Abstract
Chronic myeloid leukemia (CML) is caused by the acquisition of the tyrosine kinase BCR-ABL1 in a hemopoietic stem cell, transforming it into a leukemic stem cell (LSC) that self-renews, proliferates, and differentiates to give rise to a myeloproliferative disease. Although tyrosine kinase inhibitors (TKIs) that target the kinase activity of BCR-ABL1 have transformed CML from a once-fatal disease to a manageable one for the vast majority of patients, only ∼10% of those who present in chronic phase (CP) can discontinue TKI treatment and maintain a therapy-free remission. Strong evidence now shows that CML LSCs are resistant to the effects of TKIs and persist in all patients on long-term therapy, where they may promote acquired TKI resistance, drive relapse or disease progression, and inevitably represent a bottleneck to cure. Since their discovery in patients almost 2 decades ago, CML LSCs have become a well-recognized exemplar of the cancer stem cell and have been characterized extensively, with the aim of developing new curative therapeutic approaches based on LSC eradication. This review summarizes our current understanding of many of the pathways and mechanisms that promote the survival of the CP CML LSCs and how they can be a source of new gene coding mutations that impact in the clinic. We also review recent preclinical approaches that show promise to eradicate the LSC, and future challenges on the path to cure.
CML: the classic stem cell disease
Chronic myeloid leukemia (CML) is a classic example of a stem cell cancer and arises when the t9;22 translocation (the Philadelphia chromosome [Ph+])1-3 occurs in a hemopoietic stem cell (HSC). This event results in the constitutive expression of the fusion tyrosine kinase BCR-ABL1, transforming the HSC into the CML stem cell (referred to here as the leukemic stem cell or LSC), which then gives rise to a clonal myeloproliferative disease. Early evidence regarding the HSC origins of CML came from observations that transfusion of peripheral blood cells from CML patients into severely neutropenic recipients resulted in temporary homologous bone marrow (BM) engraftment and Ph+ progeny in the blood.4 This was later explained by the presence of high numbers of mobilized LSCs in the peripheral blood of chronic phase (CP) CML patients.5 A possible hemopoietic progenitor origin of CML was ruled out when BCR-ABL1 expression in murine hemopoietic progenitors failed to confer self-renewal capabilities to BCR-ABL1+ cells, and these cells failed to induce leukemia in mice.6 Until very recently, BCR-ABL1 expression was considered sufficient to cause a CML-like disease in mouse models using retrovirus transduction or transgene insertional mutagenesis to express the oncogene in LSC.7-9 However, issues with BCR-ABL1 copy number, high oncogene expression, and/or secondary mutations arising by retroviral or transgene insertional mutagenesis or genomic instability could theoretically contribute to leukemogenesis. In a recent knock-in model, a single copy of BCR-ABL1 expressed from the endogenous BCR locus was able to confer enhanced BM engraftment; however, this model was unable to induce leukemia.10
Although the cell of origin of CML is generally accepted to be the HSC, several studies implicate an HSC precursor cell—the multipotent hemangioblast that gives rise to both hemopoietic and endothelial cells. The BCR-ABL1 fusion can be detected in endothelial cells obtained from BM and peripheral blood of CML patients at varying frequencies.11,12 These cells show altered intracellular signaling and protein expression that may affect crosstalk between LSC and the BM microenvironment (BMM), alter immune-modulation and LSC exit from quiescence into proliferation.13,14 Collectively, these data suggest that the acquisition of BCR-ABL1 in the hemangioblast may contribute to both malignant hemopoiesis and endotheliopoiesis.
The natural history of CML
CML is a rare stem cell disease with an annual incidence of 1 to 2 cases per 100 000 individuals, peaking in the sixth and seventh decades of life.15 Data derived from atomic bomb survivors16 suggest that following a latent period of ∼7 years, the natural history of CML is for 85% to 90% of cases to present in CP, but to progress to accelerated phase and then to either myeloid or lymphoid blast crisis over a 5-year time frame.17 However, the mechanism of disease progression is complex and disease behavior is highly variable for individual patients, with some progressing within a few months and others remaining in stable CP for up to 20 years. This heterogeneity between patients may relate to the mutations subsequently acquired in the BCR-ABL1 clone,18 variations in gene expression patterns between patients,19 or the subtype of HSC in which BCR-ABL1 is first expressed—with recent evidence delineating multiple HSC subsets defined by variably fixed lineage potentialities, transcriptional profiles, and phenotypes.20-23 Furthermore, intriguing work on preleukemia24,25 and the detection of BCR-ABL1 in blood cells of normal individuals26,27 present the possibility that heterogeneity could also be driven by mutations acquired before or after BCR-ABL1, or other factors such as deregulation and skewing of lineage specification, clonal hemopoiesis, DNA damage, activation of inflammatory responses, and epigenetic alterations, all of which occur in hemopoiesis during aging.28,29 Some, or all, of these factors may also be required for, or contribute to, disease development in mouse models of CML.
LSC persistence: a bottleneck to cure
The introduction of a potent BCR-ABL1 tyrosine kinase inhibitor (TKI), imatinib, almost 2 decades ago, followed by subsequent generations of TKI (dasatinib, nilotinib, bosutinib, ponatinib) has transformed the management of CML.30,31 What was once a universally fatal disorder, unless treated with an allogeneic transplant, is now well-controlled in the outpatient setting, and overall survival has improved significantly (http://seer.cancer.gov/statfacts/html/cmyl.html), with the majority of patients requiring life-long TKI. In keeping with disease heterogeneity, patient responses to TKI are also variable. The majority of cases (50% to 70%) achieve major molecular response (MMR) in which BCR-ABL1 levels detectable by quantitative polymerase chain reaction in the blood show a 3 log10 fold reduction (ie, 0.1%, compared with a standardized baseline, reviewed elsewhere32 ). However, patient-to-patient variation in leukemic cell blood counts at diagnosis and variations in BCR-ABL1 expression between early and late stages of cell differentiation can often confound these interpretations. Approximately 10% to 20% of all patients develop even deeper molecular responses triggering dose deescalation and discontinuation/stopping trials (Stop Imatinib [STIM], Phase II Study of Withdrawal of Imatinib Therapy in Adult Patients With Chronic Phase Chronic Myeloid Leukaemia in Stable Molecular Remission [TWISTER], Dasatinib Discontinuation [DADI]), in which 50% of patients relapse within 12 months.33,34 When CP relapse occurs, the doubling time (∼9 days) for increasing disease burden mirrors the CML disease at diagnosis.35 One-quarter of CP patients fail TKI therapy,36 and approximately one-half of these cases can be explained by BCR-ABL1 kinase domain mutations,32,37 but the reason for failure in the remaining patients is unclear.
