Key Points
ATL subtypes are further classified into molecularly distinct subsets with different prognosis by genetic profiling.
PD-L1 amplifications are a strong genetic predictor for worse outcome in both aggressive and indolent ATL.
Abstract
Adult T-cell leukemia/lymphoma (ATL) is a heterogeneous group of peripheral T-cell malignancies characterized by human T-cell leukemia virus type-1 infection, whose genetic profile has recently been fully investigated. However, it is still poorly understood how these alterations affect clinical features and prognosis. We investigated the effects of genetic alterations commonly found in ATL on disease phenotypes and clinical outcomes, based on genotyping data obtained from 414 and 463 ATL patients using targeted-capture sequencing and single nucleotide polymorphism array karyotyping, respectively. Aggressive (acute/lymphoma) subtypes were associated with an increased burden of genetic and epigenetic alterations, higher frequencies of TP53 and IRF4 mutations, and many copy number alterations (CNAs), including PD-L1 amplifications and CDKN2A deletions, compared with indolent (chronic/smoldering) subtypes. By contrast, STAT3 mutations were more characteristic of indolent ATL. Higher numbers of somatic mutations and CNAs significantly correlated with worse survival. In a multivariate analysis incorporating both clinical factors and genetic alterations, the Japan Clinical Oncology Group prognostic index high-risk, older age, PRKCB mutations, and PD-L1 amplifications were independent poor prognostic factors in aggressive ATL. In indolent ATL, IRF4 mutations, PD-L1 amplifications, and CDKN2A deletions were significantly associated with shorter survival, although the chronic subtype with unfavorable clinical factors was only marginally significant. Thus, somatic alterations characterizing aggressive diseases predict worse prognosis in indolent ATL, among which PD-L1 amplifications are a strong genetic predictor in both aggressive and indolent ATL. ATL subtypes are further classified into molecularly distinct subsets with different prognosis. Genetic profiling might contribute to improved prognostication and management of ATL patients.
Introduction
Adult T-cell leukemia/lymphoma (ATL) is a distinct subtype of peripheral T-cell neoplasms associated with human T-cell leukemia virus type-1 (HTLV-1) retrovirus. Affecting 10 to 20 million HTLV-1 carriers worldwide, especially in southwest Japan, the Caribbean basin, Central and South America, intertropical Africa, Romania, and northern Iran, HTLV-1 infection is associated with a 3% to 5% risk of ATL development, with a median age at diagnosis of 50 to 70 years.1-5 The clinical and pathological presentation of ATL is highly variable, and patients are classified into 4 subtypes: acute, lymphoma, chronic, and smoldering.1-4 The chronic and smoldering ATLs are usually indolent tumors, for which combined interferon-α and zidovudine therapy or watchful waiting is the standard choice of treatment, but many patients eventually progress to the acute form of the disease.2-4 The acute and lymphoma subtypes are highly aggressive diseases associated with very poor prognosis and treated with intensive therapies, such as combination chemotherapy and molecularly targeted agents against CCR4, although allogeneic hematopoietic stem cell transplantation (HSCT) remains the only potentially curative therapy.2-4 Besides disease subtype, several clinical features, including age, performance status, and laboratory parameters, have been proposed to predict clinical course and prognosis in ATL, although only a few of them were validated in independent studies.2-4
Even though HTLV-1 infection is a hallmark of ATL and prerequisite for its diagnosis, the presence of a long latency period before tumor onset suggests that HTLV-1 infection alone is not enough for ATL development, and that additional genetic events are required.6,7 Through an integrated molecular analysis of a large number of ATL patients, we have demonstrated that typical ATL has many driver alterations, with a median of 4 mutations and 10 focal copy number alterations (CNAs) per case, which recurrently affect key components in T-cell receptor and NF-κB signaling, chemokine receptors, transcriptional and epigenetic regulators, and immune-related molecules.8 However, the relevance of these genetic alterations on disease phenotypes and clinical outcomes is largely unknown, except for the negative impacts of TP53 mutations and CDKN2A deletions.9,10 Understanding the clinical effects of these genetic alterations might improve the prognostic prediction for patients and inform the selection of specific therapies, as demonstrated in other hematologic malignanices.11-13
In the present study, we investigated possible associations between these genetic/epigenetic alterations and clinical/pathological phenotypes in a large set of ATL patients, focusing on the influence of mutations and CNAs on clinical outcome.
Methods
Patient samples
We evaluated a total of 463 patients who had been diagnosed with ATL between 1984 and 2017, of which 410 were previously reported.8 All patients were analyzed by single nucleotide polymorphism array karyotyping, and 414 were also investigated by targeted sequencing (Figure 1A). Diagnosis was based on the World Health Organization classification, and the patients were classified into acute (n = 224, 48%), lymphoma (n = 104, 22%), chronic (n = 105, 23%), and smoldering (n = 30, 6%) subtypes, according to the International Consensus Meeting proposal.1,2 HTLV-1 infection was documented in all patients. In the entire cohort, the median age was 62 years, and 53% were male. The other clinical characteristics are summarized in Table 1. This study was approved by the institutional ethics committees of the Graduate School of Medicine, Kyoto University and other participating institutes and was performed in accordance with the Declaration of Helsinki.
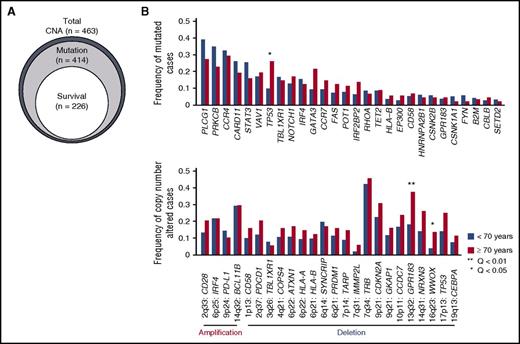
Genetic alterations associated with older age in ATL. (A) Relationship of ATL patients for which CNA, mutation, and survival data were available. (B) Comparison of frequencies of driver mutations and focal CNAs between younger (<70 years) and older (≥70 years) patients (Fisher’s exact test with Benjamini–Hochberg correction). Recurrently mutated genes (n = 28) present in >3% of ATL cases and highly significant (residual Q < 10−13) focal amplifications (n = 4) and deletions (n = 20) are shown.
Genetic alterations associated with older age in ATL. (A) Relationship of ATL patients for which CNA, mutation, and survival data were available. (B) Comparison of frequencies of driver mutations and focal CNAs between younger (<70 years) and older (≥70 years) patients (Fisher’s exact test with Benjamini–Hochberg correction). Recurrently mutated genes (n = 28) present in >3% of ATL cases and highly significant (residual Q < 10−13) focal amplifications (n = 4) and deletions (n = 20) are shown.
Clinical information of ATL patients
| . | CNA (n = 463) . | Mutation (n = 414) . | ||
|---|---|---|---|---|
| n . | % . | n . | % . | |
| Sex | ||||
| Male | 245 | 53 | 218 | 53 |
| Female | 218 | 47 | 196 | 47 |
| Subtype | ||||
| Acute | 224 | 48 | 222 | 54 |
| Lymphoma | 104 | 22 | 73 | 18 |
| Chronic | 105 | 23 | 99 | 24 |
| Smoldering | 30 | 6 | 20 | 5 |
| Age | ||||
| <70 y | 249 | 54 | 244 | 59 |
| ≥70 y | 88 | 19 | 88 | 21 |
| Not available | 126 | 27 | 82 | 20 |
| . | CNA (n = 463) . | Mutation (n = 414) . | ||
|---|---|---|---|---|
| n . | % . | n . | % . | |
| Sex | ||||
| Male | 245 | 53 | 218 | 53 |
| Female | 218 | 47 | 196 | 47 |
| Subtype | ||||
| Acute | 224 | 48 | 222 | 54 |
| Lymphoma | 104 | 22 | 73 | 18 |
| Chronic | 105 | 23 | 99 | 24 |
| Smoldering | 30 | 6 | 20 | 5 |
| Age | ||||
| <70 y | 249 | 54 | 244 | 59 |
| ≥70 y | 88 | 19 | 88 | 21 |
| Not available | 126 | 27 | 82 | 20 |
| . | Survival (n = 226) . | |||
|---|---|---|---|---|
| Aggressive subtypes (n = 152) . | Indolent subtypes (n = 74) . | |||
| n . | % . | n . | % . | |
| Sex | ||||
| Male | 83 | 55 | 30 | 41 |
| Female | 69 | 45 | 44 | 59 |
| Subtype | ||||
| Acute | 125 | 82 | — | — |
| Lymphoma | 27 | 18 | — | — |
| Chronic | — | — | 59 | 80 |
| Smoldering | — | — | 15 | 20 |
| Age | ||||
| <70 y | 107 | 70 | 55 | 74 |
| ≥70 y | 45 | 30 | 19 | 26 |
| Treatment | ||||
| CHOP/CHOP-like | 72 | 47 | — | — |
| VCAP-AMP-VECP | 61 | 40 | — | — |
| Others | 15 | 10 | — | — |
| Not available | 4 | 3 | — | — |
| HSCT during follow-up | ||||
| (–) | 119 | 78 | 59 | 80 |
| (+) | 33 | 22 | 15 | 20 |
| Anti-CCR4 antibody during follow-up | ||||
| (–) | 119 | 78 | 62 | 84 |
| (+) | 33 | 22 | 12 | 16 |
| High calcium (≥2.75 mmol/L) | ||||
| (–) | 125 | 82 | — | — |
| (+) | 27 | 18 | — | — |
| Performance status | — | — | ||
| 0-1 | 92 | 61 | — | — |
| 2-4 | 60 | 39 | — | — |
| JCOG-PI | — | — | ||
| Moderate-risk | 84 | 55 | — | — |
| High-risk | 68 | 45 | — | — |
| Low albumin (<LLN) | ||||
| (–) | — | — | 61 | 82 |
| (+) | — | — | 13 | 18 |
| High BUN (>ULN) | — | — | ||
| (–) | — | — | 71 | 96 |
| (+) | — | — | 3 | 4 |
| High LDH (>ULN) | — | — | ||
| (–) | — | — | 36 | 49 |
| (+) | — | — | 38 | 51 |
| Unfavorable factor | — | — | ||
| (–) | — | — | 30 | 41 |
| (+) | — | — | 44 | 59 |
| . | Survival (n = 226) . | |||
|---|---|---|---|---|
| Aggressive subtypes (n = 152) . | Indolent subtypes (n = 74) . | |||
| n . | % . | n . | % . | |
| Sex | ||||
| Male | 83 | 55 | 30 | 41 |
| Female | 69 | 45 | 44 | 59 |
| Subtype | ||||
| Acute | 125 | 82 | — | — |
| Lymphoma | 27 | 18 | — | — |
| Chronic | — | — | 59 | 80 |
| Smoldering | — | — | 15 | 20 |
| Age | ||||
| <70 y | 107 | 70 | 55 | 74 |
| ≥70 y | 45 | 30 | 19 | 26 |
| Treatment | ||||
| CHOP/CHOP-like | 72 | 47 | — | — |
| VCAP-AMP-VECP | 61 | 40 | — | — |
| Others | 15 | 10 | — | — |
| Not available | 4 | 3 | — | — |
| HSCT during follow-up | ||||
| (–) | 119 | 78 | 59 | 80 |
| (+) | 33 | 22 | 15 | 20 |
| Anti-CCR4 antibody during follow-up | ||||
| (–) | 119 | 78 | 62 | 84 |
| (+) | 33 | 22 | 12 | 16 |
| High calcium (≥2.75 mmol/L) | ||||
| (–) | 125 | 82 | — | — |
| (+) | 27 | 18 | — | — |
| Performance status | — | — | ||
| 0-1 | 92 | 61 | — | — |
| 2-4 | 60 | 39 | — | — |
| JCOG-PI | — | — | ||
| Moderate-risk | 84 | 55 | — | — |
| High-risk | 68 | 45 | — | — |
| Low albumin (<LLN) | ||||
| (–) | — | — | 61 | 82 |
| (+) | — | — | 13 | 18 |
| High BUN (>ULN) | — | — | ||
| (–) | — | — | 71 | 96 |
| (+) | — | — | 3 | 4 |
| High LDH (>ULN) | — | — | ||
| (–) | — | — | 36 | 49 |
| (+) | — | — | 38 | 51 |
| Unfavorable factor | — | — | ||
| (–) | — | — | 30 | 41 |
| (+) | — | — | 44 | 59 |
Clinical information of ATL patients, for which CNA, mutation, and survival data were available. JCOG-PI high-risk was defined as having high a calcium level and/or poor performance status. Unfavorable factor (+) was defined as having any of 3 clinical factors: low albumin, high BUN, and high LDH levels.
AMP, doxorubicin, ranimustine, and prednisone; CHOP, cyclophosphamide, doxorubicin, vincristine, and prednisone; LLN, lower limit of normal; ULN, upper limit of normal; VCAP, vincristine, cyclophosphamide, doxorubicin, and prednisone; VECP, vindesine, etoposide, carboplatin, and prednisone.
Single nucleotide polymorphism array karyotyping, DNA sequencing, and data analysis
The molecular status of these patients, including somatic mutations and CNAs, was analyzed as previously reported.8 Briefly, genomic copy numbers were determined for all 463 tumor samples using the Affymetrix GeneChip Human Mapping 250K NspI Array (n = 319) or the Illumina Human610-Quad BeadChip (n = 144) with the copy number analyzer for GeneChip (CNAG)/allele-specific copy-number analysis using anonymous references (AsCNAR)14,15 and allele-specific copy number analysis of tumors (ASCAT)16 software for Affymetrix and Illumina array data, respectively. Significant focal CNAs were identified using GISTIC 2.0.17 Mutation analysis was performed by deep sequencing of targeted exons using a custom SureSelect library (Agilent) designed to capture 88 genes with sufficient read coverage (supplemental Figure 1, available on the Blood Web site).8 Sequence alignment, mutation calling, and downstream data analysis were performed using the Genomon pipeline, where the overall validation rate was >99.0%.8 Detailed methods are described in the supplemental Methods. We analyzed the associations between clinical phenotypes and significant somatic alterations, namely mutations in 50 genes, 26 focal amplifications, and 51 focal deletions (supplemental Table 1). Of 463 patients, 83, 57, and 109 also had whole-exome/genome and transcriptome sequencing and methylation array data, respectively.8 Sequencing data have been deposited in the European Genome-phenome Archive under accession number EGAS00001001296.
Statistical methods
Statistical analyses were performed with R version 3.1.3 software (R Foundation for Statistical Computing). The molecular features of the aggressive and indolent subtypes were compared by using the Brunner-Munzel and Fisher’s exact tests for continuous and categorical variables, respectively, with the Benjamini–Hochberg correction (Q value). Survival analysis was performed for 226 patients who were previously untreated at the time of sampling and whose clinical information, including age, sex, subtype, and well-known clinical prognostic factors, was available. These clinical prognostic factors included treatment content and Japan Clinical Oncology Group prognostic index (JCOG-PI) based on calcium level and performance status in aggressive ATL, and albumin, blood urea nitrogen (BUN), and lactate dehydrogenase (LDH) levels in indolent ATL.2-4,18,19 Overall survival (OS) was calculated from the time of diagnosis, and observations were censored at the time of HSCT or last follow-up. For cases in which a sample was collected at the time of disease progression, OS was recalculated from that time. The mean follow-up was 25.1 months for surviving patients, and 107 patients were alive at the last follow-up. Older age was defined as ≥70 years according to the previous publication.20 Frameshift and stopgain mutations in CCR4 were not discriminated in this study because, in contrast to a previous report,21 no significant differences for OS between both mutation types were seen (supplemental Figure 2). The Kaplan-Meier method was used for estimating OS, and the log-rank test was used to assess differences in OS between patient groups. The effects of mutation status on OS were evaluated by Cox proportional hazards regression modeling and adjusted for clinical factors using the survival package in R. Model simplification was performed in a stepwise selection of variables relying on the Akaike Information Criterion using the MASS package in R, where genetic alterations having a univariate Cox P value < .10 and clinical factors were considered. All P values were calculated with the use of 2-sided tests, where P < .05 was considered statistically significant.
Results
Spectrum of mutations and CNAs in aggressive and indolent ATL
Overall, 396 (96%) of 414 and 407 (88%) of 463 ATL patients carried ≥1 mutation and CNA, respectively. When evaluated together, 402 (97%) of 414 ATL patients harbored ≥1 somatic alteration. PLCG1 (36%), PRKCB (32%), CCR4 (30%), CARD11 (24%), STAT3 (22%), VAV1 (17%), TP53 (16%), and TBL1XR1 (16%) exhibited the highest mutation frequency. TP53 mutations and 13q32 (GPR183) and 16q23 (WWOX) deletions were more frequent in older patients (≥70 years of age) (Figure 1B), although there was no significant difference in age among subtypes.
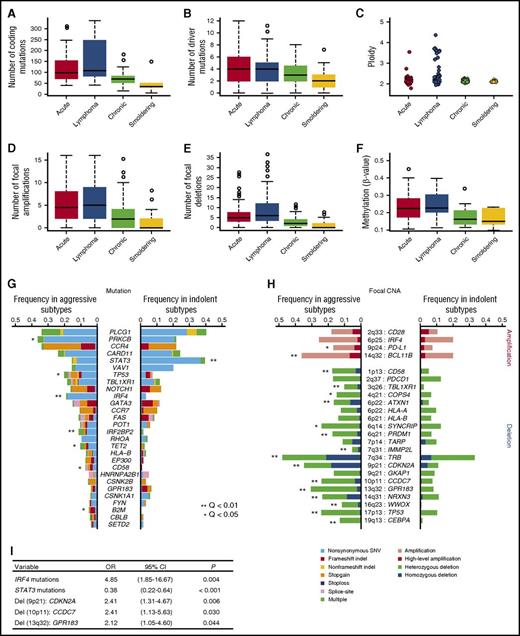
Genetic and epigenetic profiles were substantially different between aggressive and indolent diseases, suggesting a distinct molecular pathogenesis therein. Aggressive subtypes were associated with higher numbers of mutations (Figure 2A-B) as well as focal amplifications and deletions, hyperploid status (Figure 2C-E), and cytosine guanine dinucleotide island hypermethylation (Figure 2F) compared with indolent subtypes, suggesting that the accumulation of genetic and epigenetic changes drives tumor progression in ATL. With respect to the frequency distribution of each driver alteration, 7 mutations and a number of focal CNAs, including 9p24 (PD-L1) amplifications, were more common in aggressive ATL (Figure 2G-H). In accordance with previous reports, TP53 mutations as well as CDKN2A and CD58 deletions were more frequent in aggressive ATL compared with indolent ATL.9,22,23 Stepwise logistic regression analysis showed that IRF4 mutations and focal deletions involving 9p21 (CDKN2A), 10p11 (CCDC7), and 13q32 (GPR183) were independently associated with the aggressive subtypes compared with the indolent subtypes (Figure 2I).
Higher frequencies of somatic alterations in aggressive than indolent ATL. Aggressive (acute and lymphoma subtypes) ATL shows (A) a higher number of coding mutations identified in whole-exome/genome sequencing (n = 83), (B) a higher number of significant mutations in targeted capture sequencing (n = 414), (C) increased ploidy (n = 463), higher numbers of (D) significant focal amplifications and (E) deletions (n = 463), and (F) hypermethylation at promoter-associated cytosine guanine dinucleotide islands (n = 109) compared with indolent ATL (chronic and smoldering subtypes). P < .001 for all comparisons between aggressive and indolent subtypes by Brunner-Munzel test. Comparison of frequencies of (G) driver mutations and (H) focal CNAs between aggressive and indolent ATL (Fisher’s exact test with Benjamini–Hochberg correction). Recurrently mutated genes (n = 28) present in >3% of ATL cases and highly significant (residual Q < 10−13) focal amplifications (n = 4) and deletions (n = 20) are shown. (I) A multivariate logistic regression analysis identifying independent significant factors (present in >10%) for aggressive subtypes (vs indolent subtypes) in 414 ATL cases. CI, confidence interval; OR, odds ratio; SNV, single nucleotide variant.
Higher frequencies of somatic alterations in aggressive than indolent ATL. Aggressive (acute and lymphoma subtypes) ATL shows (A) a higher number of coding mutations identified in whole-exome/genome sequencing (n = 83), (B) a higher number of significant mutations in targeted capture sequencing (n = 414), (C) increased ploidy (n = 463), higher numbers of (D) significant focal amplifications and (E) deletions (n = 463), and (F) hypermethylation at promoter-associated cytosine guanine dinucleotide islands (n = 109) compared with indolent ATL (chronic and smoldering subtypes). P < .001 for all comparisons between aggressive and indolent subtypes by Brunner-Munzel test. Comparison of frequencies of (G) driver mutations and (H) focal CNAs between aggressive and indolent ATL (Fisher’s exact test with Benjamini–Hochberg correction). Recurrently mutated genes (n = 28) present in >3% of ATL cases and highly significant (residual Q < 10−13) focal amplifications (n = 4) and deletions (n = 20) are shown. (I) A multivariate logistic regression analysis identifying independent significant factors (present in >10%) for aggressive subtypes (vs indolent subtypes) in 414 ATL cases. CI, confidence interval; OR, odds ratio; SNV, single nucleotide variant.
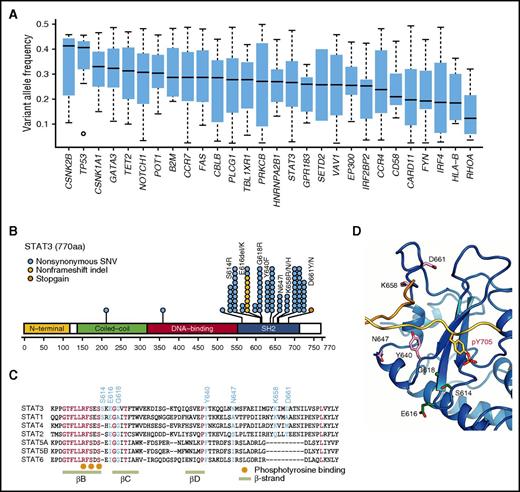
Among these, the most significant were IRF4 mutations, which predominantly affected the 3 amino acid residues located within the DNA-binding domain (K59, L70, and S114).8 IRF4 mutations showed lower allelic burden and were considered more frequently subclonal than clonal, suggesting that these mutations are more likely to be late events than founder alterations (Figure 3A). By contrast, STAT3 mutations were significantly associated with indolent ATL, being present in ∼40% of indolent cases compared with 15% of aggressive cases. Frequent STAT3 mutations have previously been reported in large granular lymphocytic leukemia (LGL), chronic lymphoproliferative disorders of natural killer (NK) cells, and anaplastic large cell lymphoma.24-26 The distribution of mutations was largely similar between ATL and the latter lymphoid malignancies. However, in addition to the hotspot mutations affecting Y640 and D661, which were frequently observed across different diseases and shown to result in an enhanced STAT3 transcriptional activity,24,26 mutations involving S614, E616, and G618 and an in-frame deletion (616_617del) were also observed frequently in ATL cases (Figure 3B). Localized in close proximity to the phosphotyrosine binding pocket within the SH2 domain (Figure 3C-D), the latter alterations were also considered gain of function.
Characteristics of IRF4 and STAT3 mutations in ATL. (A) Box plots (median and interquartile values) of variant allele frequencies of recurrent mutations (n = 28) present in >3% of ATL cases. (B) Positions and types of STAT3 mutations detected by targeted capture sequencing for 414 ATL cases. The National Center for Biotechnology Information protein reference sequence for STAT3 is NP_003141.2. (C) Amino acid sequence alignment of Homo sapiens STAT3 with other STAT proteins using the ClustalW algorithm. The mutation and evolutionally conserved residues are shown in cyan and red, respectively. The phosphotyrosine binding sites and β-strands are also shown. (D) Frequently mutated amino acid residues mapped on the STAT3 protein. All recurrently mutated residues are present around the phosphotyrosine-binding pocket.
Characteristics of IRF4 and STAT3 mutations in ATL. (A) Box plots (median and interquartile values) of variant allele frequencies of recurrent mutations (n = 28) present in >3% of ATL cases. (B) Positions and types of STAT3 mutations detected by targeted capture sequencing for 414 ATL cases. The National Center for Biotechnology Information protein reference sequence for STAT3 is NP_003141.2. (C) Amino acid sequence alignment of Homo sapiens STAT3 with other STAT proteins using the ClustalW algorithm. The mutation and evolutionally conserved residues are shown in cyan and red, respectively. The phosphotyrosine binding sites and β-strands are also shown. (D) Frequently mutated amino acid residues mapped on the STAT3 protein. All recurrently mutated residues are present around the phosphotyrosine-binding pocket.
Association of genetic features with clinical outcomes
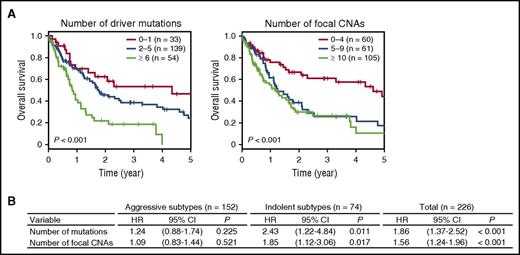
We assessed the effects of somatic lesions on the prognosis of 226 untreated ATL patients, including acute (n = 125, 55%), lymphoma (n = 27, 12%), chronic (n = 59, 26%), and smoldering (n = 15, 7%) patients, for whom survival data were available (Table 1). The number of somatic alterations significantly affected prognosis (Figure 4A). Thus, the 3-year OS rate was 54% for patients with 0 to 1 mutation, 39% for those with 2 to 5 mutations, and only 19% for those with ≥6 mutations (P < .001). Similarly, a higher number of CNAs were significantly associated with a shorter 3-year OS rate: 61%, 26%, and 27% for patients carrying 0 to 4, 5 to 9, and ≥10 CNAs, respectively (P < .001), suggesting the prognostic impact of genetic alterations on survival. We next evaluated the effects of individual alterations on clinical outcomes that were present in ≥10% of patients in the entire cohort. In a univariate analysis, somatic mutations involving PRKCB and IRF4 were significantly associated with a poor outcome (Table 2; supplemental Figure 3). In contrast, STAT3 mutations were predictive of a favorable prognosis, even when analyzed in patients with other genetic alterations (Table 2; supplemental Figures 3 and 4). In addition, focal amplifications in 9p24 (PD-L1) and 4 deletions in 6p22 (ATXN1), 6q21 (PRDM1), 9p21 (CDKN2A), and 13q32 (GPR183) were also significant predictors of an inferior survival. After adjustment for disease subtype and age, PRKCB and IRF4 mutations and PD-L1 amplifications remained significant (Table 2), suggesting a major role of these alterations in the progression and aggressiveness of ATL.
Prognostic impact of the accumulation of somatic alterations in ATL. (A) Kaplan-Meier survival curves of OS of 226 previously untreated ATL cases stratified by the cumulative numbers of driver mutations (left) and focal CNAs (right). The prognostic impact on OS was evaluated by log-rank test. (B) HRs for OS according to the number of significant mutations and focal CNAs on univariate analysis in patients with aggressive (n = 152) and indolent (n = 74) subtypes and in the entire cohort (n = 226). The prognostic impact on OS was evaluated by univariate Cox regression analysis.
Prognostic impact of the accumulation of somatic alterations in ATL. (A) Kaplan-Meier survival curves of OS of 226 previously untreated ATL cases stratified by the cumulative numbers of driver mutations (left) and focal CNAs (right). The prognostic impact on OS was evaluated by log-rank test. (B) HRs for OS according to the number of significant mutations and focal CNAs on univariate analysis in patients with aggressive (n = 152) and indolent (n = 74) subtypes and in the entire cohort (n = 226). The prognostic impact on OS was evaluated by univariate Cox regression analysis.
HRs for OS according to the presence of each genetic alteration in ATL
| Variable . | Univariate . | Adjusted . | ||||
|---|---|---|---|---|---|---|
| HR . | 95% CI . | P . | HR . | 95% CI . | P . | |
| Whole group (n = 226) | ||||||
| Age ≥70 y | 1.84 | 1.27-2.68 | .001 | |||
| Subtype: aggressive | 4.12 | 2.66-6.38 | <.001 | |||
| PRKCB mutations | 1.46 | 1.01-2.12 | .046 | 1.59 | 1.08-2.34 | .019 |
| STAT3 mutations | 0.60 | 0.38-0.95 | .028 | 1.05 | 0.64-1.72 | .837 |
| IRF4 mutations | 2.20 | 1.37-3.54 | .001 | 1.86 | 1.13-3.05 | .015 |
| Amp (9p24): PD-L1 | 2.33 | 1.47-3.71 | <.001 | 2.24 | 1.41-3.58 | .001 |
| Del (6p22): ATXN1 | 1.91 | 1.19-3.08 | .008 | 1.16 | 0.71-1.90 | .564 |
| Del (6q21): PRDM1 | 1.71 | 1.05-2.81 | .033 | 1.36 | 0.82-2.24 | .230 |
| Del (9p21): CDKN2A | 1.78 | 1.21-2.62 | .004 | 1.22 | 0.81-1.82 | .342 |
| Del (13q32): GPR183 | 2.06 | 1.36-3.13 | .001 | 1.14 | 0.73-1.79 | .562 |
| Aggressive subtype (n = 152) | ||||||
| Age ≥70 y | 1.77 | 1.16-2.70 | .009 | |||
| JCOG-PI high-risk | 3.61 | 2.30-5.67 | <.001 | |||
| Treatment | ||||||
| VCAP-AMP-VECP | 0.63 | 0.40-1.00 | .051 | |||
| Other | 1.04 | 0.52-2.05 | .917 | |||
| PRKCB mutations | 1.50 | 0.98-2.28 | .060 | 1.84 | 1.16-2.93 | .010 |
| Amp (9p24): PD-L1 | 1.72 | 1.01-2.94 | .047 | 1.75 | 1.02-3.01 | .042 |
| Del (10p11): CCDC7 | 0.59 | 0.36-0.96 | .034 | 0.74 | 0.44-1.24 | .253 |
| Indolent subtype (n = 74) | ||||||
| Age ≥70 y | 1.57 | 0.69-3.57 | .285 | |||
| Subtype: unfavorable chronic | 3.74 | 1.51-9.26 | .004 | |||
| PLCG1 mutations | 2.26 | 1.07-4.80 | .033 | 1.98 | 0.93-4.22 | .077 |
| VAV1 mutations | 2.44 | 0.96-6.24 | .062 | 1.69 | 0.65-4.38 | .282 |
| IRF4 mutations | 4.23 | 0.93-19.15 | .061 | 4.97 | 1.09-22.67 | .038 |
| Amp (9p24): PD-L1 | 5.09 | 1.93-13.42 | .001 | 4.47 | 1.68-11.87 | .003 |
| Del (7q34): TRB | 2.55 | 1.17-5.59 | .019 | 2.12 | 0.95-4.74 | .067 |
| Del (9p21): CDKN2A | 6.35 | 2.45-16.50 | <.001 | 4.26 | 1.60-11.36 | .004 |
| Variable . | Univariate . | Adjusted . | ||||
|---|---|---|---|---|---|---|
| HR . | 95% CI . | P . | HR . | 95% CI . | P . | |
| Whole group (n = 226) | ||||||
| Age ≥70 y | 1.84 | 1.27-2.68 | .001 | |||
| Subtype: aggressive | 4.12 | 2.66-6.38 | <.001 | |||
| PRKCB mutations | 1.46 | 1.01-2.12 | .046 | 1.59 | 1.08-2.34 | .019 |
| STAT3 mutations | 0.60 | 0.38-0.95 | .028 | 1.05 | 0.64-1.72 | .837 |
| IRF4 mutations | 2.20 | 1.37-3.54 | .001 | 1.86 | 1.13-3.05 | .015 |
| Amp (9p24): PD-L1 | 2.33 | 1.47-3.71 | <.001 | 2.24 | 1.41-3.58 | .001 |
| Del (6p22): ATXN1 | 1.91 | 1.19-3.08 | .008 | 1.16 | 0.71-1.90 | .564 |
| Del (6q21): PRDM1 | 1.71 | 1.05-2.81 | .033 | 1.36 | 0.82-2.24 | .230 |
| Del (9p21): CDKN2A | 1.78 | 1.21-2.62 | .004 | 1.22 | 0.81-1.82 | .342 |
| Del (13q32): GPR183 | 2.06 | 1.36-3.13 | .001 | 1.14 | 0.73-1.79 | .562 |
| Aggressive subtype (n = 152) | ||||||
| Age ≥70 y | 1.77 | 1.16-2.70 | .009 | |||
| JCOG-PI high-risk | 3.61 | 2.30-5.67 | <.001 | |||
| Treatment | ||||||
| VCAP-AMP-VECP | 0.63 | 0.40-1.00 | .051 | |||
| Other | 1.04 | 0.52-2.05 | .917 | |||
| PRKCB mutations | 1.50 | 0.98-2.28 | .060 | 1.84 | 1.16-2.93 | .010 |
| Amp (9p24): PD-L1 | 1.72 | 1.01-2.94 | .047 | 1.75 | 1.02-3.01 | .042 |
| Del (10p11): CCDC7 | 0.59 | 0.36-0.96 | .034 | 0.74 | 0.44-1.24 | .253 |
| Indolent subtype (n = 74) | ||||||
| Age ≥70 y | 1.57 | 0.69-3.57 | .285 | |||
| Subtype: unfavorable chronic | 3.74 | 1.51-9.26 | .004 | |||
| PLCG1 mutations | 2.26 | 1.07-4.80 | .033 | 1.98 | 0.93-4.22 | .077 |
| VAV1 mutations | 2.44 | 0.96-6.24 | .062 | 1.69 | 0.65-4.38 | .282 |
| IRF4 mutations | 4.23 | 0.93-19.15 | .061 | 4.97 | 1.09-22.67 | .038 |
| Amp (9p24): PD-L1 | 5.09 | 1.93-13.42 | .001 | 4.47 | 1.68-11.87 | .003 |
| Del (7q34): TRB | 2.55 | 1.17-5.59 | .019 | 2.12 | 0.95-4.74 | .067 |
| Del (9p21): CDKN2A | 6.35 | 2.45-16.50 | <.001 | 4.26 | 1.60-11.36 | .004 |
Top, HRs for OS associated with age (<70 years vs ≥70 years), subtype (aggressive vs indolent), and genetic alterations in 226 ATL patients by univariate analyses. Values adjusted for disease subtype and age are also shown. Middle, HRs for OS associated with age, JCOG-PI category (high-risk vs moderate-risk), treatment content (VCAP-AMP-VECP or others vs CHOP/CHOP-like), and genetic alterations in 152 aggressive ATL patients by univariate analyses. Values adjusted for age, JCOG-PI, and treatment content are also shown. Bottom, HRs for OS associated with age, subtype (unfavorable chronic vs favorable chronic and smoldering), and genetic alterations in 74 indolent ATL patients by univariate analyses. Values adjusted for age and subtype are also shown. Recurrently mutated genes (n = 13), as well as focal amplifications (n = 4) and deletions (n = 16) present in >10% of ATL cases were examined, and only significant alterations in univariate analyses are shown. The prognostic impact on OS was evaluated by univariate and multivariate Cox regression analyses.
Amp, amplification; AMP, doxorubicin, ranimustine, and prednisone; CI, confidence interval; Del, deletion; VCAP, vincristine, cyclophosphamide, doxorubicin, and prednisone; VECP, vindesine, etoposide, carboplatin, and prednisone.
Multivariate risk stratification of ATL patients according to genetic alterations
Finally, we evaluated the relative effects of different mutations and CNAs using Cox proportional hazards modeling with a stepwise variable selection, incorporating age, disease subtype, and molecular status as covariates. We found that disease subtype (aggressive disease), older age (≥70 years), PRKCB mutations, and PD-L1 amplifications were independently associated with a shorter OS, of which disease subtype (aggressive vs indolent) was the most significant predictor of clinical outcome of ATL patients (supplemental Table 2). Importantly, the effects of genetic alterations strongly depended on disease subtype: the number of somatic mutations and focal CNAs were significantly correlated with an inferior outcome in indolent ATL, but not in aggressive ATL (Figure 4B). In particular, PRKCB mutations were associated with a shorter OS in aggressive subtypes, but not in indolent subtypes (Table 2). By contrast, IRF4 mutations and focal deletions in CDKN2A significantly predicted a worse prognosis in indolent subtypes, but not in aggressive ones (Table 2). In addition, several clinical prognostic factors have been evaluated in aggressive and indolent diseases separately. In aggressive ATL, chemotherapy regimen and JCOG-PI were reported to influence clinical outcomes,2-4,18,19 whereas the presence of any of 3 clinical factors, including low albumin and elevated BUN and LDH levels, has been shown to identify a subset of indolent ATL showing worse OS.2-4 Indeed, these clinical factors were or tended to be associated with OS in our study (Table 2; supplemental Figure 5). Therefore, in the subsequent multivariate analyses, we stratified patients according to disease subtype, incorporating subtype-specific clinical prognostic factors.
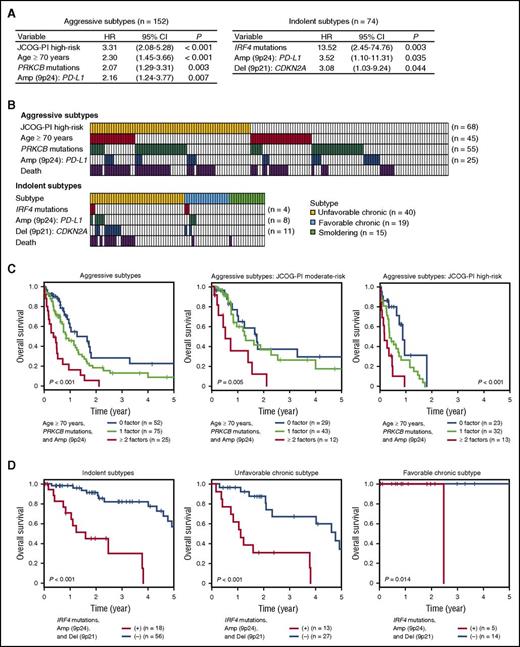
For aggressive ATL, PRKCB mutations and PD-L1 amplifications, together with JCOG-PI high-risk (high calcium level and/or poor performance status) and older age (≥70 years), were independently associated with an adverse outcome (Figure 5A). Providing that JCOG-PI high-risk was confirmed to be a strong predictor for worse prognosis, as previously reported,18 we assessed the prognostic values of other risk factors within patients stratified by the JCOG-PI. Of the 152 patients with aggressive ATL, 68 (45%) belonged to the JCOG-PI high-risk category, and 100 (65%) had ≥1 other risk factor (Figure 5B). Based on the number of these relevant risk factors (older age, PRKCB mutations, and PD-L1 amplifications) they had, patients with aggressive ATL were classified into 3 categories showing significantly different 1-year OS rates (P < .001): 58% for those with no risk factor, 45% for those with 1, and 16% for those with ≥2 risk factors (Figure 5C). This classification remained significant even when JCOG-PI moderate-risk and high-risk patients were analyzed separately (Figure 5C). Moreover, the prognostic impact of this classification was maintained when analyzed only for patients who did not undergo HSCT (supplemental Figure 6A). Thus, the evaluation of the molecular status of PRKCB mutations and PD-L1 amplifications, as well as patient age and JCOG-PI classification, would be informative in prognostication of aggressive ATL.
Multivariate risk classification of patients with aggressive and indolent ATL. (A) Cox proportional hazards model identifying independent significant risk factors for OS in 152 aggressive (left) or 74 indolent ATL (right) cases. The final Cox model resulted from a backward elimination process that considered genetic alterations having a univariate Cox P value < .10 and clinical factors (age, JCOG-PI, and treatment content in aggressive ATL, or age and subtype in indolent ATL). (B) Distribution of risk factors showing overlap between patients with JCOG-PI high-risk, older age, PRKCB mutations, and PD-L1 amplifications in aggressive ATL (top) or those with IRF4 mutations, PD-L1 amplifications, and CDKN2A deletions in indolent ATL (bottom). Patients who died within 1 year in aggressive ATL and within 3 years in indolent ATL are also shown. (C) Kaplan-Meier survival curves of 152 aggressive ATL cases and those categorized as JCOG-PI moderate-risk (n = 84) and high-risk (n = 68), stratified by the number of risk factors (older age, PRKCB mutations, and PD-L1 amplifications). (D) Kaplan-Meier survival curves of 74 indolent (chronic and smoldering), 40 unfavorable chronic, and 19 favorable chronic ATL cases, stratified by the presence of risk factors (IRF4 mutations, PD-L1 amplifications, and CDKN2A deletions). The chronic subtype was classified into unfavorable and favorable subtypes based on the presence of clinical prognostic factors (low albumin, high BUN, and high LDH levels). The prognostic impact on OS was evaluated by log-rank test. CI, confidence interval.
Multivariate risk classification of patients with aggressive and indolent ATL. (A) Cox proportional hazards model identifying independent significant risk factors for OS in 152 aggressive (left) or 74 indolent ATL (right) cases. The final Cox model resulted from a backward elimination process that considered genetic alterations having a univariate Cox P value < .10 and clinical factors (age, JCOG-PI, and treatment content in aggressive ATL, or age and subtype in indolent ATL). (B) Distribution of risk factors showing overlap between patients with JCOG-PI high-risk, older age, PRKCB mutations, and PD-L1 amplifications in aggressive ATL (top) or those with IRF4 mutations, PD-L1 amplifications, and CDKN2A deletions in indolent ATL (bottom). Patients who died within 1 year in aggressive ATL and within 3 years in indolent ATL are also shown. (C) Kaplan-Meier survival curves of 152 aggressive ATL cases and those categorized as JCOG-PI moderate-risk (n = 84) and high-risk (n = 68), stratified by the number of risk factors (older age, PRKCB mutations, and PD-L1 amplifications). (D) Kaplan-Meier survival curves of 74 indolent (chronic and smoldering), 40 unfavorable chronic, and 19 favorable chronic ATL cases, stratified by the presence of risk factors (IRF4 mutations, PD-L1 amplifications, and CDKN2A deletions). The chronic subtype was classified into unfavorable and favorable subtypes based on the presence of clinical prognostic factors (low albumin, high BUN, and high LDH levels). The prognostic impact on OS was evaluated by log-rank test. CI, confidence interval.
Multivariate modeling of the patients with indolent ATL showed that IRF4 mutations, PD-L1 amplifications, and CDKN2A deletions were among the independent predictors of a poor prognosis, although the unfavorable chronic subtype was marginally not significant (hazard ratio [HR], 0.96-6.78; P = .060) (Figure 5A). Nearly one-fourth of the patients (n = 18, 24%) carried ≥1 of these driver alterations, accounting for 59% of the total deaths (within 3 years) in the patients with indolent ATL (Figure 5B). All of these alterations were more frequent in aggressive ATL (Figure 2G-H), among which IRF4 mutations and CDKN2A deletions were independently enriched in aggressive ATL in the multivariate logistic regression model (Figure 2I). More importantly, based on these risk factors, patients with indolent ATL can be classified into 2 categories showing remarkably different prognosis, where the 3-year OS rate was 82% vs 30% for patients without and with risk factors (P < .001) (Figure 5D). In addition, for patients with the unfavorable chronic subtype, the presence of these factors highlighted a subset with significantly shorter OS (3-year OS rate, 67% vs 31%). The prognostic power of this classification remained significant when evaluated only in patients who did not receive HSCT (supplemental Figure 6B). Our results suggest that patients with indolent ATL harboring a genetic feature of the aggressive subtypes might clinically and biologically represent a distinct subset with a worse prognosis, and would be better managed if they were considered to have aggressive disease.
Discussion
On the basis of large-scale genotyping data, we are the first to demonstrate the clinical effects of genetic abnormalities, pointing to the significance of genetic profiling in terms of better classification and prognostication in ATL. Among the recurrent alterations, PD-L1 amplifications were a powerful predictor for an adverse outcome in both aggressive and indolent ATL. Conspicuously, a molecular profile pathognomonic of aggressive subtypes (IRF4 mutations, PD-L1 amplifications, and CDKN2A deletions) significantly predicted a poor prognosis in indolent ATL. These data enable us to identify a subset of patients who would likely benefit from more intensive treatment, such as combined chemotherapy and/or allogeneic HSCT. The identification of the same molecular alterations recognized by different analyses on disease subtype and patient survival confirms the malignant phenotype conferred by these alterations and their biological significance as a poor prognostic indicator. Taken together, these findings suggest that somatic alterations in specific genes help account for the clinical heterogeneity of ATL and that the identification of these abnormalities would improve the prediction of prognosis in ATL patients.
Through a comprehensive analysis of molecular profiles in ATL, we found that IRF4 and TP53 mutations and many focal CNAs, such as PD-L1 amplifications and CDKN2A deletions, were characteristic of aggressive subtypes, suggesting that acquisition of these somatic alterations contributes to acute transformation of indolent ATL. By contrast, significantly enriched in the indolent ATL subtypes, STAT3 mutations suggest a distinct molecular pathogenesis between aggressive and indolent forms. In addition, frequently observed in other mature T- or NK-cell neoplasms, including T-LGL and chronic lymphoproliferative disorders of NK cells, STAT3 mutations are characterized by indolent clinical and biological behavior.24,25 Therefore, activating STAT3 mutations are implicated in the slowly progressive expansion of clonal mature T and NK cells.
Among the significant predictors of a poor prognosis in ATL, PD-L1 amplifications are of particular therapeutic value, because they might be a plausible target of immune checkpoint blockade using anti-PD1/PD-L1 antibodies, which has demonstrated excellent efficacy in a variety of human malignancies.27,28 Importantly, we have recently reported that elevated PD-L1 expression in ATL is strongly associated with a 3′-UTR truncation of the PD-L1 gene, which is caused by a variety of structural variations, frequently accompanied by gene amplifications affecting the relevant locus.29 Currently, the relationship between PD-L1 amplifications and PD-L1 3′-UTR–involving structural variations remains to be elucidated. However, given that an excellent response has been obtained for Hodgkin lymphoma, which frequently harbors PD-L1 amplifications, targeting PD-L1 expression using anti–PD-1/PD-L1 antibodies should be a promising therapeutic strategy for ATL.30,31 Because HTLV-1–derived proteins, such as tax, are highly immunogenic, escape from immune surveillance is thought to be crucial for ATL cells.6,7 Represented by PD-L1 aberrations, the genetic alterations associated with immune modulation are likely to play a critical role in ATL progression and therapeutic resistance, although their precise roles, including those against antitumor immunity in HSCT, need to be clarified.
IRF4 mutations, more frequently found in subclones, were the strongest indicator of worse prognosis in indolent ATL. Along with the observations that IRF4 (also known as MUM1) expression is associated with inferior outcome in a variety of non-Hodgkin lymphomas, including ATL,32,33 these results indicate the pivotal role of IRF4 in clonal evolution and therapeutic resistance not only in B-cell lymphomas, but also in ATL. These mutations are also an attractive therapeutic target, because lenalidomide was reported to kill activated B-cell–like diffuse large B-cell lymphoma cells through the downregulation of IRF4.34 In addition, a recent clinical trial of lenalidomide monotherapy has shown meaningful antitumor activity in patients with relapsed or recurrent aggressive ATL.35 Therefore, lenalidomide may overcome the poor prognosis conferred by IRF4 mutations.
In conclusion, based on comprehensive genetic profiling, we have demonstrated that the known subtypes of ATL could be further classified into genetically and biologically distinct subsets of tumors characterized by discrete sets of genetic lesions and substantially different prognosis. Our results suggest that molecular profiling using next-generation sequencing and/or microarrays could potentially improve the prediction of prognosis in ATL patients and better guide therapy options, such as early intervention with combined chemotherapy and/or allogeneic HSCT in indolent ATL. Further independent studies, particularly those in other ethnic populations, are warranted to validate the clinical utility of molecular profiling in ATL.
Presented in abstract form at the 57th annual meeting of the American Society of Hematology, Orlando, FL, 6 December 2015.
The data reported in this article have been deposited in the European Genome-phenome Archive (accession number EGAS00001001296).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the Joint Study on Predisposing Factors of ATL Development investigators for sample collection and Miki Sagou, Maki Nakamura, and Hitomi Higashi for technical assistance. The supercomputing resources were provided by the Human Genome Center, The Institute of Medical Science, The University of Tokyo.
This work was supported by Grant-in-Aid from the Japan Agency for Medical Research and Development (Practical Research for Innovative Cancer Control [15Ack0106014h0002, 17ck0106261h0001], Research on Development of New Drugs [17ak0101064h0001], and Medical Research and Development Programs Focused on Technology Transfer [15im0210102h0001]), Grant-in-Aid for Scientific Research (KAKENHI 22134006, 15H05909, 16H06249, and 16H06277), and National Cancer Center Research and Development Funds (26-A-6).
Authorship
Contribution: K.K. and S.O. designed the study, had access to the raw data, analyzed the data, and wrote the manuscript; Y. Shiraishi, K.C., H.T., and S. Miyano developed sequence data processing pipelines; Y.N., A.S.-O., M.S., Y.O., K.A., H.S., Y. Shiozawa, T.Y., Y. Sato, K.Y., and T.S. performed sequencing data analysis; R.I. and O.N. performed structural analysis; M.I., J.-i.Y., A.K., T.K., M.Y., K.N., M.H., H.I., Y.I., W.M., K. Shide, Y.K., T.H., T.N., K. Ishiyama, S. Miyawaki, K.T., Y.M., A.T.-K., K. Ishitsuka, A.U., K. Shimoda, M.M., and T.W. managed the patients and prepared the samples; S.O. had full access to all the data in the study and had final responsibility for the decision to submit for publication; and all authors reviewed the manuscript during its preparation.
Conflict-of-interest disclosure: K.K. and S.O. have a patent application pending related to findings from this study. The remaining authors declare no competing financial interests.
Correspondence: Seishi Ogawa, Department of Pathology and Tumor Biology, Graduate School of Medicine, Kyoto University, Yoshida Konoe-cho, Sakyo-ku, Kyoto 6068501, Japan; e-mail: sogawa-tky@umin.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal