7.01 | <.001 | 3.39 | 1.90-6.06 | <.001 |
| TP53 mutation | 1.36 | 0.64-2.89 | .42 | 1.40 | 0.70-2.79 | .35 |
| p50high | 0.77 | 0.35-1.69 | .51 | 0.71 | 0.33-1.57 | .40 |
| p65high | 1.90 | 0.81-4.46 | .14 | 1.48 | 0.65-3.36 | .35 |
| p-STAT3high | 0.73 | 0.28-1.89 | .51 | 1.20 | 0.54-2.69 | .65 |
| MYC-R+ | 3.01 | 1.06-3.98 | .038 | 2.46 | 1.03-3.04 | .043 |
| Mychigh† | 2.18 | 1.14-4.16 | .018 | 1.67 | 0.90-3.08 | .10 |
| CD37+* | 0.21 | 0.078-0.54 | .001 | 0.18 | 0.073-0.43 | <.001 |
| CD37− | 4.86 | 1.84-12.82 | .001 | 5.64 | 2.32-13.74 | <.001 |
| In ABC-DLBCL | ||||||
| Female sex | 0.86 | 0.51-1.46 | .58 | 0.94 | 0.57-1.55 | .80 |
| B symptoms | 1.57 | 0.93-2.66 | .098 | 1.52 | 0.91-2.54 | .11 |
| Tumor size ≥5 cm | 1.31 | 0.77-2.24 | .12 | 1.18 | 0.71-1.94 | .53 |
| IPI >2 | 2.75 | 1.65-4.56 | <.001 | 2.76 | 1.58-4.82 | <.001 |
| p50high | 1.87 | 1.10-3.16 | .02 | 1.73 | 1.07-2.81 | .027 |
| Survivinhigh | 1.61 | 0.89-2.88 | .11 | 1.47 | 0.85-2.55 | .17 |
| CXCR4high | 2.17 | 1.30-3.61 | .003 | 1.90 | 1.11-3.24 | .019 |
| p63+ | 0.58 | 0.34-1.00 | .049 | 0.62 | 0.38-1.00 | .048 |
| CD37+* | 0.39 | 0.23-0.67 | .001 | 0.40 | 0.23-0.67 | .001 |
| CD37− | 2.57 | 1.50-4.40 | .001 | 2.53 | 1.50-4.44 | .001 |
| In CD37+GCB-DLBCL | ||||||
| Female sex | 0.25 | 0.016-4.08 | .33 | 0.17 | 0.014-2.07 | .17 |
| B symptoms | 2.13 | 0.17-27.3 | .56 | 1.59 | 0.14-17.9 | .71 |
| Tumor size ≥5 cm | 2.61 | 0.31-22.2 | .38 | 1.44 | 0.24-8.62 | .69 |
| IPI >2 | 2.08 | 0.14-30.5 | .60 | 2.01 | 0.14-28.0 | .60 |
| TP53 mutation | 1.37 | 0.095-19.8 | .82 | 3.17 | 0.35-28.8 | .31 |
| Mychigh | 20.0 | 1.87-213.6 | .013 | 14.1 | 1.39-143 | .025 |
| CXCR4high | 5.45 | 0.61-48.7 | .13 | 6.65 | 0.85-51.8 | .071 |
Multivariate survival analysis for CD37 expression adjusting clinical factors and molecular biomarkers associated with CD37+/− in overall DLBCL, GCB-DLBCL, and ABC-DLBCL, and multivariate analysis in CD37+ GCB-DLBCL. The cutoffs for high/positive expression of p50, p65, p-STAT3, Myc, CXCR4, survivin, and p63 were ≥20%, ≥50%, ≥50%, ≥70%, ≥20%, >25%, and >5%, respectively. P values designating statistical significance, and the HR and 95% CI for CD37 status, are highlighted in bold.
Two different models were used to include either CD37+ or CD37− as 1 variant separately.
Two different Cox regression models were used to include either MYC-R+ or Mychigh as 1 variant separately. MYC rearrangement was not included as a variant in the multivariate analysis in CD37+ GCB-DLBCL because of limited case numbers.
Impressively, in patients with GCB-DLBCL, the HR of CD37− was even higher than that of IPI >2 (for OS, 4.86 vs 3.66; for PFS, 5.64 vs 3.39), and the IPI lost prognostic significance in CD37+ GCB-DLBCL (Table 3). Moreover, in both CD37+ and CD37− GCB-DLBCL subsets, all biomarkers except Mychigh lost significance as independent factors for poorer survival. Differently, in CD37+ ABC-DLBCL, IPI >2, TP53 mutations, Mychigh, p63−, and survivinhigh had significant independent adverse impact, and in CD37− ABC-DLBCL, IPI >2, p50high, p63−, and CXCR4high were significant independent adverse prognostic factors (supplemental Table 1).
Molecularly adjusted IPI for R-CHOP significantly improves risk stratification in DLBCL
Because CD37 status and GCB/ABC cell of origin showed remarkable prognostic significance independent of the IPI, we tested whether combining these 2 risk factors with the IPI improved the prognostic prediction. The IPI did separate the discovery cohort into 4 groups but had limited power to identify high-risk patients (15.2% of the discovery cohort; P = .073 [OS] and .017 [PFS] compared with intermediate-high-risk patients) (Figure 4A). We added the scores for CD37 status (add 1 point if CD37−) and cell of origin (add 1 point if ABC) to the IPI, resulting in a molecularly adjusted IPI for R-CHOP (M-IPI-R) score for each patient. This M-IPI-R could redistribute patients into 4 groups (low risk, score 0-1 [16.6%]; intermediate risk, score 2-3 [42.7%]; high risk, score 4-5 [34.6%]; and very-high risk, score 6-7 [6.2%]), and showed significantly improved stratification power compared with the traditional IPI and cell-of-origin–adjusted IPI (supplemental Figure 5A) scores in separating high-risk and very-high-risk patients from intermediate-risk patients (P < .0001) (Figure 4B). The 5-year OS rates for high-risk and very-high-risk groups were 40.19% and 18.06%, respectively, compared with 38.38% for the high-risk group identified by the traditional IPI.
Comparison of risk stratification in patients with DLBCL by the traditional IPI and adjusted IPI scores. (A) Risk stratification of DLBCL groups by the traditional IPI. (B) Risk stratification of DLBCL by the M-IPI-R, defined by each patient’s IPI score plus risk scores for CD37 status (CD37+, 0; CD37−, 1 or 2 as indicated) and GCB/ABC cell of origin (GCB, 0; ABC, 1). (C) Risk stratification of DLBCL by the IPI+IHC, defined by each patient’s IPI score plus risk scores for CD37 status (CD37+, 0; CD37−, 3), and Myc and Bcl-2 protein levels (low [IHC <70%], 0; high [IHC ≥70%], 1).
Comparison of risk stratification in patients with DLBCL by the traditional IPI and adjusted IPI scores. (A) Risk stratification of DLBCL groups by the traditional IPI. (B) Risk stratification of DLBCL by the M-IPI-R, defined by each patient’s IPI score plus risk scores for CD37 status (CD37+, 0; CD37−, 1 or 2 as indicated) and GCB/ABC cell of origin (GCB, 0; ABC, 1). (C) Risk stratification of DLBCL by the IPI+IHC, defined by each patient’s IPI score plus risk scores for CD37 status (CD37+, 0; CD37−, 3), and Myc and Bcl-2 protein levels (low [IHC <70%], 0; high [IHC ≥70%], 1).
Within the M-IPI-R–defined risk groups, GCB/ABC was not prognostic anymore but CD37− status still showed significant adverse impact (supplemental Figure 5B). Therefore, we assigned 1 additional point for CD37− into the M-IPI-R, and found neither CD37+/− nor GCB/ABC had further prognostic significance within the newly defined risk groups. This version of M-IPI-R, which may have fully adjusted the risk conferred by CD37−, could refine the stratification into 5 different risk groups (Figure 4B): low risk, score 0-1 (11.5%); low-intermediate risk, score 2-3 (33.3%); intermediate risk, score 4 (20.6%); high risk, score 5-6 (28.7%); and very-high risk, score 7-8 (5.9%). The 5-year OS rates for the high-risk and very-high-risk groups were 37.33% and 19.64%, respectively.