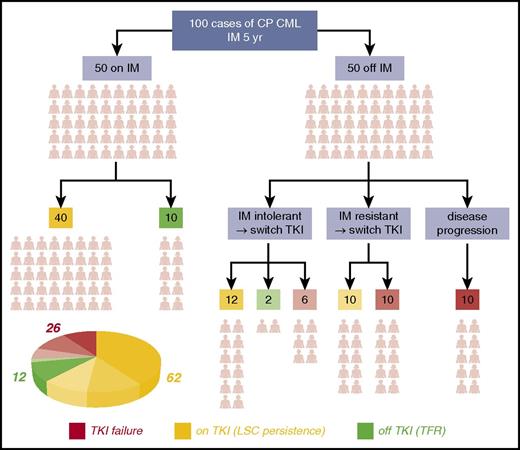
Ironically, the earliest evidence of CML LSC38 predated the introduction of TKI; this was followed by definitive evidence of a deeply, but reversibly, quiescent subpopulation of leukemic cells in patients with CML.39 In the subsequent years, the consensus view has emerged that virtually all CP patients on TKI therapy and in MMR are not cured of CML and show signs of residual disease burden from the presence of LSC in the BM (termed “LSC persistence”). In a typical cohort of 100 CP CML patients who undertake TKI therapy over a 5-year period, almost two-thirds will have this “LSC persistence” phenotype (Figure 1). Researchers have consistently detected BCR-ABL1+ primitive cells in the BM of TKI-treated patients in MMR, which are capable of growth in colony-forming cell and long-term culture initiation cell assays, even in patients in deep molecular response with no detectable BCR-ABL1 transcripts by quantitative polymerase chain reaction.40-43 The most recent of these studies has shown that, although LSCs are not always detectable in cases of very deep molecular response, most likely from technical limitations, some patients with no detectable LSCs can subsequently relapse after TKI discontinuation.43 Others have shown that the LSCs that persist in patients in MMR express BCR-ABL1 at lower levels than the LSCs at the point of diagnosis. Furthermore, murine BM cells engineered to express low levels of BCR-ABL1 levels were far less sensitive to imatinib, whereas those expressing higher levels were prone to de novo mutations.44 These findings point to LSC persistence as a “low mutator” phenotype, perhaps explaining why the majority of these patients do not develop drug resistance or progress to BC. The eradication of the LSC remains a challenge in the majority of CML patients, a significant bottleneck to cure, and an area of intensive research.
The CP CML patients’ journey through TKI therapy for 5 years. The schematic shows the clinical outcome for a typical 100 CP CML patients with respect to response to imatinib (IM) over the course of 5 years and the decision-tree leading to discontinuing TKI or switching to second- or third-generation TKIs for various reasons. Outcomes were compiled based on data obtained from various sources.31,143-146 By the end of year 5: 12 (green segment of the pie chart) of these 100 patients will typically be off TKI and in therapy-free remission (TFR), more than one-quarter (26) (red segment of the pie chart) will have failed TKI therapy (even after drug switching or through disease progression to accelerated phase or blast crisis), and the majority (62) (amber segment of the pie chart) will remain on long-term TKI therapy but have residual disease resulting from LSC persistence in their BM.
The CP CML patients’ journey through TKI therapy for 5 years. The schematic shows the clinical outcome for a typical 100 CP CML patients with respect to response to imatinib (IM) over the course of 5 years and the decision-tree leading to discontinuing TKI or switching to second- or third-generation TKIs for various reasons. Outcomes were compiled based on data obtained from various sources.31,143-146 By the end of year 5: 12 (green segment of the pie chart) of these 100 patients will typically be off TKI and in therapy-free remission (TFR), more than one-quarter (26) (red segment of the pie chart) will have failed TKI therapy (even after drug switching or through disease progression to accelerated phase or blast crisis), and the majority (62) (amber segment of the pie chart) will remain on long-term TKI therapy but have residual disease resulting from LSC persistence in their BM.
General features of the LSC
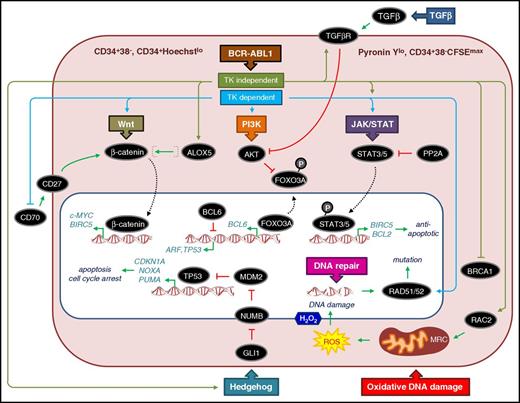
At the time of CP diagnosis, BCR-ABL1− cells coexist with BCR-ABL1+ cells and enriched CD34+ populations require dual-fluorescent in situ hybridization to determine the proportion of cells that carry Ph+ (usually >90% BCR-ABL1+). The more primitive LSC fraction can be purified by fluorescence-activated cell sorting (FACS) in a variety of ways, giving rise to overlapping, primitive, quiescent populations (Figure 2). Phenotypically and functionally, we define CP CML LSCs as those primitive stem/progenitor cells that show a higher capacity to engraft in immunocompromised mice than bulk CD34+ cells,45 have stem cell properties (self-renewal), are resistant to apoptosis,46,47 are prone to genomic instability,48,49 and have impaired DNA damage responses.50-52 Because BCR-ABL1 drives survival and proliferation, it is somewhat of a paradox that CML LSCs express BCR-ABL1 but can also be quiescent39 —a feature that may enable them to become refractory to TKI-induced apoptosis. However, TKIs also exert a potent antiproliferative effect on CML CD34+ cells and LSCs to induce quiescence46,53 ; and subsequent evidence has shown that TKIs exert additional effects to subvert a number of pathways to promote survival (see the following section).
General features and critical pathways that contribute to CP CML LSCs being quiescent, refractory to apoptosis, and prone to DNA damage. Typically LSCs represent 1% to 5% of the bulk CML CD34+ cells, are enriched by FACS as CD34+CD38−, and show more variable levels of Ph+ cells than bulk CML CD34+ cells. Some researchers also include Lin−/CD90+/CD45RA− cells as part of the CD34+CD38− LSC definition.112 Other FACS approaches can also be used to isolate LSCs by using Hoechst, Pyronin Y, and carboxyfluorescein succinimidyl ester (CFSE) intracellular staining in combination with CD34 to identify quiescent/undivided cells.39,46,47,147 CD34+CD38− CML cells from patients at diagnosis that retain high levels of CFSE (CFSEmax) or are CD34+ and both Hoechstlo and Pyronin Ylo, and survive exposure to TKI, are often considered surrogate in vitro models for the TKI-resistant cells found in patients with LSC persistence. The schematic diagram of the LSC shows key (but not exhaustive) pathways and components and whether the published evidence points to TKI-dependent (blue) or independent (olive green) mechanisms of regulation. Dotted lines denote translocation of components from the cytoplasm (light red) to the nucleus (white). TK, tyrosine kinase. Activation and repression are denoted according to convention. Specific details of each pathway are described in the text.
General features and critical pathways that contribute to CP CML LSCs being quiescent, refractory to apoptosis, and prone to DNA damage. Typically LSCs represent 1% to 5% of the bulk CML CD34+ cells, are enriched by FACS as CD34+CD38−, and show more variable levels of Ph+ cells than bulk CML CD34+ cells. Some researchers also include Lin−/CD90+/CD45RA− cells as part of the CD34+CD38− LSC definition.112 Other FACS approaches can also be used to isolate LSCs by using Hoechst, Pyronin Y, and carboxyfluorescein succinimidyl ester (CFSE) intracellular staining in combination with CD34 to identify quiescent/undivided cells.39,46,47,147 CD34+CD38− CML cells from patients at diagnosis that retain high levels of CFSE (CFSEmax) or are CD34+ and both Hoechstlo and Pyronin Ylo, and survive exposure to TKI, are often considered surrogate in vitro models for the TKI-resistant cells found in patients with LSC persistence. The schematic diagram of the LSC shows key (but not exhaustive) pathways and components and whether the published evidence points to TKI-dependent (blue) or independent (olive green) mechanisms of regulation. Dotted lines denote translocation of components from the cytoplasm (light red) to the nucleus (white). TK, tyrosine kinase. Activation and repression are denoted according to convention. Specific details of each pathway are described in the text.
BCR-ABL1 kinase-independent survival
To understand why LSCs were refractory to the effects of TKI, we exposed CML CD34+ cells to high concentrations of dasatinib for 12 days, and subjected them, in parallel, to BCR-ABL1 knockdown. These in vitro studies were complemented in vivo using the inducible transgenic SCL-tTA/BCR-ABL model.9 BCR-ABL1 expression was induced in mice to lead to the development of CML-like disease, then switched off to determine whether the LSC population required BCR-ABL1 for survival, and then induced for a second time to see whether the LSCs were still functional and could again drive the development of CML-like disease. In the in vitro studies, functional BCR-ABL1+ LSCs persisted in culture despite evidence for complete kinase inhibition and significant BCR-ABL1 knockdown. In the mouse model, CML-like disease reoccurred following the second induction of BCR-ABL1. This work demonstrated that LSC survival is not dependent on BCR-ABL1 kinase activity54 and suggested that BCR-ABL1 may have nonkinase-mediated functions that modulate signaling pathways to promote LSC survival. These conclusions were further supported by others who used imatinib to fully inhibit BCR-ABL1 kinase activity in both LSCs and quiescent cells.55 Taken together, these studies concluded that CML LSCs were not “oncogene-addicted” and that targeting of BCR-ABL1 kinase activity alone would not eliminate them. Furthermore, this work has led investigators worldwide to search for LSC-selective, BCR-ABL1 kinase-independent targets and pathways that might offer potential for improved targeting of LSCs in CML. To date, several mechanisms, pathways, and drug-able targets have been proposed to contribute to the TKI-resistant LSC phenotype (Figures 2 and 3; Tables 1 and 2).
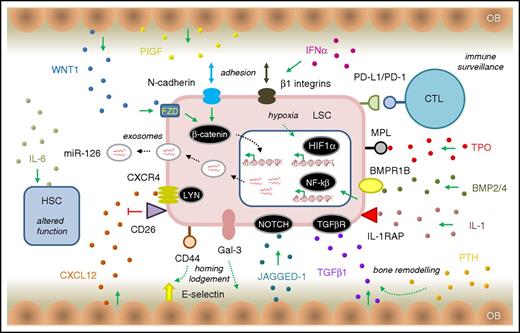
LSC survival signaling in the CP CML BMM. The schematic diagram of the BMM shows key (but not exhaustive) pathway components that mediate signaling between the LSC (light red) and other BMM cell types. HSC is shown in blue. OB, osteoblast cells (tan). Ligands involved in various signaling pathways are shown as small, colored spheres. IL-1/IL-1RAP regulates NFKβ signaling in LSCs and can be blocked using a monoclonal antibody to IL-1RAP.135 MPL, the thrombopoietin (TPO) receptor, regulates JAK/STAT signaling and CML patients with high MPL expression on their LSCs have reduced sensitivity to BCR-ABL1 kinase inhibition with TKI, but a higher sensitivity to JAK inhibitors.139 Leukemic progenitor expansion is driven by exposure of LSC, overexpressing BMPR1B, to BMP2 and BMP4.127 The CML BMM is also thought to overexpress the NOTCH ligand JAGGED-1, implicating NOTCH signaling in LSC quiescence.138 LSCs stimulate the production of placental growth factor (PIGF) by BM stromal cells that work in a positive feedback loop to increase angiogenesis of the BM and promote CML cell proliferation through FLT1 (VEGFR1) signaling.140 Stimulation of BM osteoblasts with parathyroid hormone (PTH) resulted in bone remodelling and production of TGF-β1, eradicated LSCs by stimulating TGF-β signaling142 (the opposite effect to other reports of TGF-β signaling in LSCs58,122 ). Similarly, others have shown that expansion of the osteoblast layer of the CML BMM can contribute to creating a hostile environment for HSCs; these effects are mediated by TPO, CCL3, and direct cell–cell interactions that alter TGF-β, NOTCH, and pro-inflammatory signaling in the remodelled osteoblasts.148 Other abbreviations are as described in the text. Other features are as described in Figures 2 and 3.
LSC survival signaling in the CP CML BMM. The schematic diagram of the BMM shows key (but not exhaustive) pathway components that mediate signaling between the LSC (light red) and other BMM cell types. HSC is shown in blue. OB, osteoblast cells (tan). Ligands involved in various signaling pathways are shown as small, colored spheres. IL-1/IL-1RAP regulates NFKβ signaling in LSCs and can be blocked using a monoclonal antibody to IL-1RAP.135 MPL, the thrombopoietin (TPO) receptor, regulates JAK/STAT signaling and CML patients with high MPL expression on their LSCs have reduced sensitivity to BCR-ABL1 kinase inhibition with TKI, but a higher sensitivity to JAK inhibitors.139 Leukemic progenitor expansion is driven by exposure of LSC, overexpressing BMPR1B, to BMP2 and BMP4.127 The CML BMM is also thought to overexpress the NOTCH ligand JAGGED-1, implicating NOTCH signaling in LSC quiescence.138 LSCs stimulate the production of placental growth factor (PIGF) by BM stromal cells that work in a positive feedback loop to increase angiogenesis of the BM and promote CML cell proliferation through FLT1 (VEGFR1) signaling.140 Stimulation of BM osteoblasts with parathyroid hormone (PTH) resulted in bone remodelling and production of TGF-β1, eradicated LSCs by stimulating TGF-β signaling142 (the opposite effect to other reports of TGF-β signaling in LSCs58,122 ). Similarly, others have shown that expansion of the osteoblast layer of the CML BMM can contribute to creating a hostile environment for HSCs; these effects are mediated by TPO, CCL3, and direct cell–cell interactions that alter TGF-β, NOTCH, and pro-inflammatory signaling in the remodelled osteoblasts.148 Other abbreviations are as described in the text. Other features are as described in Figures 2 and 3.
Candidate therapeutic targets in CP CML LSCs
| Target or survival factor . | Pathway . | Exemplar inhibitors/activators . | Reference . | CML clinical trial . |
|---|---|---|---|---|
| ALOX15 | β-catenin; PI3K/AKT signaling | PD146176 | 106 | No |
| ALOX5 | Wnt/β-catenin signaling | Zileuton | 69 | Yes |
| Autophagy | Autophagy | Chloroquine; bafilomycin A1 | 113 | Yes |
| As2O3 | ||||
| 114 | ||||
| 115 | ||||
| BCL6 | PI3K/AKT/FOX3a/BCL6 signaling | RI-BPI | 56 | No |
| BLK | MYC/PAX5/BLK/CDKN1B signaling | NA | 116 | No |
| CCN | Wnt/Ca2+/NFAT signaling | Cyclosporin A | 68 | No |
| CD25 | JAK/STAT signaling | BEZ235 | 111 | No |
| CD70/CD27 | Wnt/β-catenin signaling | αCD70 mAb | 67 | No |
| c-MYC and TP53 | apoptosis | CPI-203; RITA/RG7388 | 98 | No |
| EZH2 | Histone H3 K27 trimethylation | GSK 343; GSK126 | 101 | No |
| EPZ-6438 (tazemetostat) | 100 | |||
| Farnesyl transferases | RAS signaling; protein farnesylation | BMS-214662 | 117 | Yes |
| FOX03A | TGF-β/AKT/FOXO3a/BCL6 signaling | NA | 57 | No |
| 58 | ||||
| GSK3β | Wnt/β-catenin signaling | SB216763 | 118 | No |
| HDACs | Histone acetylation | LBH589 | 119 | Yes |
| JAK2 | JAK/STAT signaling | Ruxolitinib | 74 | Yes |
| PML | Apoptosis; mTOR repression | As2O3 | 120 | Yes |
| PP2A | JAK/STAT/β-catenin signaling | 1,9-dideoxy-forskolin; FTY720 | 76 | No |
| 77 | ||||
| PPARγ | STAT5/HIF2α/CITED2 | Pioglitazone | 102 | Yes |
| 103 | ||||
| RAD52 | DNA repair | RAD52 F79 peptide aptamer | 82 | No |
| SIRT1 | Deacetylation of p53 | Tenovin-6 | 121 | No |
| SMO | Hedgehog signaling | Cyclopamine; LDE225 | 60 | Yes |
| 61 | ||||
| 62 | ||||
| STAT3 | JAK/STAT signaling | BP-5087 | 80 | No |
| TGF-β RI, ALK5 | TGF-β/AKT/FOXO3a/BCL6 signaling | Ly364947; EW-7197 | 58 | No |
| 122 |
| Target or survival factor . | Pathway . | Exemplar inhibitors/activators . | Reference . | CML clinical trial . |
|---|---|---|---|---|
| ALOX15 | β-catenin; PI3K/AKT signaling | PD146176 | 106 | No |
| ALOX5 | Wnt/β-catenin signaling | Zileuton | 69 | Yes |
| Autophagy | Autophagy | Chloroquine; bafilomycin A1 | 113 | Yes |
| As2O3 | ||||
| 114 | ||||
| 115 | ||||
| BCL6 | PI3K/AKT/FOX3a/BCL6 signaling | RI-BPI | 56 | No |
| BLK | MYC/PAX5/BLK/CDKN1B signaling | NA | 116 | No |
| CCN | Wnt/Ca2+/NFAT signaling | Cyclosporin A | 68 | No |
| CD25 | JAK/STAT signaling | BEZ235 | 111 | No |
| CD70/CD27 | Wnt/β-catenin signaling | αCD70 mAb | 67 | No |
| c-MYC and TP53 | apoptosis | CPI-203; RITA/RG7388 | 98 | No |
| EZH2 | Histone H3 K27 trimethylation | GSK 343; GSK126 | 101 | No |
| EPZ-6438 (tazemetostat) | 100 | |||
| Farnesyl transferases | RAS signaling; protein farnesylation | BMS-214662 | 117 | Yes |
| FOX03A | TGF-β/AKT/FOXO3a/BCL6 signaling | NA | 57 | No |
| 58 | ||||
| GSK3β | Wnt/β-catenin signaling | SB216763 | 118 | No |
| HDACs | Histone acetylation | LBH589 | 119 | Yes |
| JAK2 | JAK/STAT signaling | Ruxolitinib | 74 | Yes |
| PML | Apoptosis; mTOR repression | As2O3 | 120 | Yes |
| PP2A | JAK/STAT/β-catenin signaling | 1,9-dideoxy-forskolin; FTY720 | 76 | No |
| 77 | ||||
| PPARγ | STAT5/HIF2α/CITED2 | Pioglitazone | 102 | Yes |
| 103 | ||||
| RAD52 | DNA repair | RAD52 F79 peptide aptamer | 82 | No |
| SIRT1 | Deacetylation of p53 | Tenovin-6 | 121 | No |
| SMO | Hedgehog signaling | Cyclopamine; LDE225 | 60 | Yes |
| 61 | ||||
| 62 | ||||
| STAT3 | JAK/STAT signaling | BP-5087 | 80 | No |
| TGF-β RI, ALK5 | TGF-β/AKT/FOXO3a/BCL6 signaling | Ly364947; EW-7197 | 58 | No |
| 122 |
Column 1 indicates key survival factors/drug targets intrinsic to the LSC. Column 2 lists the roles of these factors/targets in cellular pathways (if known or as proposed by investigators). Column 3 shows exemplar compounds that have been identified as having potential therapeutic value for each factor/target or pathway (if known). Column 4 provides the references in which the factors/targets are described. Column 5 indicates whether factors/targets or compounds have been evaluated in clinical trials.
As2O3, arsenic trioxide; NA, not available.
Candidate therapeutic targets implicated in CP CML LSC/BMM interactions
| Target or survival factor . | Pathway . | Exemplar inhibitors/activators . | Reference . | CML clinical trial . |
|---|---|---|---|---|
| β1-integrins | Cell adhesion; proliferation | IFN-α | 87 | Yes |
| 86 | ||||
| CD26 | CXCL12/CXCR4 signaling | Gliptins | 90 | No |
| CD44 | E-selectin ligands; homing, engraftment | NA | 83 | No |
| CXCR4 | CXCL12/CXCR4 axis | AMD3465; AMD3100; plerixafor | 123 | Yes |
| 124 | ||||
| 125 | ||||
| 126 | ||||
| BMP2/4 | BMP signaling | NA | 127 | No |
| Galectin-3 | BM lodgement | NA | 85 | No |
| HIF1α | Hypoxia response | NA | 128, 129 | No |
| IFN-α | Immune surveillance; cytokine-mediated proliferation | Interferon alfa-2a | 130 | Yes |
| 131 | ||||
| 132 | ||||
| 133 | ||||
| 134 | ||||
| IL-1RAP/IL-1 | NFKβ/AKT signaling | IL-1RAP mAb (mAb81.2) | 110 | Yes |
| 135 | ||||
| 136 | ||||
| IL-6 | IL-6Rα signaling from the microenvironment | αIL-6 mAb | 92 | No |
| LYN | CXCR4/CXCL12 signaling in lipid rafts | Methyl-β-cyclodextrin; PP2 | 89 | No |
| MHC-II/CIITA | JAK/STAT signaling | Ruxolitinib, IFN-γ | 97 | Yes |
| miR-126 | Exosomal transfer | 2-O-Me-miR-126 | 137 | No |
| N-cadherins | N-cadherin/Wnt/β-catenin signaling | ICG001 | 88 | No |
| JAGGED-1 | NOTCH signaling | NA | 138 | No |
| MPL | JAK/STAT signaling; engraftment | NA | 139 | No |
| PD-1/PD-L1 | Immune surveillance | αPD-L1 mAb | 95, 96 | Yes |
| αPD-1 mAb | ||||
| PIGF | VEGF signaling: BM angiogenesis; proliferation; metabolism | 5D11D4 | 140 | No |
| Selectins | BM homing; engraftment | GMI-1271 | 84, 141 | No |
| TGF-β1 | Osteoblast → LSC TGF-β signaling | Parathyroid hormone | 142 | No |
| Target or survival factor . | Pathway . | Exemplar inhibitors/activators . | Reference . | CML clinical trial . |
|---|---|---|---|---|
| β1-integrins | Cell adhesion; proliferation | IFN-α | 87 | Yes |
| 86 | ||||
| CD26 | CXCL12/CXCR4 signaling | Gliptins | 90 | No |
| CD44 | E-selectin ligands; homing, engraftment | NA | 83 | No |
| CXCR4 | CXCL12/CXCR4 axis | AMD3465; AMD3100; plerixafor | 123 | Yes |
| 124 | ||||
| 125 | ||||
| 126 | ||||
| BMP2/4 | BMP signaling | NA | 127 | No |
| Galectin-3 | BM lodgement | NA | 85 | No |
| HIF1α | Hypoxia response | NA | 128, 129 | No |
| IFN-α | Immune surveillance; cytokine-mediated proliferation | Interferon alfa-2a | 130 | Yes |
| 131 | ||||
| 132 | ||||
| 133 | ||||
| 134 | ||||
| IL-1RAP/IL-1 | NFKβ/AKT signaling | IL-1RAP mAb (mAb81.2) | 110 | Yes |
| 135 | ||||
| 136 | ||||
| IL-6 | IL-6Rα signaling from the microenvironment | αIL-6 mAb | 92 | No |
| LYN | CXCR4/CXCL12 signaling in lipid rafts | Methyl-β-cyclodextrin; PP2 | 89 | No |
| MHC-II/CIITA | JAK/STAT signaling | Ruxolitinib, IFN-γ | 97 | Yes |
| miR-126 | Exosomal transfer | 2-O-Me-miR-126 | 137 | No |
| N-cadherins | N-cadherin/Wnt/β-catenin signaling | ICG001 | 88 | No |
| JAGGED-1 | NOTCH signaling | NA | 138 | No |
| MPL | JAK/STAT signaling; engraftment | NA | 139 | No |
| PD-1/PD-L1 | Immune surveillance | αPD-L1 mAb | 95, 96 | Yes |
| αPD-1 mAb | ||||
| PIGF | VEGF signaling: BM angiogenesis; proliferation; metabolism | 5D11D4 | 140 | No |
| Selectins | BM homing; engraftment | GMI-1271 | 84, 141 | No |
| TGF-β1 | Osteoblast → LSC TGF-β signaling | Parathyroid hormone | 142 | No |
Column 1 indicates key survival factors that act through LSC/BMM interactions. Column 2 shows the roles of these survival factors in cellular pathways (if known or as proposed by investigators). Column 3 lists exemplar compounds that have been identified as having potential therapeutic value for each target (if known). Column 4 provides the references in which the targets were described. Column 5 indicates whether targets or compounds have been evaluated in clinical trials.
miR, microRNA; NA, not applicable; VEGF, vascular endothelial growth factor.
PI3K/AKT/FOXO signaling
BCR-ABL1 has been shown to upregulate phosphatidylinositol 3-kinase (PI3K)/AKT signaling, and AKT-mediated phosphorylation of FOXO transcription factors results in their cytoplasmic localization, where they are inactive (Figure 2). One important consequence of TKI exposure is inhibition of BCR-ABL1 and downregulation of PI3K/AKT signaling in the LSC (kinase-dependent), leading in turn to relocalization of FOXO1 and FOXO3a from the cytoplasm to the nucleus, where they modulate expression of CCND1, ATM, CDKN1C, and BCL6, causing a G1 arrest56,57 and may fuel an antiapoptotic phenotype. The transcriptional repressor BCL6, a FOXO3A target, likely plays an important role in this process by repressing the tumor suppressors p53 and ARF. In this respect, TKI exposure permits FOXO3A-mediated upregulation of BCL6, resulting in a protective, pro-survival effect. Others have shown that the PI3K signaling axis in LSC is also under the control of transforming growth factor β (TGF-β) signaling (kinase-independent) and that blocking this pathway reversed the effects of FOXO nuclear translocation.58,59 However, the precise mechanism of how this occurs is not fully understood and may not be completely cell-autonomous.
Hedgehog signaling
Several studies have implicated the hedgehog pathway in the maintenance (self-renewal) and proliferation of the LSC,60-62 where Smoothened (SMO) is a critical mediator. Hedgehog binding to Patched activates SMO that in turn activates the transcription factor GLI1. This leads to reductions of NUMB expression and increased MDM2-mediated degradation of the p53 protein (Figure 2). This has the effect of suppressing apoptotic responses and/or cell-cycle arrest through repression of p53 targets. SMO deletion or pharmacological inhibition in mouse models of CML blocked this pathway and led to loss of LSCs.60,61 However, TKI treatment alone was unable to block this pathway, suggesting that hedgehog signaling was kinase-independent. More recently, similar results were obtained using SMO inhibitors in human CML samples in vitro and in vivo using xenografts in immunocompromised nonobese diabetic severe combined immunodeficiency γ mice.62
Canonical and noncanonical Wnt signaling
β-catenin is a central mediator of both canonical and non-Wnt signaling and has a dual role in regulating cell-to-cell contact through tight junctions and acting as a transcriptional regulator when translocated to the nucleus (Figure 2). In the absence of Wnt signaling, cytoplasmic β-catenin is ultimately phosphorylated by GSK3β and targeted for degradation by an axin-mediated multimeric complex. Nuclear β-catenin is required for self-renewal and survival of normal HSCs63 ; therefore, it is not surprising that it has also been shown to be a key mediator of LSC survival. Loss of β-catenin in a murine model of CML impaired the development of the disease by inhibiting LSC self-renewal,64 and genetic and pharmacological inhibition of β-catenin activity synergized with TKI to target the loss of LSC.65 Several alternative Wnt-regulated pathways have been implicated in CML LSC. TKI exposure induced the upregulation of CD70 ligand-induced CD27 signaling66,67 resulting in β-catenin nuclear translocation and activation of Wnt target genes, including NOTCH, and c-MYC (kinase-dependent). TKI exposure also induces a noncanonical Wnt signaling mediated through NFAT signaling, which reduces levels of the pro-survival cytokine interleukin-4 (IL-4)68 (kinase-dependent). Fatty acid metabolism was demonstrated to be important in LSC when arachidonate 5-lipoxygenase, encoded by ALOX5, was shown to be upregulated in LSCs in a kinase-independent manner,69 where it is thought to regulate β-catenin levels. Inhibition of ALOX5, through genetic deletion or by pharmacological inhibition in mouse models, targeted the loss of LSCs, implicating this component as an important mediator of LSC survival.
JAK/STAT signaling
The Janus kinases family of intracellular nonreceptor kinases play important roles in regulating cytokine-mediated signal transduction via the JAK/STAT pathway (Figure 2). Activation of STAT5 was demonstrated in primary CML and CML cell lines 20 years ago70 and involves its phosphorylation and translocation to the nucleus where it regulates transcription. Subsequent evidence has also shown that a single null mutation in the STAT5a isoform can attenuate CML-like disease in mouse models71 and knockdown can impair Ph+ myeloid colony formation from CML patient samples.72 Modulating JAK2 activity in human and mouse cell lines reduces BCR-ABL1 and STAT5 signaling,73 and pharmacological inhibition using ruxolitinib resulted in the loss of LSCs both in vitro and in vivo,74 implicating JAK2 as an upstream mediator of a CML JAK/STAT signaling cascade in LSCs. However, BCR-ABL1 has also been implicated in the direct activation of STAT575 (kinase-dependent), suggesting that JAK2 may not be necessary for CML disease maintenance. Furthermore, the reactivation of the tumor suppressor and serine-threonine phosphatase PP2A, through either knockdown or pharmacological inhibition of its repressor SET, has been shown to inhibit BCR-ABL1 and STAT5 activation in CML blast crisis.76 The scenario, however, is different in LSCs, where BCR-ABL1 exerts kinase-independent roles to recruit JAK2 to modulate JAK/STAT signaling77,78 (see also the following section). Activation of STAT3 has also been implicated in the JAK/STAT cascade, where it exerts a protective effect on CML cells upon exposure to TKI.79 Inhibition of STAT3 in combination with TKI-induced synthetic lethality to target the loss of LSC.80
Genomic instability, DNA damage, and repair
Whether TKI-induced quiescence contributes to LSC persistence in patients is still an open question. Two possible beneficial consequences of TKI treatment would be to reduce the turnover and expansion of LSCs in patients and enhance a “low mutator” phenotype.44 However, a more cautionary interpretation of these possible benefits has come from examining the mechanisms and pathways that contribute to genomic instability in LSCs. BCR-ABL1 kinase activity leads to increased levels of reactive oxygen species (ROS),48,49,81 including H2O2, and these lead to oxidative DNA damage, including point mutations and double-stranded breaks. In this regard, the RAC2 GTPase has been shown to alter the function of the mitochondrial respiratory chain complex to generate ROS and DNA damage in LSCs, as evidenced by the accumulation of chromosomal aberrations and clinically relevant BRC-ABL1 kinase domain mutations.49 This effect was also observed under hypoxia, the conditions that LSCs are exposed to in the BMM, and during exposure to TKIs in which RAC2 levels were unaffected, thus demonstrating a kinase-independent pathway. Inhibition of RAC2 or disruption of the mitochondrial respiratory chain complex reduced the level of genomic instability. Similarly, high ROS levels and associated genomic damage were recapitulated using the transgenic SCL-tTA/BCR-ABL model,48 in which both BCR-ABL1 kinase domain mutations and various base pair additions/deletions in genes linked to progression to blast crisis were identified in LSCs in both TKI-naïve and TKI-treated mice. Evidence as to why such DNA damage is tolerated in LSCs has also emerged. BCR-ABL1 can inhibit mismatch repair to protect cells from apoptosis50 and can stimulate single-strand annealing, homologous recombination repair, and nonhomologous end-joining, all of which are error-prone in BCR-ABL1–expressing cells.52,81 Furthermore, LSCs are dependent on the alternative RAD52-RAD51 pathway of homologous recombination repair to deal with double-stranded breaks rather than BRCA1/2-RAD51 because of the kinase-independent downregulation of BRCA1.82 Although we are unable to reconcile these data with a “low mutator” phenotype,44 they point to the LSC as a potent source of clinically relevant mutations and argue that CML is constantly evolving at the molecular level even in CP, countering the clinical view that it is a disease of 3 distinct phases.
The LSC BMM
Although the pathways described here have ostensibly been studied as primarily cell-intrinsic or cell-autonomous, it is likely that some, if not all, are regulated through interactions between the CML LSCs and the BMM—and several of these interactions have been identified (Figure 3; Table 2), some of which mediate TKI resistance.
LSC adhesion within the BMM is likely to contribute to homing and lodgement—critical steps in LSC engraftment subsequent to transplantation. CD44, expressed on LSCs, is a ligand for e-selectins; lack of CD44 reduced homing and engraftment of LSCs.83 Similarly, a critical role for selectins and their ligands in engraftment has also been shown,84 and e-selectins can be blocked pharmacologically to reduce the number of LSCs. The lectin galectin-3 mediates resistance to TKIs through binding β-galactosides on stromal cells and overexpression-activated AKT signaling and increased lodgement of LSCs in the BM.85 β1-integrins mediate adhesion of LSCs to BM stromal cells, a process likely to be regulated by interferon-α (IFN-α).86,87 TKI-induced upregulation of N-cadherin in LSCs, and adhesion to mesenchymal stem cells led to increased canonical Wnt signaling and protection of the LSC from apoptosis.88 The CXCL12 ligand and its receptor CXCR4 has been linked to intracellular LYN signaling in LSCs,89 and the CXCL12/CXCR4 axis is regulated through CXCL12 cleavage by CD26.90 Reduced homing capacity of LSCs has also been attributed to alterations of the CXCL12/CXCR4 signaling pathway as a result of increased granulocyte colony-stimulating factor levels that conferred a selective growth disadvantage to normal HSCs.91 LSCs also exert other molecular and phenotypic effects on HSCs through extrinsic IL-6 signaling in the CML BMM.92,93 Indeed, a variety of ligand receptor–mediated signaling pathways regulate CML LSCs in the BMM (Figure 3; Table 2).
It is likely that LSCs also avoid eradication by modulation of host immune surveillance in the BMM (reviewed in detail elsewhere94 ). In this respect, cytotoxic T lymphocytes (CTLs) are unable to elicit an appropriate immune response against CML cells through CTL exhaustion; and this is believed to be mediated by the interaction of the PD-1 receptor expressed on CTLs with its inhibitory ligand PD-L1 expressed on CML cells. PD-L1 is expressed on patient-derived CML cells95 and on LSCs in mouse models of CML.96 Blockade of the PD-1/PD-L1 interaction in combination with T-cell immunotherapy was able to trigger the loss of LSCs and prevent development of CML-like disease.96 Our recent work has demonstrated that cytokine-mediated downregulation of MHC-II expression may be an alternative way that LSCs evade immune surveillance; treatment with ruxolitinib or IFN-γ can reverse this effect in vitro and enhance proliferation of responder CD4+CD69+ T cells in mixed lymphocyte reactions.97 These examples represent exciting areas of research that could lead to new immune therapy-based therapeutic approaches.
New therapies to target LSC: recent approaches
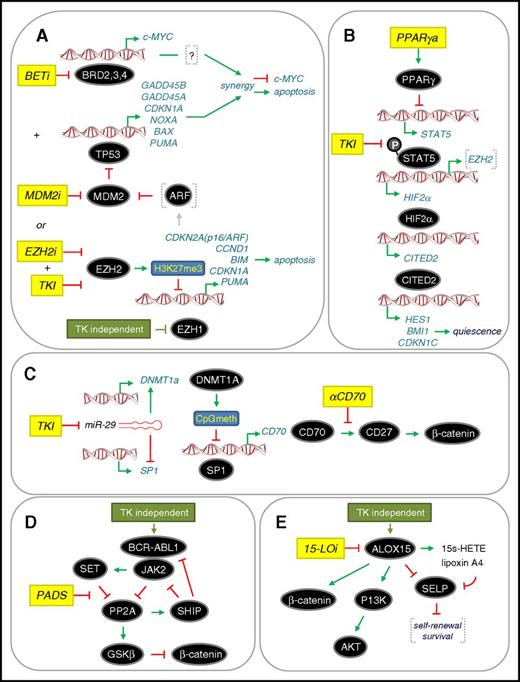
The many examples summarized here illustrate the scope of potentially drug-able targets that have been identified in CML to eradicate LSC (Tables 1 and 2). Disappointingly, drugs against these targets have yet to be implemented in the clinic as standard of care. In the past 3 to 4 years, additional drug-able targets and pathway have been identified, whereas others previously identified have been further elaborated in preclinical studies (Figure 4). Our analysis of global proteomics and transcriptomics in drug-naïve primary patient material (bulk CD34+ cells and LSCs) pointed toward a dependency of CML cells on a p53 and c-MYC regulated network.98 This provided a rationale to use a combination of MDM2 and BET inhibitors (MDM2i and BETi, respectively) to target the synergistic eradication of LSCs through upregulation of the p53 apoptotic pathway and downregulation of c-MYC by both drugs (Figure 4A). Given that BETi acts generally as a transcriptional repressor, how its effects lead to upregulation of apoptosis in CML LSCs is not fully understood, although this appears to be a common phenomenon of BETi in preclinical cancer studies.99 We have also used global epigenetic and transcriptomic analysis of drug-naïve primary patient material to reveal that misregulation of the PRC2 complex (including kinase-independent downregulation of EZH1 in LSCs) results in the functional dependency of LSCs on EZH2 and its biochemical readout H3K27me3. Using murine models, others have also reported that CML LSCs are dependent on EZH2.100 Combining an EZH2 inhibitor (EZH2i) with TKI was highly effective at eradicating the LSC population.101 Our data support a model whereby apoptosis is induced in CML LSCs through upregulation of EZH2 targets upstream of p53 (such as ARF), which could lead to increased p53 levels, or through upregulation of p53 target genes directly, which are normally repressed by EZH2 activity (Figure 4A).
Recent therapeutic approaches to target the eradication of CP CML LSC. (A) Dual targeting of c-MYC and TP53 (p53) or combined treatment with TKI and EZH2 inhibitor (EZH2i).98,100,101 Both approaches converge on upregulating p53-mediated apoptosis through different mechanisms. BETi and MDM2i lead to synergistic repression of c-MYC transcription and upregulation of p53 target genes. A dependency on EZH2 for LSC survival is accompanied by a TKI-independent downregulation of EZH1. (B) Inhibition of STAT5 upstream of the HIF2α-CITED2 pathway that governs LSC quiescence. Combining a PPARγ activator (PPARγa) with TKI102,103 inhibits STAT5 transcription and STAT5 phosphorylation, respectively, and downregulates HIF2α-CITED2 leading to LSC exit from quiescence. (C) Inhibition of noncanonical Wnt/β-catenin signaling mediated by CD70/CD27. TKI upregulates the Wnt/β-catenin pathway by inhibiting miR-29 expression, facilitating both increased CD70 expression and CD70/CD27 receptor/ligand interaction. Treatment with a monoclonal antibody that blocks the CD70/CD27 interaction (αCD70) in a TKI background blocks the pathway.66,67 (D) Activation of PP2A to inhibit a novel CML network driven by JAK2-β-catenin signaling. PP2A activating drugs (PADs) disrupt the PP2A-SET interaction, thereby allowing PP2A reactivation, which inhibits BCR-ABL1 recruitment of JAK2 (TKI-independent) and impairs β-catenin signaling through GSK-3β activation.77 (E) Inhibition of ALOX15 to inhibit β-catenin and PI3K/AKT signaling. Knockdown of ALOX15 or treatment with a 15-LO inhibitor (15-LOi), which blocks ALOX15 enzymatic activity, reduced LSC survival in association with reduced PI3K/AKT and β-catenin levels. This “kill” phenotype was rescued by loss of p-selectin (SELP), which is thought to negatively regulate LSC self-renewal and survival.106 Activation and repression are denoted according to convention. Drug treatments are shown in yellow. Further details are described in the text.
Recent therapeutic approaches to target the eradication of CP CML LSC. (A) Dual targeting of c-MYC and TP53 (p53) or combined treatment with TKI and EZH2 inhibitor (EZH2i).98,100,101 Both approaches converge on upregulating p53-mediated apoptosis through different mechanisms. BETi and MDM2i lead to synergistic repression of c-MYC transcription and upregulation of p53 target genes. A dependency on EZH2 for LSC survival is accompanied by a TKI-independent downregulation of EZH1. (B) Inhibition of STAT5 upstream of the HIF2α-CITED2 pathway that governs LSC quiescence. Combining a PPARγ activator (PPARγa) with TKI102,103 inhibits STAT5 transcription and STAT5 phosphorylation, respectively, and downregulates HIF2α-CITED2 leading to LSC exit from quiescence. (C) Inhibition of noncanonical Wnt/β-catenin signaling mediated by CD70/CD27. TKI upregulates the Wnt/β-catenin pathway by inhibiting miR-29 expression, facilitating both increased CD70 expression and CD70/CD27 receptor/ligand interaction. Treatment with a monoclonal antibody that blocks the CD70/CD27 interaction (αCD70) in a TKI background blocks the pathway.66,67 (D) Activation of PP2A to inhibit a novel CML network driven by JAK2-β-catenin signaling. PP2A activating drugs (PADs) disrupt the PP2A-SET interaction, thereby allowing PP2A reactivation, which inhibits BCR-ABL1 recruitment of JAK2 (TKI-independent) and impairs β-catenin signaling through GSK-3β activation.77 (E) Inhibition of ALOX15 to inhibit β-catenin and PI3K/AKT signaling. Knockdown of ALOX15 or treatment with a 15-LO inhibitor (15-LOi), which blocks ALOX15 enzymatic activity, reduced LSC survival in association with reduced PI3K/AKT and β-catenin levels. This “kill” phenotype was rescued by loss of p-selectin (SELP), which is thought to negatively regulate LSC self-renewal and survival.106 Activation and repression are denoted according to convention. Drug treatments are shown in yellow. Further details are described in the text.
Two groups have shown that activators of the peroxisome proliferator-activated receptor γ (PPARγ) have increased antileukemic activities in combination with TKI.102,103 Quiescence of LSCs is regulated by a pathway involving the receptor PPARγ, STAT5, HIF2α, and CITED2, a master regulator of blood stem cell quiescence (Figure 4B). Activators of PPARγ result in transcriptional downregulation of STAT5, whereas TKIs block phosphorylation of STAT5, with the combined effects of both drugs significantly down-regulating this pathway and causing LSCs to exit quiescence where they were eradicated by TKIs.104 Recently, EZH2 has been shown to be activated by STAT5 in CML cells105 suggesting possible crosstalk between the effects of PPARy activators and those of EZH2i.
The TKI-mediated upregulation of CD70 has been further examined to provide a clear rationale for inhibiting noncanonical Wnt/β-catenin signaling in LSCs.67 Upon exposure to TKIs, the microRNA miR-29 is downregulated, the consequence of which is upregulation of CD70 through the opposing roles of miR-29 on SP1 and DNMT1a regulation (Figure 4C). Thus, antibody-based blockade of the interaction between CD70 and CD27 resulted in a potent loss of LSCs in the presence of TKIs.67 Two other routes for inhibiting β-catenin signaling in LSCs have also recently been deduced. In the first, BCR-ABL1 interacts directly with JAK2 in a kinase-independent manner to activate a JAK2/β-catenin survival/self-renewal pathway that results in inhibition of PP2A and activation of β-catenin (Figure 4D). Use of PP2A-activating drugs reversed these effects, resulting in GSKβ-dependent degradation of β-catenin and eradication of LSCs.77 In the second, another enzyme in fatty acid metabolism arachidonate 15-lipoxygenase (15-LO encoded by ALOX15) has been implicated in the kinase-independent upregulation of β-catenin, although the exact mechanism is unclear. However, pharmacological inhibition of 15-LO in combination with nilotinib on human LSCs in vitro appeared synergistic.106 In addition, the p-selectin SELP appears to be a key downstream target of 15-LO, which is normally repressed to promote LSC survival. Further preclinical studies and mechanistic studies are required to provide a clearer rationale for taking 15-LO inhibitors into clinical trials, as has been done with zileuton, which inhibits 5-LO.69
Future challenges
We know little about how TKI-resistant LSC clones evolve in patients in MMR and the degree of intra- and interpatient heterogeneity that is likely to exist—not only at the DNA level, but also with respect to the many pathways we have identified by studying diagnostic drug-naïve LSCs for many years. This is because (1) the TKI-resistant LSCs are extremely rare in the BM of these CML patients and (2) the LSCs cannot be selectively isolated from the normal HSCs that reconstitute normal hemopoiesis in the BM subsequent to TKI therapy. Surrogate in vivo analysis has also been problematic because the majority of CML primary samples do not engraft well in commonly used immunodeficient mice strains. These issues most likely underpin the failure of many promising new drugs to deliver results in clinical trials. However, recent advances in tracking individual normal and malignant clones in xenograft models using bar-coding,21,107 the development of humanized xenograft models,108 an explosion of single-cell technologies,20,109 and the identification of a number of leukemia-specific cell surface markers, make the analysis of individual LSCs or LSC clones much more accessible. Furthermore, several groups have identified markers that discriminate LSCs from HSCs (CD26,90 IL-1RAP,110 CD25,111 and CD93112 ), but how these will perform in samples from patients in MMR has yet to be determined. For those of us intent on curing CML, this new era of game-changing technologies provides some tantalizing prospects that will enable us to finally stem the tide on drug-resistant LSC.
Authorship
Contribution: T.L.H. and D.V. reviewed the literature and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tessa Holyoake, R314 Level 3 Paul O'Gorman Leukaemia Research Centre, Institute of Cancer Sciences, University of Glasgow, Gartnavel General Hospital, Glasgow, United Kingdom G12 0YN; e-mail: tessa.holyoake@glasgow.ac.uk; and David Vetrie, Room 311 Wolfson Wohl Cancer Research Centre, Institute of Cancer Sciences, University of Glasgow, Garscube Estate, Glasgow, United Kingdom G61 1QH; e-mail: david.vetrie@glasgow.ac.uk.