TO THE EDITOR:
Although frequently effective1,2 and usually benign, high-dose (2 g/kg) intravenous immunoglobulin (IVIG) therapy can result in marked red blood cell (RBC) hemolysis, which in some cases is life threatening in severity.3-5 The mechanism by which this hemolysis occurs is not completely understood but appears to involve the binding of isohemagglutinins within the product (anti-A, anti-B, and anti-A,B) to the cognate antigen on recipient RBCs,6,7 with these opsonized RBCs subject to antibody-dependent cell-mediated phagocytosis. This process has been reproduced in vitro through the use of a modified monocyte monolayer assay4 and is the likely reason why virtually all patients who develop verified IVIG-associated hemolysis are non–blood group O recipients of high-dose therapy.4,5 Nonetheless, only a subset of this high-risk subgroup will hemolyze, suggesting that there may be other factors influencing susceptibility.4,8-11
Three plausible risk factors to examine for IVIG-associated hemolysis are ABO zygosity, ABH secretor status, and Fcγ receptor (FcγR) polymorphisms. Specifically, because there is a general correlation between the extent of hemolysis and antigen density, 7 individuals who are genetically AA, BB, or AB would conceivably be more prone to IVIG-associated hemolysis than would those who are AO or BO, respectively. Conversely, because there is some evidence that the incidence of IVIG-mediated hemolysis varies with isohemagglutinin titer,6 the presence of neutralizing substances in the plasma of non–blood group O individuals may interfere with erythrocyte binding of isohemagglutinins (eg, the soluble A and B substances in so-called secretors with functional FUT212 ), thereby protecting against hemolysis. Finally, because IVIG-associated hemolysis is presumably triggered by the binding of the Fcγ portion of RBC-bound immunoglobulin G (IgG)–class isohemagglutinins to the FcγRs expressed by monocytes and macrophages,4,7,11 both the number (as defined by FcγR gene copy number) and relative avidity (as defined by specific FcγR genetic polymorphisms) of these receptors might modulate susceptibility to IVIG-mediated hemolysis.
A total of 42 non–blood group O patients who received high-dose IVIG (≥2 g/kg adjusted for lean body mass13 ) had samples collected for genetic testing. The 42 were derived from 3 cohorts: 4 patients from an initial prospective hemovigilance pilot (patients 1-4); a larger, prospective, multicenter, observation study with 31 patients (ClinicalTrials.gov; NCTO2259478) (n = 31/78 patients; patients 5-36); and a passive hemovigilance jurisdiction with 6 patients (patients 37-42) (for more details, see supplemental Data, Materials, and Methods, available on the Blood Web site). The protocol was reviewed and approved by each hospital’s research ethics board. All patients gave written informed consent. The definition of hemolysis was based on the Canadian IVIG Hemolysis Pharmacovigilance Group guidelines14 (see supplemental Data, Materials, and Methods for a complete definition of hemolysis as it is used in this study and supplemental Table 1 for a list of patients studied and clinical data). DNA isolation and genotyping for ABO was done in Lund (Sweden), as has been described.15-20 Secretor FUT2 genotyping was done in Lund (Sweden) (see supplemental Data, Materials, and Methods). Genotyping for Fcγ receptor copy number and polymorphisms was performed in Amsterdam (The Netherlands), as has been described.21-23
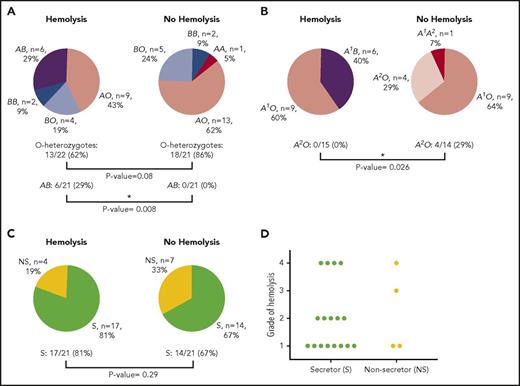
Of the 42 patients studied, zygosity analysis showed a trend toward mitigation of IVIG-associated hemolysis by the O allele. In AA (n = 1 as A1/A2), BB (n = 4), and A1B (n = 6) versus AO (n = 22) and BO (n = 9), the odds ratio for AO and BO groups evading hemolysis was 3.7 (95% CI: 0.8-16.7); 13 out of 21 patients (62%) were AO or BO among those with hemolysis versus a somewhat higher proportion of 18 of 21 (86%) among those patients without hemolysis (P = .080) (Figure 1A-B). In turn, the group-specific incidence of hemolysis was highest for group A1B (6 of 6, or 100%), in comparison with group A (genotype AO or AA; 9 of 23, or 39%; P = .0078) or group B (genotype BO or BB; 6 of 13, or 46%; P = .024). Of the group A (genotypes AA and AO) recipients (n = 23), 0 of 15 (0%) hemolysis patients had a group A2 allele, and 4 of 14 (29%) of those without hemolysis did (P = .026), which is consistent with a lower antigen density of group A2 in comparison with group A1.24 It could not be determined with any accuracy, given the small sample size studied, whether patients with phenotype A1B (genotype A1B) had more severe hemolysis than did those with genotypes A1O or A1A2 (we had no A1A1 patients) (significance was P = .439 when restricting the comparison with only patients who hemolyzed). An interesting finding was that 11 patients who had a positive direct antiglobulin test (weak to 2+) and a positive eluate did not have other evidences of hemolysis, suggesting that other factors such as an activated mononuclear phagocyte system could be involved in those patients who did hemolyze with similar serologic findings.