To develop a more applicable immunohistochemistry-based index, we tested various biomarkers and found combining CD37−, Mychigh, and Bcl-2high risk factors with the IPI, defined as IPI plus immunohistochemistry (IPI+IHC), showed the strongest stratification power without redundancy (supplemental Figure 5C, 1 point for CD37−; and Figure 4C, 3 points for CD37−). In Figure 4C, the 5-year OS rates for high-risk (score 7-8, 16.3%) and very-high-risk (score 9-10, 3.5%) groups were 24.21% and 7.81%, respectively.
GEP analysis suggests roles of CD37 in immune signaling
We compared gene expression profiles of CD37+ and CD37− groups in the discovery DLBCL cohort. Surprisingly, no significant CD37 gene signatures were identified in GCB-DLBCL although CD37 status had remarkable prognostic significance. In overall and ABC-DLBCL only, weak CD37 signatures were identified (Table 4; supplemental Table 2), but these signatures were also heterogeneously expressed within both the CD37+ and CD37− subgroups (Figure 5A).
GEP analysis between CD37+ and CD37− DLBCL groups
| Function categories . | Upregulated . | Downregulated . |
|---|---|---|
| CD37−vs CD37+DLBCL (FDR <0.05) | ||
| Immune responses, inflammation, host defense | FCGR1B ↑ LILRB2 ↑ SERPING1 ↑ IRF1 ↑ TLR2 ↑ SAMHD1 ↑ IFITM1 ↑ IFITM3 ↑ PYCARD ↑ HEBP1 ↑ | |
| Signaling | FNIP2 ↑ RCAN1 ↑ SERPINA1 ↑ | CD37 ↓ MS4A1/CD20 ↓ VSNL1 ↓ LPAR6 ↓ |
| Differentiation | FNDC3B ↑ | AICDA ↓ BCL11A ↓ FOXP1 ↓ XBP1 ↓ PAX5 ↓ |
| Gene expression, translation, cell cycle | EIF1 ↑ | CDK5R1 ↓ METTL2A/B ↓ ZNF581 ↓ ZNF765 ↓ |
| Apoptosis, DNA damage response | DRAM1 ↑ | C9orf102 ↓ |
| Metabolism, protein repair and degradation, oxidative stress, lipid movement, transport, intracellular membrane trafficking | GCH1 ↑ MT1G ↑ MT1P2 ↑ PAM ↑ MT1E ↑ C11orf67 ↑ MT1E/MT1H/MT1M ↑ UBR1 ↑ APOL6 ↑ RCN1 ↑ PCMT1 ↑ MSRB2 ↑ NUB1 ↑ RAB6A ↑ ARF4 ↑ RAB1B ↑ AQP9 ↑ CDIPT ↑ C12orf62 ↑ DPYD ↑ | ZDHHC21 ↓ GOLGA8A/B ↓ |
| Adhesion, regulation of actin, cytoskeleton, exocytosis, and extracellular matrix | RAB27A ↑ AOC3 ↑ GPR124 ↑ NINJ1 ↑ WDR1 ↑ CRISPLD2 ↑ RHOU ↑ | DMD ↓ |
| Unknown function, RNA gene | ANKRD22 ↑ MOP-1 ↑ CNIH4 ↑ PLEKHO2 ↑ FTHP1 ↑ C4orf32 ↑ FAM167B ↑ PWWP2B ↑ | FAM129C ↓ NAPSB ↓ SNORA74A ↓ BRWD2 ↓ FAM108C1 ↓ LOC283887 ↓ RAB7L1 ↓ LOC133874 ↓ |
| CD37−vs CD37+ABC-DLBCL (FDR <0.05) | ||
| Immune responses, inflammation, host defense | LILRB2 ↑ C1S ↑ AIF1 ↑ IFITM1 ↑ IFITM3 ↑ SERPING1 ↑ HLA-C ↑ LST1 ↑ B2M ↑ ISG15 ↑ | |
| Signaling | SRC ↑ STAT4 ↑ CD63 ↑ SEMA4D ↑ SECTM1 ↑ VDR ↑ RCAN1 ↑ ACVRL1 ↑ PREX1 ↑ PROCR ↑ TNIP2 ↑ RALB ↑ | CD37 ↓ |
| Differentiation | AICDA ↓ BCL11A ↓ FOXP1 ↓ IKZF1 ↓ PAX5 ↓ | |
| Gene expression, translation | TCF7L2 ↑ EIF4E3 ↑ EIF1 ↑ C14orf4 ↑ KLF3 ↑ | TBL1XR1 ↓ HELLS ↓ RPS2 ↓ EBF1 ↓ ZNF587 ↓ |
| Apoptosis, autophagy, DNA repair | DRAM1 ↑ C19orf40 ↑ | C9orf102 ↓ |
| Metabolism, ion channel, transport, trafficking, protein degradation | FAM26F ↑ GCH1 ↑ MT1P2 ↑ MT1E ↑ STOM ↑ MT1E/MT1H/MT1M ↑ ACSL1 ↑ AAK1 ↑ VAMP5 ↑ RAB13 ↑ RCN1 ↑ RNF217 ↑ RAB6A ↑ MGST2 ↑ BLVRA ↑ C14orf133 ↑ PDXK ↑ GMPPA ↑ HTATIP2 ↑ HEBP1 ↑ CHP1 ↑ | C7orf68 ↓ GOLGA8B ↓ RIMKLB ↓ DPY19L2 ↓ UBE3C ↓ DNAJB7 ↓ |
| Adhesion, regulation of actin, cytoskeleton, exocytosis, and extracellular matrix | RAB27A ↑ PECAM1 ↑ RHOU ↑ MYOF ↑ DYSF ↑ CCDC80 ↑ AOC3 ↑ CTNNA1 ↑ GPR124 ↑ | RALGPS2 ↓ |
| Unknown function, RNA gene | ANKRD22 ↑ GLIPR2 ↑ NKG7 ↑ CNIH4 ↑ SAMD3 ↑ TMEM176A ↑ SLFN12 ↑ PLEKHO2 ↑ C21orf33/PWP2 ↑ | FAM129C ↓ C13orf18 ↓ NAPSB ↓ SNORA68 ↓ SNORA74A ↓ BRWD2 ↓ FAM108C1 ↓ SNORD104 ↓ FLJ43663 ↓ SNHG12 ↓ PMS2L1 ↓ LOC284513 ↓ |
| PI3Khigh/CD37−vs PI3Khigh/CD37+ABC-DLBCL (FDR <0.01) | ||
| Immune responses, inflammation, host defense | FCGR1B ↑ GBP1 ↑ FPR2 ↑ TRBC1 ↑ IFITM1 ↑ FCGR3B ↑ FCGR1A/C ↑ C3AR1 ↑ CD14 ↑ | |
| Signaling | SECTM1 ↑ TNFSF10 ↑ SEMA4D ↑ APLP2 ↑ | |
| Differentiation, transcription factors | BCL11B ↑ KLF3 ↑ | |
| Cell cycle | CCND1 ↑ PTP4A2 ↑ | |
| Apoptosis | DRAM1 ↑ EPB41L3 ↑ | |
| Ion channel, transport, trafficking, protein degradation, metabolism | MT1H ↑ FAM26F ↑ MT1G ↑ MT2A ↑ MT1X ↑ CPD ↑ MT1E/MT1H/MT1M ↑ SLC8A1 ↑ MT1E ↑ AAK1 ↑ LYST ↑ APOL6 ↑ RAB1A ↑ | |
| Adhesion | PECAM1 ↑ | |
| RNA regulation, unknown function | ANKRD22 ↑ ZC3H12C ↑ GLIPR2 ↑ | SNORD104 ↓ |
| CD37−ABC vs CD37−GCB (FDR <0.01, fold change >2) | ||
| Immune responses, inflammation | TNFRSF13B ↑ S100A8 ↑ GZMB ↑ CLECL1 ↑ LILRB2 ↑ FCRL5 ↑ CCL8 ↑ | IGHG1 ↓ |
| Signaling | IGHM ↑ | MME ↓ STAP1 ↓ |
| Transcription regulation, DNA replication, microRNA | BATF ↑ IRF4 ↑ MIR155HG ↑ | MYBL1 ↓ ELL3/SERINC4 ↓ STAG3 ↓ SSBP2 ↓ BCL6 ↓ |
| Metabolism, ion channel | FUT8 ↑ FAM26F ↑ DHRS9 ↑ | |
| Adhesion, cytoskeleton, extracellular matrix | TUBB4 ↑ CD44 ↑ | CILP ↓ KANK1 ↓ PCDHGB7 ↓ MARCKSL1 ↓ |
| Unknown function | ANKRD22 ↑ C1orf186 ↑C13orf18 ↑ MPEG1 ↑ | FLJ42418 ↓C17orf99 ↓ TMEM108 ↓ TEX9 ↓ CRNDE ↓ CCDC85A ↓ BTNL9 ↓ KIAA0746 ↓ |
| CD37+ABC vs CD37+GCB (FDR <0.01, fold change >2.18) | ||
| Immune responses | TNFRSF13B ↑ CLECL1 ↑ FCRL5 ↑ GZMB ↑ | IGHG1 ↓ HLA-DOB ↓ LRMP ↓ CXCL14 ↓ |
| Signaling | IGHM ↑ BLNK ↑ | MME ↓ STAP1 ↓ PTPRS ↓ SPRED2 ↓ PDE4D ↓ RRAS2 ↓ FNDC1 ↓ |
| Differentiation | AICDA ↑ | LMO2 ↓ |
| Transcription regulation, DNA replication, microRNA | FOXP1 ↑ PARP15 ↑ BATF ↑ TCF4 ↑ IRF4 ↑ MIR155HG ↑ | MYBL1 ↓ STAG3 ↓ HOPX ↓TOX ↓ SSBP2 ↓ REL ↓ |
| Cell cycle | NEK6 ↓ | |
| Metabolism, transport | XK ↑ P2RX5 ↑ FUT8 ↑ SLC2A13 ↑ | SLC12A8 ↓ PLEKHF2 ↓ GALNT14 ↓ SULF1 ↓ LHPP ↓ |
| Adhesion, cytoskeleton, extracellular matrix, migration | TUBB4 ↑ | CILP ↓ POSTN ↓ MARCKSL1 ↓ DPT ↓ PCDHGB7 ↓ RAPH1 ↓ CPNE3 ↓ COL5A1 ↓ KANK1 ↓ PTK2 ↓ |
| Unknown function | C13orf18 ↑ FAM129C ↑ C1orf186 ↑ TBC1D27 ↑ MPEG1 ↑ | C17orf99 ↓ FLJ42418 ↓ CCDC144B ↓ TMEM108 ↓ BTNL9 ↓ PRO1483 ↓ CCDC85A ↓ KIAA0746 ↓ |
| Function categories . | Upregulated . | Downregulated . |
|---|---|---|
| CD37−vs CD37+DLBCL (FDR <0.05) | ||
| Immune responses, inflammation, host defense | FCGR1B ↑ LILRB2 ↑ SERPING1 ↑ IRF1 ↑ TLR2 ↑ SAMHD1 ↑ IFITM1 ↑ IFITM3 ↑ PYCARD ↑ HEBP1 ↑ | |
| Signaling | FNIP2 ↑ RCAN1 ↑ SERPINA1 ↑ | CD37 ↓ MS4A1/CD20 ↓ VSNL1 ↓ LPAR6 ↓ |
| Differentiation | FNDC3B ↑ | AICDA ↓ BCL11A ↓ FOXP1 ↓ XBP1 ↓ PAX5 ↓ |
| Gene expression, translation, cell cycle | EIF1 ↑ | CDK5R1 ↓ METTL2A/B ↓ ZNF581 ↓ ZNF765 ↓ |
| Apoptosis, DNA damage response | DRAM1 ↑ | C9orf102 ↓ |
| Metabolism, protein repair and degradation, oxidative stress, lipid movement, transport, intracellular membrane trafficking | GCH1 ↑ MT1G ↑ MT1P2 ↑ PAM ↑ MT1E ↑ C11orf67 ↑ MT1E/MT1H/MT1M ↑ UBR1 ↑ APOL6 ↑ RCN1 ↑ PCMT1 ↑ MSRB2 ↑ NUB1 ↑ RAB6A ↑ ARF4 ↑ RAB1B ↑ AQP9 ↑ CDIPT ↑ C12orf62 ↑ DPYD ↑ | ZDHHC21 ↓ GOLGA8A/B ↓ |
| Adhesion, regulation of actin, cytoskeleton, exocytosis, and extracellular matrix | RAB27A ↑ AOC3 ↑ GPR124 ↑ NINJ1 ↑ WDR1 ↑ CRISPLD2 ↑ RHOU ↑ | DMD ↓ |
| Unknown function, RNA gene | ANKRD22 ↑ MOP-1 ↑ CNIH4 ↑ PLEKHO2 ↑ FTHP1 ↑ C4orf32 ↑ FAM167B ↑ PWWP2B ↑ | FAM129C ↓ NAPSB ↓ SNORA74A ↓ BRWD2 ↓ FAM108C1 ↓ LOC283887 ↓ RAB7L1 ↓ LOC133874 ↓ |
| CD37−vs CD37+ABC-DLBCL (FDR <0.05) | ||
| Immune responses, inflammation, host defense | LILRB2 ↑ C1S ↑ AIF1 ↑ IFITM1 ↑ IFITM3 ↑ SERPING1 ↑ HLA-C ↑ LST1 ↑ B2M ↑ ISG15 ↑ | |
| Signaling | SRC ↑ STAT4 ↑ CD63 ↑ SEMA4D ↑ SECTM1 ↑ VDR ↑ RCAN1 ↑ ACVRL1 ↑ PREX1 ↑ PROCR ↑ TNIP2 ↑ RALB ↑ | CD37 ↓ |
| Differentiation | AICDA ↓ BCL11A ↓ FOXP1 ↓ IKZF1 ↓ PAX5 ↓ | |
| Gene expression, translation | TCF7L2 ↑ EIF4E3 ↑ EIF1 ↑ C14orf4 ↑ KLF3 ↑ | TBL1XR1 ↓ HELLS ↓ RPS2 ↓ EBF1 ↓ ZNF587 ↓ |
| Apoptosis, autophagy, DNA repair | DRAM1 ↑ C19orf40 ↑ | C9orf102 ↓ |
| Metabolism, ion channel, transport, trafficking, protein degradation | FAM26F ↑ GCH1 ↑ MT1P2 ↑ MT1E ↑ STOM ↑ MT1E/MT1H/MT1M ↑ ACSL1 ↑ AAK1 ↑ VAMP5 ↑ RAB13 ↑ RCN1 ↑ RNF217 ↑ RAB6A ↑ MGST2 ↑ BLVRA ↑ C14orf133 ↑ PDXK ↑ GMPPA ↑ HTATIP2 ↑ HEBP1 ↑ CHP1 ↑ | C7orf68 ↓ GOLGA8B ↓ RIMKLB ↓ DPY19L2 ↓ UBE3C ↓ DNAJB7 ↓ |
| Adhesion, regulation of actin, cytoskeleton, exocytosis, and extracellular matrix | RAB27A ↑ PECAM1 ↑ RHOU ↑ MYOF ↑ DYSF ↑ CCDC80 ↑ AOC3 ↑ CTNNA1 ↑ GPR124 ↑ | RALGPS2 ↓ |
| Unknown function, RNA gene | ANKRD22 ↑ GLIPR2 ↑ NKG7 ↑ CNIH4 ↑ SAMD3 ↑ TMEM176A ↑ SLFN12 ↑ PLEKHO2 ↑ C21orf33/PWP2 ↑ | FAM129C ↓ C13orf18 ↓ NAPSB ↓ SNORA68 ↓ SNORA74A ↓ BRWD2 ↓ FAM108C1 ↓ SNORD104 ↓ FLJ43663 ↓ SNHG12 ↓ PMS2L1 ↓ LOC284513 ↓ |
| PI3Khigh/CD37−vs PI3Khigh/CD37+ABC-DLBCL (FDR <0.01) | ||
| Immune responses, inflammation, host defense | FCGR1B ↑ GBP1 ↑ FPR2 ↑ TRBC1 ↑ IFITM1 ↑ FCGR3B ↑ FCGR1A/C ↑ C3AR1 ↑ CD14 ↑ | |
| Signaling | SECTM1 ↑ TNFSF10 ↑ SEMA4D ↑ APLP2 ↑ | |
| Differentiation, transcription factors | BCL11B ↑ KLF3 ↑ | |
| Cell cycle | CCND1 ↑ PTP4A2 ↑ | |
| Apoptosis | DRAM1 ↑ EPB41L3 ↑ | |
| Ion channel, transport, trafficking, protein degradation, metabolism | MT1H ↑ FAM26F ↑ MT1G ↑ MT2A ↑ MT1X ↑ CPD ↑ MT1E/MT1H/MT1M ↑ SLC8A1 ↑ MT1E ↑ AAK1 ↑ LYST ↑ APOL6 ↑ RAB1A ↑ | |
| Adhesion | PECAM1 ↑ | |
| RNA regulation, unknown function | ANKRD22 ↑ ZC3H12C ↑ GLIPR2 ↑ | SNORD104 ↓ |
| CD37−ABC vs CD37−GCB (FDR <0.01, fold change >2) | ||
| Immune responses, inflammation | TNFRSF13B ↑ S100A8 ↑ GZMB ↑ CLECL1 ↑ LILRB2 ↑ FCRL5 ↑ CCL8 ↑ | IGHG1 ↓ |
| Signaling | IGHM ↑ | MME ↓ STAP1 ↓ |
| Transcription regulation, DNA replication, microRNA | BATF ↑ IRF4 ↑ MIR155HG ↑ | MYBL1 ↓ ELL3/SERINC4 ↓ STAG3 ↓ SSBP2 ↓ BCL6 ↓ |
| Metabolism, ion channel | FUT8 ↑ FAM26F ↑ DHRS9 ↑ | |
| Adhesion, cytoskeleton, extracellular matrix | TUBB4 ↑ CD44 ↑ | CILP ↓ KANK1 ↓ PCDHGB7 ↓ MARCKSL1 ↓ |
| Unknown function | ANKRD22 ↑ C1orf186 ↑C13orf18 ↑ MPEG1 ↑ | FLJ42418 ↓C17orf99 ↓ TMEM108 ↓ TEX9 ↓ CRNDE ↓ CCDC85A ↓ BTNL9 ↓ KIAA0746 ↓ |
| CD37+ABC vs CD37+GCB (FDR <0.01, fold change >2.18) | ||
| Immune responses | TNFRSF13B ↑ CLECL1 ↑ FCRL5 ↑ GZMB ↑ | IGHG1 ↓ HLA-DOB ↓ LRMP ↓ CXCL14 ↓ |
| Signaling | IGHM ↑ BLNK ↑ | MME ↓ STAP1 ↓ PTPRS ↓ SPRED2 ↓ PDE4D ↓ RRAS2 ↓ FNDC1 ↓ |
| Differentiation | AICDA ↑ | LMO2 ↓ |
| Transcription regulation, DNA replication, microRNA | FOXP1 ↑ PARP15 ↑ BATF ↑ TCF4 ↑ IRF4 ↑ MIR155HG ↑ | MYBL1 ↓ STAG3 ↓ HOPX ↓TOX ↓ SSBP2 ↓ REL ↓ |
| Cell cycle | NEK6 ↓ | |
| Metabolism, transport | XK ↑ P2RX5 ↑ FUT8 ↑ SLC2A13 ↑ | SLC12A8 ↓ PLEKHF2 ↓ GALNT14 ↓ SULF1 ↓ LHPP ↓ |
| Adhesion, cytoskeleton, extracellular matrix, migration | TUBB4 ↑ | CILP ↓ POSTN ↓ MARCKSL1 ↓ DPT ↓ PCDHGB7 ↓ RAPH1 ↓ CPNE3 ↓ COL5A1 ↓ KANK1 ↓ PTK2 ↓ |
| Unknown function | C13orf18 ↑ FAM129C ↑ C1orf186 ↑ TBC1D27 ↑ MPEG1 ↑ | C17orf99 ↓ FLJ42418 ↓ CCDC144B ↓ TMEM108 ↓ BTNL9 ↓ PRO1483 ↓ CCDC85A ↓ KIAA0746 ↓ |
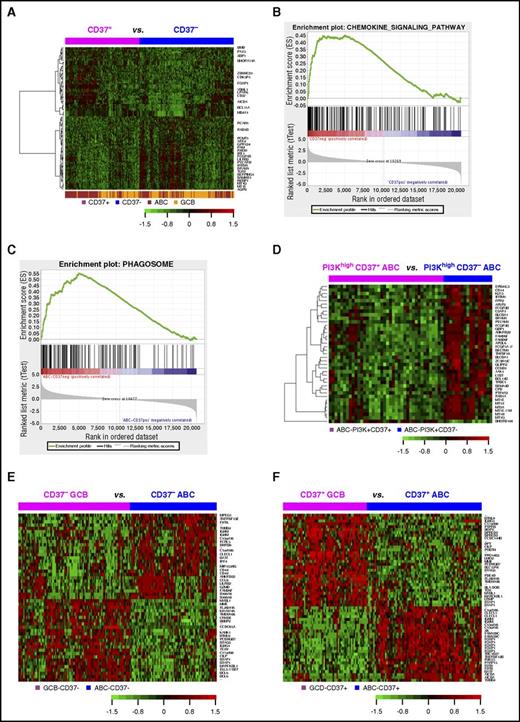
GEP analysis in DLBCL. (A) Heatmap for GEP comparison between CD37+ and CD37− DLBCL (FDR, <0.05). (B) The KEGG chemokine signaling pathway gene set was enriched in the CD37− DLBCL group with an enrichment score of 0.45 (FDR, 0.09). (C) The KEGG phagosome gene set was enriched in CD37− ABC-DLBCL with an enrichment score of 0.55 (FDR, 0.006). (D) Heatmap for GEP comparison between CD37+ ABC-DLBCL and CD37− ABC-DLBCL groups with high (≥70%) PI3K expression (FDR, <0.01). (E) Heatmap for genes differentially expressed between GCB and ABC subtypes of CD37− DLBCL with more than twofold difference (FDR, <0.01). (F) Heatmap for genes differentially expressed between GCB and ABC subtypes of CD37+ DLBCL with >2.4-fold difference (FDR, < 0.01).
GEP analysis in DLBCL. (A) Heatmap for GEP comparison between CD37+ and CD37− DLBCL (FDR, <0.05). (B) The KEGG chemokine signaling pathway gene set was enriched in the CD37− DLBCL group with an enrichment score of 0.45 (FDR, 0.09). (C) The KEGG phagosome gene set was enriched in CD37− ABC-DLBCL with an enrichment score of 0.55 (FDR, 0.006). (D) Heatmap for GEP comparison between CD37+ ABC-DLBCL and CD37− ABC-DLBCL groups with high (≥70%) PI3K expression (FDR, <0.01). (E) Heatmap for genes differentially expressed between GCB and ABC subtypes of CD37− DLBCL with more than twofold difference (FDR, <0.01). (F) Heatmap for genes differentially expressed between GCB and ABC subtypes of CD37+ DLBCL with >2.4-fold difference (FDR, < 0.01).
Further gene set enrichment analysis showed that 48 and 125 gene sets were enriched in CD37− DLBCL and in CD37− ABC-DLBCL (false discovery rate [FDR] <0.25; supplemental Tables 3-4), respectively, whereas no Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were enriched in CD37+ DLBCL or CD37+ ABC-DLBCL significantly. Most gene sets enriched in CD37− DLBCL and CD37− ABC-DLBCL were related to infection (eg, pertussis, Salmonella, Helicobacter pylori, and prion disease) and immune signaling, including NOD-like receptor signaling, phagosome, chemokine signaling (Figure 5B-C), osteoclast differentiation, Toll-like receptor signaling, and tumor necrosis factor (TNF) signaling pathway among the top enriched gene sets.
Interestingly, a more distinct CD37 gene signature was identified in the PI3Khigh but not PI3Klow ABC-DLBCL subset (Figure 5D), suggesting PI3K-involved CD37 signaling in ABC-DLBCL.10 With FDR <0.05, 26 genes were upregulated in CD37+/PI3Khigh compared with CD37−/PI3Khigh ABC-DLBCL, including BCL11A, PAX5, TCF4, and CLECL1 which functions as a T-cell costimulatory molecule. Paradoxically, tumor suppressors EPB41L3 and BCL11B, DRAM1 which is critical for p53-mediated apoptosis,34 and SAMSN1 (a negative regulator of B-cell activation and proliferation) were downregulated in CD37+/PI3Khigh ABC-DLBCL. Conversely, 225 genes were upregulated in CD37−/PI3Khigh compared with CD37+/PI3Khigh ABC-DLBCL, including many related to immune signaling. Of note, CD163 (3.27-fold) is a marker of M2 (tumor-promoting) macrophage,35 CD14 (1.53-fold) is a marker for macrophage activation, SERPING1 inhibits C1 complex in complement activation, LILRB2 (1.83-fold) encodes a leukocyte immunoglobulin-like receptor that negatively regulates MHC-I–mediated antigen presentation and immune responses leading to tolerance development,36,37 and CD300A (1.98-fold) inhibits the antitumor activities by natural killer and mast cells38,39 (Table 4; supplemental Table 5).
Different from CD37+/− status only showing a weak GEP signature, GCB/ABC cell of origin showed remarkable GEP signatures in both the CD37+ DLBCL and CD37− DLBCL subsets (Figure 5E-F): 2346 significant transcripts in CD37+ DLBCL and 1383 transcripts in CD37− DLBCL with FDR <0.01 (Table 4, more than twofold difference). In both CD37− and CD37+ DLBCL subsets, ABC compared with GCB cell of origin had significant upregulation of TNFRSF13B/TACI (receptor for TNFSF13/APRIL and TNFSF13B/BAFF), IGHM, CLECL1, MIR155HG, IRF4, BATF, and CCL8. Additionally, in the CD37+ subset, ABC- compared with GCB-DLBCL had significant upregulation of FOXP1 and AICDA, whereas downregulation of HLA-DOB, LRMP (which plays a role in the delivery of peptides to MHC-I), and LMO2. In the CD37− subset, ABC compared with GCB cell of origin had significant LILRB2 upregulation.
CD37 may play important roles in the costimulatory and PD-1 pathways
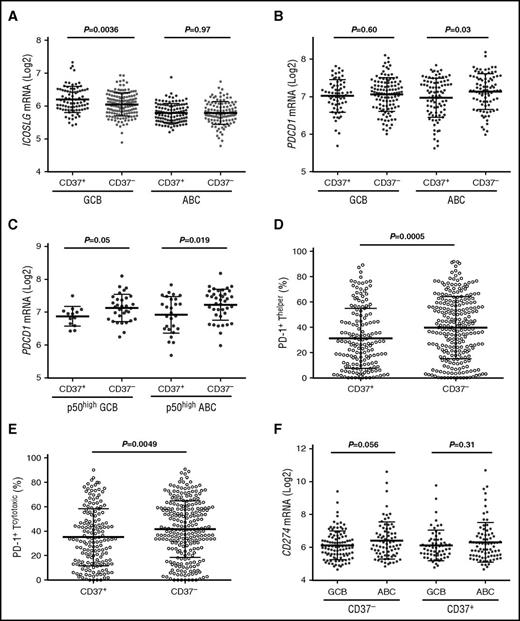
We compared the expression of important immune genes between CD37− and CD37+ patients and within the CD37− and CD37+ subsets (Table 5). In GCB-DLBCL, CD37+ status, which robustly predicted favorable clinical outcomes, correlated with ICOSLG (encoding ICOSL/ICOSLG,40 the ligand for the inducible T-cell costimulator [ICOS]) upregulation, whereas CD37 loss in ABC-DLBCL correlated with PDCD1/PD-1 upregulation (engagement of programmed cell death protein 1 [PD-1] on T cells with its ligand [PD-L1] on tumor cells inhibits T-cell antitumor responses) (Figure 6A-B). In p50high DLBCL, CD37− correlated with PD-1 upregulation in both GCB- and ABC-DLBCL (Figure 6C). Immunohistochemical analysis for PD-1 expression (Z.Y.X.-M. and K.H.Y., unpublished data) further confirmed the significant association of CD37 loss with PD-1 overexpression in DLBCL (Figure 6D-E), with significant or borderline P values in the ABC and GCB subsets.
Differential expression of genes involved in immune responses
| Genes involved in immune responses or differentiation . | CD37− vs CD37+ in GCB/ABC DLBCL . | p50high* vs p50low in CD37− GCB/ABC DLBCL . | CXCR4high* vs CXCR4low in CD37+ GCB/ABC DLBCL . | ABC vs GCB in CD37+ DLBCL . |
|---|---|---|---|---|
| ICOSLG | ↓ in GCB (P = .0036) | ↓ (P < .0001) | ||
| CD58 | ↓ in GCB (P = .022) | ↓ in CD37+ ABC (P = .056) | ↓ (P = .0009) | |
| FCGR2A (CD32A) | ↑ in both GCB (P = .016) and ABC (P = .044) | ↑ in both CD37− GCB (P = .0078) and CD37− ABC (P = .016) | ↓ in CD37+ ABC (P = .02) | ↑ (P = .0022) |
| FCGR1B (CD64) | ↑ in both GCB (P = .01) and ABC (P = .0007) | ↑ in both CD37− GCB (P = .0003) and CD37− ABC (P = .0094) | ↑ (P = .0005) | |
| C1QB | ↑ in both GCB (P = .0001) and ABC (P = .0027) | ↑ in both CD37− GCB (P < .0001) and CD37− ABC (P = .0007) | ↑ (P < .0001) | |
| CD8A | ↑ in ABC (P = .0014) | ↑ in both CD37− GCB (P = .017) and CD37− ABC (P = .039) | ||
| CD8B | ↑ in both GCB (P = .068) and in ABC (P = .0012) | ↑ in both CD37− GCB (P = .0059) and CD37− ABC (P = .038) | Trends of ↓ in CD37+ GCB (P = .08) and CD37+ ABC (P = .068) | |
| PDCD1 (PD-1) | ↑ in ABC (P = .03); ↑ in p50high GCB (P = .05) | ↑ in CD37− ABC (P = .03) | ↓ in CD37+ GCB (P = .014) and trend of ↓ in CD37+ ABC (P = .069) | |
| CD274 (PD-L1) | ↑ in CD37− GCB (P = .013) | ↑ in CD37+ ABC (P = .018) | ||
| PDCD1LG2 (PD-L2) | Trend of ↑ in ABC (P = .09) | ↑ in CD37− GCB (P = .0025) | ||
| CTLA4 | ↑ in both CD37− GCB (P = .027) and CD37− ABC (P = .0032) | ↓ in CD37+ ABC (P = .0005); trend of ↓ in CD37+ GCB (P = .086) | ||
| HAVCR2 (TIM3) | ↑ in both CD37− GCB (P = .0027) and CD37− ABC (P = .0001) | |||
| CIITA | ↓ in GCB (P = .014) | ↓ (P < .0001) | ||
| HLA-A-G | (↑ in overall CD37− ABC vs CD37+ ABC, but it is due to the association with p50 and CXCR4) | ↑ in both CD37− GCB (P = .0056) and CD37− ABC (P = .0003) | ↓ in both CD37+ GCB (P = .0073) and CD37+ ABC (P = .025) | |
| LILRB2 (LIR2) | ↑ in both GCB (P = .0014) and ABC (P < .0001) | ↑ in both CD37− GCB (P = .0001) and CD37− ABC (P = .0065) | ↓ in CD37+ GCB (P = .0017) | ↑ (P < .0001) |
| LILRA3 (LIR4) | ↑ in ABC (P = .0021) | ↑ in CD37− GCB (P = .001) | ↑ (P = .0028) | |
| LILRA1 (LIR6) | ↑ in GCB (P = .028) | ↑ in CD37− GCB (P = .0062) | ↑ (P = .0002) | |
| CASP1 | ↑ in ABC (P = .018) | ↑ in both CD37− GCB (P = .041) and CD37− ABC (P = .029) | ↓ in CD37+ ABC (P = .0056) | ↑ (P = .023) |
| CASP7 | ↑ in GCB (P = .02) | ↑ in CD37− ABC (P = .031) | ↑ (P < .0001) | |
| CASP8 | ↑ in GCB (P = .023) | ↑ (P < .0001) | ||
| CASP10 | ↑ in ABC (P = .018); trends of ↑ in GCB (P = .068) | ↓ in CD37+ GCB (P = .0097) | ||
| CXCL12 | ↑ in ABC (P = .0072) | ↑ in both CD37− GCB (P = .025) and CD37− ABC (P = .0007) | ↓ in CD37+ ABC (P = .0002) | |
| IL6R | ↑ in both CD37− GCB (P = .009) and CD37− ABC (P = .0001) | ↓ in CD37+ ABC (P = .049) | ||
| IL10RB | ↑ in ABC (P = .038) | ↑ in CD37− GCB (P = .0005) | ↑ in CD37+ ABC (P = .034) | |
| IL21R | ↓ in GCB (P = .0036) | |||
| IRF1 | ↑ in both GCB (P = .025) and ABC (P = .0005) | ↑ in CD37− ABC (P = .0004) | ↑ (P = .044) | |
| STAT1 | ↑ in ABC (P = .017) | ↑ in both CD37− GCB (P = .0068) and CD37− ABC (P = .0006) | ||
| IFIT3 | ↑ in both GCB (P = .0071) and ABC (P = .0011) | ↑ in CD37− GCB (P < .0001) | ↓ in CD37+ GCB (P = .047) | ↑ (P = .011) |
| CARD16 | ↑ in ABC (P = .0022) | ↓ in CD37+ ABC (P = .022) | ||
| TCL1A | ↓ in ABC (P = .0071) | Trend of ↓ in CD37− ABC (P = .076) | ↑ in CD37+ GCB (P = .036) | |
| AICDA | ↓ in ABC (P < .0001) | ↑ in both CD37+ GCB (P = .028) and CD37+ ABC (P < .0001) | ↑ (P < .0001) | |
| FOXP1 | ↓ in ABC (P < .0001) | ↑ in CD37+ GCB (P = .03) | ↑ (P < .0001) | |
| BCL11A | ↓ in both GCB (P = .0017) and ABC (P < .0001) | ↓ in both CD37− GCB (P = .0058) and CD37− ABC (P = .017) | ↑ in both CD37+ GCB (P = .04) and CD37+ ABC (P = .0006) | |
| LMO2 | ↓ in GCB (P = .0007) | ↓ (P < .0001) | ||
| POU2F2 (OTF2) | ↓ in ABC (P = .0001) | ↑ (P < .0001) |
| Genes involved in immune responses or differentiation . | CD37− vs CD37+ in GCB/ABC DLBCL . | p50high* vs p50low in CD37− GCB/ABC DLBCL . | CXCR4high* vs CXCR4low in CD37+ GCB/ABC DLBCL . | ABC vs GCB in CD37+ DLBCL . |
|---|---|---|---|---|
| ICOSLG | ↓ in GCB (P = .0036) | ↓ (P < .0001) | ||
| CD58 | ↓ in GCB (P = .022) | ↓ in CD37+ ABC (P = .056) | ↓ (P = .0009) | |
| FCGR2A (CD32A) | ↑ in both GCB (P = .016) and ABC (P = .044) | ↑ in both CD37− GCB (P = .0078) and CD37− ABC (P = .016) | ↓ in CD37+ ABC (P = .02) | ↑ (P = .0022) |
| FCGR1B (CD64) | ↑ in both GCB (P = .01) and ABC (P = .0007) | ↑ in both CD37− GCB (P = .0003) and CD37− ABC (P = .0094) | ↑ (P = .0005) | |
| C1QB | ↑ in both GCB (P = .0001) and ABC (P = .0027) | ↑ in both CD37− GCB (P < .0001) and CD37− ABC (P = .0007) | ↑ (P < .0001) | |
| CD8A | ↑ in ABC (P = .0014) | ↑ in both CD37− GCB (P = .017) and CD37− ABC (P = .039) | ||
| CD8B | ↑ in both GCB (P = .068) and in ABC (P = .0012) | ↑ in both CD37− GCB (P = .0059) and CD37− ABC (P = .038) | Trends of ↓ in CD37+ GCB (P = .08) and CD37+ ABC (P = .068) | |
| PDCD1 (PD-1) | ↑ in ABC (P = .03); ↑ in p50high GCB (P = .05) | ↑ in CD37− ABC (P = .03) | ↓ in CD37+ GCB (P = .014) and trend of ↓ in CD37+ ABC (P = .069) | |
| CD274 (PD-L1) | ↑ in CD37− GCB (P = .013) | ↑ in CD37+ ABC (P = .018) | ||
| PDCD1LG2 (PD-L2) | Trend of ↑ in ABC (P = .09) | ↑ in CD37− GCB (P = .0025) | ||
| CTLA4 | ↑ in both CD37− GCB (P = .027) and CD37− ABC (P = .0032) | ↓ in CD37+ ABC (P = .0005); trend of ↓ in CD37+ GCB (P = .086) | ||
| HAVCR2 (TIM3) | ↑ in both CD37− GCB (P = .0027) and CD37− ABC (P = .0001) | |||
| CIITA | ↓ in GCB (P = .014) | ↓ (P < .0001) | ||
| HLA-A-G | (↑ in overall CD37− ABC vs CD37+ ABC, but it is due to the association with p50 and CXCR4) | ↑ in both CD37− GCB (P = .0056) and CD37− ABC (P = .0003) | ↓ in both CD37+ GCB (P = .0073) and CD37+ ABC (P = .025) | |
| LILRB2 (LIR2) | ↑ in both GCB (P = .0014) and ABC (P < .0001) | ↑ in both CD37− GCB (P = .0001) and CD37− ABC (P = .0065) | ↓ in CD37+ GCB (P = .0017) | ↑ (P < .0001) |
| LILRA3 (LIR4) | ↑ in ABC (P = .0021) | ↑ in CD37− GCB (P = .001) | ↑ (P = .0028) | |
| LILRA1 (LIR6) | ↑ in GCB (P = .028) | ↑ in CD37− GCB (P = .0062) | ↑ (P = .0002) | |
| CASP1 | ↑ in ABC (P = .018) | ↑ in both CD37− GCB (P = .041) and CD37− ABC (P = .029) | ↓ in CD37+ ABC (P = .0056) | ↑ (P = .023) |
| CASP7 | ↑ in GCB (P = .02) | ↑ in CD37− ABC (P = .031) | ↑ (P < .0001) | |
| CASP8 | ↑ in GCB (P = .023) | ↑ (P < .0001) | ||
| CASP10 | ↑ in ABC (P = .018); trends of ↑ in GCB (P = .068) | ↓ in CD37+ GCB (P = .0097) | ||
| CXCL12 | ↑ in ABC (P = .0072) | ↑ in both CD37− GCB (P = .025) and CD37− ABC (P = .0007) | ↓ in CD37+ ABC (P = .0002) | |
| IL6R | ↑ in both CD37− GCB (P = .009) and CD37− ABC (P = .0001) | ↓ in CD37+ ABC (P = .049) | ||
| IL10RB | ↑ in ABC (P = .038) | ↑ in CD37− GCB (P = .0005) | ↑ in CD37+ ABC (P = .034) | |
| IL21R | ↓ in GCB (P = .0036) | |||
| IRF1 | ↑ in both GCB (P = .025) and ABC (P = .0005) | ↑ in CD37− ABC (P = .0004) | ↑ (P = .044) | |
| STAT1 | ↑ in ABC (P = .017) | ↑ in both CD37− GCB (P = .0068) and CD37− ABC (P = .0006) | ||
| IFIT3 | ↑ in both GCB (P = .0071) and ABC (P = .0011) | ↑ in CD37− GCB (P < .0001) | ↓ in CD37+ GCB (P = .047) | ↑ (P = .011) |
| CARD16 | ↑ in ABC (P = .0022) | ↓ in CD37+ ABC (P = .022) | ||
| TCL1A | ↓ in ABC (P = .0071) | Trend of ↓ in CD37− ABC (P = .076) | ↑ in CD37+ GCB (P = .036) | |
| AICDA | ↓ in ABC (P < .0001) | ↑ in both CD37+ GCB (P = .028) and CD37+ ABC (P < .0001) | ↑ (P < .0001) | |
| FOXP1 | ↓ in ABC (P < .0001) | ↑ in CD37+ GCB (P = .03) | ↑ (P < .0001) | |
| BCL11A | ↓ in both GCB (P = .0017) and ABC (P < .0001) | ↓ in both CD37− GCB (P = .0058) and CD37− ABC (P = .017) | ↑ in both CD37+ GCB (P = .04) and CD37+ ABC (P = .0006) | |
| LMO2 | ↓ in GCB (P = .0007) | ↓ (P < .0001) | ||
| POU2F2 (OTF2) | ↓ in ABC (P = .0001) | ↑ (P < .0001) |
Differential expression of genes involved in immune responses or differentiation between CD37− and CD37+ patients, between p50high/CD37− and p50low/CD37− patients (CD37− was associated with p50high), between CXCR4high/CD37+ and CXCR4low/CD37+ patients (CD37+ was associated with CXCR4high in ABC-DLBCL), and between CD37+ ABC-DLBCL and CD37+ GCB-DLBCL (CD37+ was associated with ABC cell of origin). The cutoffs for p50high and CXCR4high were ≥20%. The P values were obtained by the unpaired Student t test (2-tailed).
Comparison of ICOSLG, PDCD1, and PD-1 expression in CD37+and CD37−DLBCL. (A) CD37 positivity was associated with significantly higher levels of ICOSLG in GCB-DLBCL. (B) CD37 loss correlated with PDCD1 (PD-1) upregulation in ABC-DLBCL. (C) In p50high (≥20% nuclear expression) DLBCL, CD37 loss correlated with PD-1 upregulation in both GCB- and ABC-DLBCL. (D-E) CD37 loss in DLBCL correlated with significantly higher levels of PD-1 receptor expression on both cytotoxic and helper T cells. (F) The ABC subtype had significantly higher CD274 (PD-L1) levels compared with the GCB subtype, which was more apparent in the CD37− DLBCL subset.
Comparison of ICOSLG, PDCD1, and PD-1 expression in CD37+and CD37−DLBCL. (A) CD37 positivity was associated with significantly higher levels of ICOSLG in GCB-DLBCL. (B) CD37 loss correlated with PDCD1 (PD-1) upregulation in ABC-DLBCL. (C) In p50high (≥20% nuclear expression) DLBCL, CD37 loss correlated with PD-1 upregulation in both GCB- and ABC-DLBCL. (D-E) CD37 loss in DLBCL correlated with significantly higher levels of PD-1 receptor expression on both cytotoxic and helper T cells. (F) The ABC subtype had significantly higher CD274 (PD-L1) levels compared with the GCB subtype, which was more apparent in the CD37− DLBCL subset.
ABC vs GCB cell of origin correlated with ICOSLG downregulation (P < .0001) and IL10RA/IL1041 and LILRB/A upregulation in both CD37− and CD37+ subsets, and with CD274 and PDCD1LG2 (encoding the PD-1 ligands PD-L1 and PD-L2,42 respectively) upregulation in the CD37− subset (also in overall DLBCL) (Figure 6F). Immune dysregulation was also found in subsets with TP53 mutations, Mychigh, p50high, p-STAT3high, FOXP1high, MUM1/IRF4high, or CXCR4high (eg, upregulation of PD-L1/L2, CTLA4, TIM3, LILRB2, IL6R, and IL10RB, whereas downregulation of ICOSLG, CD58, and MHC-I/II) (Figure 7; supplemental Figure 6).
A hypothetic model illustrating the pivotal role of CD37 status for R-CHOP outcome in DLBCL and the important molecular mechanisms for R-CHOP resistance in CD37−DLBCL and CD37+ABC-DLBCL. (A) Antiapoptotic and immune escape mechanisms existed in both CD37+ and CD37− DLBCL before R-CHOP treatment. Comparably, CD37+ DLBCL had higher frequencies of ABC cell of origin and CXCR4 overexpression, whereas CD37− DLBCL had higher frequencies of TP53 mutations and nuclear p50, STAT3, and Myc (only in GCB-DLBCL) overexpression. (B) CD37 positivity independently predicted favorable outcome, likely because CD37+ DLBCL is sensitive to R-CHOP owing to the increased CD20 and ICOSLG whereas decreased PD-1 expression (depicted by green 1, 3, and 2, respectively), as well as CD20-independent CD37 function in enhancing antibody-dependent cellular cytotoxicity (ADCC) and apoptosis upon CD20-rituximab ligation (*). This favorable impact can be hindered by ICOSLG and MHC-II downregulation, upregulation of PD-L1, AICDA, LILRA/B, IL10/IL10RA, and antiapoptotic pathways downstream of the chronic active BCR signaling in ABC-DLBCL. (C) CD37 loss robustly predicted poor survival. Rituximab efficacy is limited due to decreased CD20 levels (depicted by yellow 1, with postulated reasons of attenuated BCR, cytokine, and trogocytosis) and loss of CD37-rituximab signaling. The significantly worse prognosis is also contributed by 1 increased PD-1 (highlighted by yellow 2), ICOSLG downregulation (highlighted by yellow 3), and frequent TP53 mutations, Myc, STAT3, or p50 overexpression in CD37− DLBCL (which were probably oncogenic drivers acquired during lymphomagenesis and further escaped from immune surveillance by various mechanisms as depicted). Illustrated immune escape mechanisms include upregulation of PD-L1/L2, LILRB/A, TIM3, CTLA4, and the IL6/IL10 pathway, and downregulation of MHC-I/II, CIITA, and costimulatory molecules CD58 and CD40. The model is based on our biomarker correlation, GEP, and survival analysis, except the speculated CD37 functions during R-CHOP treatment as denoted by an asterisk (*).
A hypothetic model illustrating the pivotal role of CD37 status for R-CHOP outcome in DLBCL and the important molecular mechanisms for R-CHOP resistance in CD37−DLBCL and CD37+ABC-DLBCL. (A) Antiapoptotic and immune escape mechanisms existed in both CD37+ and CD37− DLBCL before R-CHOP treatment. Comparably, CD37+ DLBCL had higher frequencies of ABC cell of origin and CXCR4 overexpression, whereas CD37− DLBCL had higher frequencies of TP53 mutations and nuclear p50, STAT3, and Myc (only in GCB-DLBCL) overexpression. (B) CD37 positivity independently predicted favorable outcome, likely because CD37+ DLBCL is sensitive to R-CHOP owing to the increased CD20 and ICOSLG whereas decreased PD-1 expression (depicted by green 1, 3, and 2, respectively), as well as CD20-independent CD37 function in enhancing antibody-dependent cellular cytotoxicity (ADCC) and apoptosis upon CD20-rituximab ligation (*). This favorable impact can be hindered by ICOSLG and MHC-II downregulation, upregulation of PD-L1, AICDA, LILRA/B, IL10/IL10RA, and antiapoptotic pathways downstream of the chronic active BCR signaling in ABC-DLBCL. (C) CD37 loss robustly predicted poor survival. Rituximab efficacy is limited due to decreased CD20 levels (depicted by yellow 1, with postulated reasons of attenuated BCR, cytokine, and trogocytosis) and loss of CD37-rituximab signaling. The significantly worse prognosis is also contributed by 1 increased PD-1 (highlighted by yellow 2), ICOSLG downregulation (highlighted by yellow 3), and frequent TP53 mutations, Myc, STAT3, or p50 overexpression in CD37− DLBCL (which were probably oncogenic drivers acquired during lymphomagenesis and further escaped from immune surveillance by various mechanisms as depicted). Illustrated immune escape mechanisms include upregulation of PD-L1/L2, LILRB/A, TIM3, CTLA4, and the IL6/IL10 pathway, and downregulation of MHC-I/II, CIITA, and costimulatory molecules CD58 and CD40. The model is based on our biomarker correlation, GEP, and survival analysis, except the speculated CD37 functions during R-CHOP treatment as denoted by an asterisk (*).
Discussion
In this study, we demonstrated the robust prognostic value of CD37 status in a large cohort of DLBCL. Integrating assessment of CD37 status and GCB/ABC cell of origin into the IPI calculation (M-IPI-R) markedly improved the predicative power of IPI in R-CHOP–treated DLBCL patients. However, currently GCB/ABC classification is not yet the standard of care, although it may be in the future.43 Respecting this, IHC for Myc and Bcl-2 may be an alternative for GCB/ABC determination in the M-IPI-R.
The pivotal role of CD37+/− status for prognosis has 2 aspects. First, CD37 positivity is an independent predictor of favorable outcome in R-CHOP–treated DLBCL patients. CD37 ligation experiments have shown that CD37 can function as a death receptor in B cells.10 Upon cross-ligation, CD37 becomes associated with SHP1, LYN, and SYK recruiting PI3Kγ, and transduces signaling favoring cellular death.10 Considering the somehow R-CHOP–specific but CD20-independent prognostic significance of CD37 expression (Figure 2), and the lack of a prominent CD37+ GEP signature at diagnosis, we speculated that CD37 might act as a “molecular facilitator” of rituximab action during R-CHOP treatment, especially for antibody-dependent cellular cytotoxicity, cross-linking, aggregation in lipid rafts thereby transactivating tyrosine kinases, apoptosis induction, and long-term T-cell responses (Figure 7B).44-46 Remarkably, such potential CD37-rituximab signaling could abolish the adverse impact of many prognostic factors and even that of high IPI scores in GCB-DLBCL, but not in ABC-DLBCL, however, even though there was no significant difference in CD20 mRNA levels between GCB-DLBCL and ABC-DLBCL (supplemental Figure 2B). The enhanced antitumor effect by CD37+ might have been dampened in ABC-DLBCL by ICOSLG downregulation, upregulation of TNFRSF13B/TACI, PD-L1, AICDA, IL10/IL10RA, and LILRA/B, increased FOXP1 and IRF4 levels which suppress MHC-II expression, chronic active BCR signaling, and apoptosis-suppressive mechanisms (eg, DRAM1, EPB41L3, and BCL11B were downregulated in CD37+/PI3Khigh ABC-DLBCL).47,48 Of note, TNFRSF13B/TACI suppresses ICOSLG and BIM (proapoptotic) expression.49
Second, CD37 loss independently predicted significantly worse survival in DLBCL. Increased PD-1, but decreased ICOSLG and CD20 expression,50-56 may contribute to the poor clinical outcome of CD37− patients (Figure 7C). CD20 downregulation could be due to cell of origin57,58 of CD37− DLBCL cells or increased cytokines which was reported to suppress CD20 expression.59 In addition, many FCGR genes were upregulated in CD37− DLBCL especially in p50high/CD37− DLBCL, suggesting the potential for trogocytosis (the transfer of rituximab-CD20 complexes from tumor cells to Fc receptor–bearing cells53 ) and R-CHOP resistance. Previous studies have demonstrated that CD37 has a role in T-cell–B-cell interaction and humoral responses, which can be observed under suboptimal costimulatory conditions,11 and that Cd37−/− mice develop germinal center–derived lymphomas.16 In this study, CD37− DLBCL was enriched with gene sets related to infection and immune signaling and presence of oncogenic drivers (eg, TP53 mutations and MYC rearrangements), which might result from loss of CD37’s antitumor function during lymphomagenesis (potentially also after R-CHOP treatment).
These results also have therapeutic implications. For CD37+ ABC-DLBCL with higher CD37 (but not CD20) expression, anti-CD37 antibody alone or combined with rituximab may have higher efficacy. Clinical outcomes may be further improved by combining with Bruton tyrosine kinase inhibitors that inhibit BCR and CXCR4 signaling,60 PD-L1 inhibitors, ICOS agonists, and immunoregulatory lenalidomide.28,40,61 For CD37− DLBCL with immunosuppressive mechanisms and decreased CD20 levels, PD-1 blockade and ICOS agonists may be effective, as well as other targeted agents in p50high, Mychigh, p-STAT3high, or ABC subgroups of CD37− DLBCL (Figure 7).62,63 Notably, Hodgkin and Reed-Sternberg cells which are thought to originate from proapoptotic germinal center B cells but are rescued by acquired survival mechanisms, also show loss of CD20, CD37, and BCR and demonstrate particular sensitivity to immune-checkpoint blockade.64-66
In summary, in this study, we established CD37−/+ status as a robust biomarker and introduced 2 novel prognostic indices, M-IPI-R and IPI+IHC, for risk prediction in R-CHOP–treated DLBCL. Whether these indices can be useful prognostic tools in the clinic needs to be validated by future prospective studies. GEP analysis indicates that novel strategies are needed, especially immunotherapies for CD37− DLBCL and anti-CD37 antibodies for CD37+ ABC-DLBCL. Our findings from human samples are also valuable for understanding DLBCL pathogenesis and heterogeneity and have important clinical implications.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health National Cancer Institute grants (R01CA138688 and R01CA187415) (Y.L., K.H.Y.). A.B.v.S. was supported by The Netherlands Organization for Scientific Research (NWO-ALW VIDI grant 864.11.006) and the Dutch Cancer Society (KUN2014-6845). K.H.Y. was also supported by The University of Texas MD Anderson Cancer Center Lymphoma Moonshot Program, an Institutional Research Grant Award, an MD Anderson Lymphoma Specialized Programs of Research Excellence (SPORE) Research Development Program Award, and an MD Anderson Myeloma SPORE Research Developmental Program Award. The work was also partially supported by National Institutes of Health, National Cancer Institute grants (P50CA136411 and P50CA142509) and MD Anderson Cancer Center Support grant CA016672.
Authorship
Contribution: Z.Y.X.-M. and K.H.Y. conceived and designed the study; Z.Y.X.-M., K.J.J., C.M.d.W., M.v.d.B., A.B.v.S., and K.H.Y. performed research; Z.Y.X.-M., L.L., J.C.B., A.T., G.C.M., C.V., J.W., K.D., A.C., A.O., Y.Z., G.B., K.L.R., E.D.H., W.W.L.C., J.H., M.P., A.J.M.F., B.M.P., M.B.M., J.N.W., M.W., F.B.H., M.A.P., J.H.v.K., L.J.M., Y.L., A.B.v.S., and K.H.Y. provided study thought, materials, key reagents, and technology; Z.Y.X.-M., A.T., C.V., K.D., A.C., A.O., Y.Z., G.B., K.L.R., E.D.H., W.W.L.C., J.H.v.K., J.H., M.P., A.J.M.F., B.M.P., M.B.M., J.N.W., M.A.P., and K.H.Y. collected and assembled data under an approved institutional review board and material transfer agreement; Z.Y.X.-M., L.L., J.C.B., Y.L., A.B.v.S., and K.H.Y. analyzed and interpreted data; Z.Y.X.-M. and K.H.Y. wrote the manuscript; and all authors gave final approval of the manuscript.
Conflict-of-interest disclosure: K.H.Y. receives research support from Roche Molecular System, Gilead Sciences Pharmaceutical, Seattle Genetics, Dai Sanyo Pharmaceutical, Adaptive Biotechnology, Incyte Pharmaceutical, and HTG Molecular Diagnostics. E.D.H. is a consultant for HTG Molecular Diagnostics. B.M.P. joins speakers bureaus for Celgene and Amgen, and is a consultant for Celgene. The remaining authors declare no competing financial interests.
Correspondence: Ken H. Young, Department of Hematopathology, The University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd, Houston, TX 77030-4009; e-mail: khyoung@mdanderson.org.
References
Author notes
Z.Y.X.-M. and L.L. contributed equally to this study.



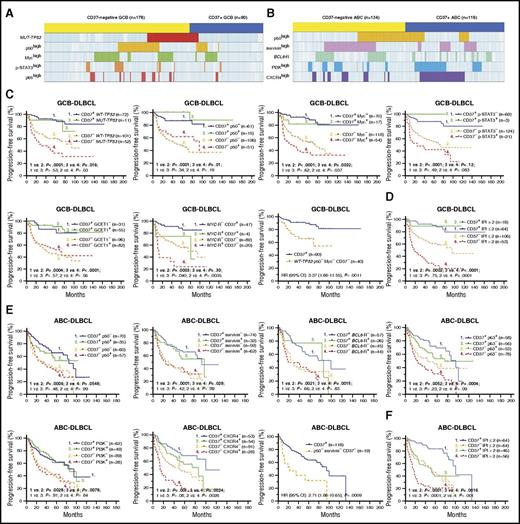
![Figure 4. Comparison of risk stratification in patients with DLBCL by the traditional IPI and adjusted IPI scores. (A) Risk stratification of DLBCL groups by the traditional IPI. (B) Risk stratification of DLBCL by the M-IPI-R, defined by each patient’s IPI score plus risk scores for CD37 status (CD37+, 0; CD37−, 1 or 2 as indicated) and GCB/ABC cell of origin (GCB, 0; ABC, 1). (C) Risk stratification of DLBCL by the IPI+IHC, defined by each patient’s IPI score plus risk scores for CD37 status (CD37+, 0; CD37−, 3), and Myc and Bcl-2 protein levels (low [IHC <70%], 0; high [IHC ≥70%], 1).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/128/26/10.1182_blood-2016-05-715094/5/m_blood715094f4.jpeg?Expires=1767717258&Signature=gEMZDQJehPNIDOCNbt0t34XQ~AcmWNN3gQkIlQ4K2l2c1~NoDvobGPCv4Aa-oCntVt06nXPBNuYZDHo0Gcq-CTVoZ~QA2jtRBncLTR18NjFG4xAvXCzUeDWLCh6x1LydjRavZn~Eac2Wsr52FbcY9M0lE39OAqVOWYtJ3anH6~WmeYUhuatCsrB3pVCvV--VzSZesxIM~o9FrgmE0I42GnZEdk-B9JxVuXS4N4JYOTca2lnAsm2GffZTiWfMCfv25NfO-yz4Vg55jaR75swjeHv-mXl~hmByhcucHktDwTlxvfuy4tI6paUfru9fMUtKIjbeTOwarDSE5QNlVEh44A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)