ABO and FUT2 (secretor) genotyping results. (A) ABO results of patients who hemolyzed in comparison with those who did not hemolyze following IVIG therapy. AB represents A1B genotype. (B) Genotyping results for blood group A patients who hemolyzed in comparison with those who did not hemolyze following IVIG therapy. (C) FUT2 genotyping for secretor status of patients who hemolyzed in comparison with those patients who did not hemolyze following IVIG therapy. (D) Severity of hemolysis following IVIG therapy for secretors in comparison with nonsecretors. Because both compensated hemolysis and grade 1 hemolysis are mild, they are grouped together. P values were determined by Fisher’s exact test. Asterisk represents a significant difference. S, secretor; NS, nonsecretor.
ABO and FUT2 (secretor) genotyping results. (A) ABO results of patients who hemolyzed in comparison with those who did not hemolyze following IVIG therapy. AB represents A1B genotype. (B) Genotyping results for blood group A patients who hemolyzed in comparison with those who did not hemolyze following IVIG therapy. (C) FUT2 genotyping for secretor status of patients who hemolyzed in comparison with those patients who did not hemolyze following IVIG therapy. (D) Severity of hemolysis following IVIG therapy for secretors in comparison with nonsecretors. Because both compensated hemolysis and grade 1 hemolysis are mild, they are grouped together. P values were determined by Fisher’s exact test. Asterisk represents a significant difference. S, secretor; NS, nonsecretor.
Secretor status, as defined by the presence of a functional FUT2 gene product, showed no correlation with hemolysis, which occurred in 17 of 31 secretors (55%) and 4 of 11 (36%) nonsecretors (P = .29) (Figure 1C). There also did not appear to be any difference in hemolytic severity based on secretor status, with grade 4 hemolysis in 4 of 17 (24%) secretors and 1 of 4 (25%) nonsecretors (P = .95) versus compensated or grade 1 hemolysis in 8 of 17 (47%) secretors and 2 of 4 (50%) nonsecretors (P = .91) (Figure 1D). Thus, we conclude that the presence of soluble A/B antigen does not protect against hemolysis (Table 1).
ABO and FUT2 (secretor) genotyping of hemolyzing and nonhemolyzing patients
| Patient . | Age . | Sex . | Hemolysis (Y/N) . | Hemolysis grade . | Genotype FUT2 . | Secretor status . | Phenotype ABO, RhD . | Genotype ABO* . |
|---|---|---|---|---|---|---|---|---|
| 1 | 21 | M | Compensated† | 385AA 428GG | S | A pos | A1.01/O.01.02 | |
| 2 | 54 | F | N | 385AA 428AA | NS | B pos | B.01/B.01 | |
| 3 | 51 | F | Y | 2 | 385AA 428AG | S | A pos | A1.01/O.01.01 |
| 4 | 50 | M | N | 385AA 428GG | S | B pos | B.01/O.01.01 | |
| 5 | 77 | F | N | 385AA 428GG | S | A pos | A1.01/O.01.01 | |
| 6 | 73 | M | Y | 1 | 385AA 428AG | S | A pos | A1.01/O.01.02 |
| 7 | 34 | F | N | 385AA 428AA | NS | A pos | A1.02 /O.01.02 | |
| 8 | 27 | M | N | 385AA 428GG | S | A pos | A1.01/O.01.01 | |
| 9 | 39 | F | Y | 1 | 385AA 428AG | S | A pos | A1.01/O.01.01 |
| 10 | 53 | F | N | 328AA 428GG | S | B pos | B.01/O.01.01 | |
| 11 | 74 | M | N | 385AA 428AA | NS | A pos | A1.01/O.01.02 | |
| 12 | 58 | F | Y | 1 | 328AA 428GG | S | AB pos | A1.01/B.01 |
| 13 | 64 | M | N | 328AA 428AG | S | A pos | A2.01/O.01.01 | |
| 14 | 70 | M | N | 385AA 428AA | NS | A pos | A2.01/O.01.02 | |
| 15 | 60 | M | N | 385AA 428GG | S | B pos | B.01/O.01.01 | |
| 16 | 60 | M | Y | 3 | 385AA 428AA | NS | A pos | A1.01/O.01.01 |
| 17 | 76 | M | N | 385AA 428AG | S | A pos | A1.01/O.01.02 | |
| 18 | 48 | F | N | 385AA 428AG | S | A neg | A1.01/A2.01 | |
| 19 | 72 | F | Y | 2 | 385AA 428AG | S | AB pos | A1.01/B.01 |
| 20 | 46 | F | Y | 4 | 385AA 428GG | S | A pos | A1.01/O.01.01 |
| 21 | 54 | M | N | 385AA 428AA | NS | A pos | A2.01/O.01.01 | |
| 22 | 52 | F | N | 385AA 428AG | S | Apos | A1.01/O.01.01 | |
| 23 | 29 | F | Y | 1 | 385AT 428AG | NS‡ | B pos | B.01/O.01.01 |
| 24 | 60 | F | N | 385AA 428AG | S | A neg | A2.01/O.01.01 | |
| 25 | 43 | M | N | 385AA 428GG | S | A pos | A1.01/O.01.02 | |
| 26 | 62 | F | N | 385AA 428AG | S | B pos | B.01/O.01.01 | |
| 27 | 81 | M | Y | 4 | 385AA 428AG | S | AB pos | A1.01/B.01 |
| 28 | 63 | M | N | 385AA 428AA | NS | A pos | A1.01/O.01.01 | |
| 29 | 53 | M | Y | 2 | 385AA 428AG | S | B pos | B.01/B.01 |
| 30 | 28 | F | Y | 1 | 385AA 428AG | S | B pos | B.01/B.01 |
| 31 | 26 | F | Y | 1 | 385AA 428AA | NS | B pos | B.01/O.01.02 |
| 32 | 63 | F | Y | 2 | 385AA 428AG | S | B pos | B.01/O.01.02 |
| 33 | 75 | F | N | 385AA 428AA | NS | A pos | A1.01/O.01.01 | |
| 34 | 56 | M | N | 385AA 428GG | S | B pos | B.01/O.01.02 | |
| 35 | 80 | F | Y | 2 | 385AA 428GG | S | A pos | A1.01/O.01.02 |
| 36 | 11 | M | N | 385AA 428AG | S | B pos | B.01/B.01 | |
| 37 | 63 | F | Y | 1 | 385AA 428AG | S | A pos | A1.01/O.01.01 |
| 38 | 19 | F | Y | 1 | 385AA 428AG | S | A1B pos | A1.01/B.01 |
| 39 | 59 | F | Y | 1 | 385AA 428GG | S | B pos | B.01/O.01.01 |
| 40 | 63 | M | Y | 4 | 385AA 428AA | NS | A1B pos | A1.01/B.01 |
| 41 | 82 | F | Y | 4 | 385AA 428AG | S | A1pos | A1.02/O.01.01 |
| 42 | 59 | F | Y | 4 | 385AA 428AG | S | A1Bpos | A1.01/B.01 |
| Patient . | Age . | Sex . | Hemolysis (Y/N) . | Hemolysis grade . | Genotype FUT2 . | Secretor status . | Phenotype ABO, RhD . | Genotype ABO* . |
|---|---|---|---|---|---|---|---|---|
| 1 | 21 | M | Compensated† | 385AA 428GG | S | A pos | A1.01/O.01.02 | |
| 2 | 54 | F | N | 385AA 428AA | NS | B pos | B.01/B.01 | |
| 3 | 51 | F | Y | 2 | 385AA 428AG | S | A pos | A1.01/O.01.01 |
| 4 | 50 | M | N | 385AA 428GG | S | B pos | B.01/O.01.01 | |
| 5 | 77 | F | N | 385AA 428GG | S | A pos | A1.01/O.01.01 | |
| 6 | 73 | M | Y | 1 | 385AA 428AG | S | A pos | A1.01/O.01.02 |
| 7 | 34 | F | N | 385AA 428AA | NS | A pos | A1.02 /O.01.02 | |
| 8 | 27 | M | N | 385AA 428GG | S | A pos | A1.01/O.01.01 | |
| 9 | 39 | F | Y | 1 | 385AA 428AG | S | A pos | A1.01/O.01.01 |
| 10 | 53 | F | N | 328AA 428GG | S | B pos | B.01/O.01.01 | |
| 11 | 74 | M | N | 385AA 428AA | NS | A pos | A1.01/O.01.02 | |
| 12 | 58 | F | Y | 1 | 328AA 428GG | S | AB pos | A1.01/B.01 |
| 13 | 64 | M | N | 328AA 428AG | S | A pos | A2.01/O.01.01 | |
| 14 | 70 | M | N | 385AA 428AA | NS | A pos | A2.01/O.01.02 | |
| 15 | 60 | M | N | 385AA 428GG | S | B pos | B.01/O.01.01 | |
| 16 | 60 | M | Y | 3 | 385AA 428AA | NS | A pos | A1.01/O.01.01 |
| 17 | 76 | M | N | 385AA 428AG | S | A pos | A1.01/O.01.02 | |
| 18 | 48 | F | N | 385AA 428AG | S | A neg | A1.01/A2.01 | |
| 19 | 72 | F | Y | 2 | 385AA 428AG | S | AB pos | A1.01/B.01 |
| 20 | 46 | F | Y | 4 | 385AA 428GG | S | A pos | A1.01/O.01.01 |
| 21 | 54 | M | N | 385AA 428AA | NS | A pos | A2.01/O.01.01 | |
| 22 | 52 | F | N | 385AA 428AG | S | Apos | A1.01/O.01.01 | |
| 23 | 29 | F | Y | 1 | 385AT 428AG | NS‡ | B pos | B.01/O.01.01 |
| 24 | 60 | F | N | 385AA 428AG | S | A neg | A2.01/O.01.01 | |
| 25 | 43 | M | N | 385AA 428GG | S | A pos | A1.01/O.01.02 | |
| 26 | 62 | F | N | 385AA 428AG | S | B pos | B.01/O.01.01 | |
| 27 | 81 | M | Y | 4 | 385AA 428AG | S | AB pos | A1.01/B.01 |
| 28 | 63 | M | N | 385AA 428AA | NS | A pos | A1.01/O.01.01 | |
| 29 | 53 | M | Y | 2 | 385AA 428AG | S | B pos | B.01/B.01 |
| 30 | 28 | F | Y | 1 | 385AA 428AG | S | B pos | B.01/B.01 |
| 31 | 26 | F | Y | 1 | 385AA 428AA | NS | B pos | B.01/O.01.02 |
| 32 | 63 | F | Y | 2 | 385AA 428AG | S | B pos | B.01/O.01.02 |
| 33 | 75 | F | N | 385AA 428AA | NS | A pos | A1.01/O.01.01 | |
| 34 | 56 | M | N | 385AA 428GG | S | B pos | B.01/O.01.02 | |
| 35 | 80 | F | Y | 2 | 385AA 428GG | S | A pos | A1.01/O.01.02 |
| 36 | 11 | M | N | 385AA 428AG | S | B pos | B.01/B.01 | |
| 37 | 63 | F | Y | 1 | 385AA 428AG | S | A pos | A1.01/O.01.01 |
| 38 | 19 | F | Y | 1 | 385AA 428AG | S | A1B pos | A1.01/B.01 |
| 39 | 59 | F | Y | 1 | 385AA 428GG | S | B pos | B.01/O.01.01 |
| 40 | 63 | M | Y | 4 | 385AA 428AA | NS | A1B pos | A1.01/B.01 |
| 41 | 82 | F | Y | 4 | 385AA 428AG | S | A1pos | A1.02/O.01.01 |
| 42 | 59 | F | Y | 4 | 385AA 428AG | S | A1Bpos | A1.01/B.01 |
Only the last 4 patients were phenotyped for A1/A2.
F, female; M, male; N, no; neg, negative; NS = nonsecretor; pos, positive; RhD, rhesus factor; S = secretor; Y, yes. Bold indicates patients that hemolyzed.
ABO allele terminology here is according to the International Society of Blood Transfusion (www.isbt-web.org) and the Blood Group Antigen Facts Book.26
Compensated hemolysis is determined by positive direct antiglobulin test, Hgb drop < 10 g/L, and reticulocytosis.
Likely partial secretor (included in the nonsecretor group because very little ABO substance is expected in body fluids).
The relevant single nucleotide polymorphisms (SNPs) and copy number variation (CNV) at the FCGR2/3 locus have been described in detail previously.25 Because of the high-sequence homology in the FCGR2/3 locus, FCGR2C, FCGR3A, and FCGR3B are subject to gene duplications or deletions. Of all 42 individuals, the CNV of the low-affinity FcγRs was determined, as well as the SNPs of these genes (ie, FCGR2A H131R, FCGR2B I232T, FCGR3A V158F, FCGR3B NA1NA2, and FCGR3B SH). The frequencies of the different SNPs and of the open reading frame (ORF) for FCGR2C in the “hemolysis” and the “nonhemolysis” groups were identical (P = .49-1.00; see supplemental Table 2). There was thus no link between the CNV and polymorphisms of low-affinity FcγRs (as defined by the FCGR2/3 locus) and IVIG-associated hemolysis. To explore whether relative IgG receptor avidity might modulate the relative severity of hemolysis, we compared hemolytic grade specifically in FCGR2A 131H homozygotes with that in heterozygotes; no difference in mean hemolytic grade was observed between these 2 groups (results not shown).
On the basis of the results reported herein, we can conclude the following regarding the mechanism of high-dose IVIG-associated hemolysis1 : patients phenotyped as A1B (genotype A1B) are at high risk for IVIG-associated hemolysis, with heterozygosity for the O allele and patients genotyped as A2 exhibiting protection (unfortunately, we did not have any A1A1 homozygous patients in the cohort, so we cannot conclude whether they are at the same high risk as are A1B patients).2 IVIG-associated hemolysis is not related to secretor/nonsecretor status.3 IVIG-associated hemolysis is not a result of genetic determinants of FcγR expression or the known qualitative polymorphisms that may influence the ability of monocyte-macrophages to phagocytose anti-A/anti-B opsonized RBCs.
Although our results suggest that a clinical approach to minimizing the risk of IVIG-associated hemolysis may be to avoid this therapy in those who are blood group A1B or known to be homozygous AA or BB, mass genotyping is still impractical, and proof of efficacy for IVIG substitutes has yet to be established in the relevant conditions. However, if patients who phenotype as blood group AB are to be treated with high-dose IVIG, it would be prudent to monitor these patients more closely for hemolysis.
The online version of this article contains a data supplement.
Acknowledgments
This study was supported by grants from the Canadian Blood Services (XT00113) (D.R.B. and J.P.), the Knut and Alice Wallenberg Foundation (2014.0312) (M.L.O.), and the Swedish Research Council (2014-71X-14251) (M.L.O.), and by governmental ALF grants to the university health care in Region Skåne, Sweden (M.L.O.). Funding is also provided through general resources provided, in part, by Health Canada (D.R.B.).
The views expressed herein do not necessarily represent the view of the federal government of Canada.
Authorship
Contribution: D.R.B., M.L.O., J.P., and C.C.-G. wrote the paper; J.R.S. developed the FUT2 assay; Å.H., J.R.S., and C.W.B. performed genotyping; Å.H., J.R.S., M.L.O., C.W.B., T.W.K., J.P., and C.C.-G. analyzed data and edited the manuscript; D.S., M.B., T.N.T., E.B.-M., and B.B. managed samples; N.P., L.S.R., K.W., C.A., and J.A. managed clinical data and sample collection; J.P., C.C.-G., N.S., K.P., W.L., Y.L., L.L., J.C., V.L., and B.H. provided samples and clinical data.
Conflict-of-interest disclosure: D.R.B. declares research contracts from Grifols and CSL Behring. The remaining authors declare no competing financial interests.
Correspondence: Donald R. Branch, Canadian Blood Services, 67 College St, Toronto, ON M5G 2M1, Canada; e-mail: don.branch@utoronto.ca.
References
Author notes
M.L.O., C.C.-G., and J.P. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal