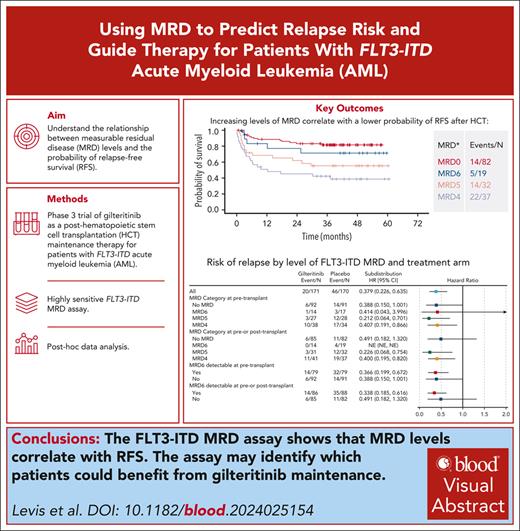
Key Points
FLT3-ITD MRD correlates with RFS.
FLT3-ITD MRD can identify which patients benefit from gilteritinib maintenance.
Visual Abstract
BMT CTN (Blood and Marrow Transplant Clinical Trials Network) 1506 (“MORPHO”) was a randomized study of gilteritinib compared with placebo as maintenance therapy after hematopoietic cell transplantation (HCT) for patients with FLT3-ITD–mutated acute myeloid leukemia (AML). A key secondary end point was to determine the impact on survival of before and/or after HCT measurable residual disease (MRD), as determined using a highly sensitive assay for FLT3-ITD mutations. Generally, gilteritinib maintenance therapy was associated with improved relapse-free survival (RFS) for participants with detectable peri-HCT MRD, whereas no benefit was evident for those lacking detectable MRD. We conducted a post hoc analysis of the data and found that the level of MRD detected with this approach correlated remarkably with RFS and relapse risk, and that MRD detectable at any level negatively affected RFS. In the placebo arm, 42.2% of participants with detectable FLT3-ITD MRD relapsed compared with 13.4% of those without detectable MRD. We found that 14.8% of participants had multiple FLT3-ITD clones detected as MRD and had worse survival irrespective of treatment arm. Finally, we examined the kinetics of FLT3-ITD clonal relapse or eradication and found that participants on the placebo arm with detectable MRD relapsed rapidly after HCT, often within a few weeks. MRD-positive participants on the gilteritinib arm relapsed either with FLT3 wild-type clones (assessed by capillary electrophoresis), after cessation of gilteritinib with persistent MRD, or on progression of multiclonal disease. These data demonstrate the potential of FLT3-ITD MRD to guide therapy with gilteritinib for this subtype of AML. This trial was registered at www.clinicaltrials.gov as #NCT02997202.
Introduction
Attainment of a morphologic complete remission (CR) according to the International Working Group criteria has long represented an early benchmark for therapeutic efficacy in the management of acute myeloid leukemia (AML).1 More recently, assessment of measurable residual disease (MRD) in morphologic remission has emerged as a potential new tool for guiding treatment decisions.2 However, the detection of MRD is more often prognostic than actionable.3
Internal tandem duplication (ITD) mutations of FLT3-ITD are one of the most common mutations in AML.4 They are associated with an aggressive phenotype and, depending on the context of comutations, they generally confer a worse prognosis.5 As the field of AML and MRD has evolved, FLT3-ITD mutations have not traditionally been regarded as useful for MRD monitoring because they are considered late events in leukemogenesis and detecting them at low levels can be technically challenging.6,7 Fortunately, next-generation sequencing (NGS)–based approaches have recently been shown in multiple studies to reliably detect low levels of FLT3-ITD mutations in remission blood or bone marrow samples.8-14 Retrospective studies using banked DNA samples for FLT3-ITD MRD analysis have confirmed the prognostic utility of FLT3-ITD MRD in multiple clinical settings, including before hematopoietic cell transplantation (HCT).14 Some of these studies have also revealed the presence of multiple FLT3-ITD clones (eg, ITD mutations of different lengths in the same patient sample) in a substantial fraction of tested samples.9,10,14 The clinical significance of these multiclonal FLT3-ITD mutations remains unclear.
FLT3 inhibitors are now a standard of care in the treatment of newly diagnosed and relapsed or refractory FLT3-ITD AML.15,16 However, no prospective data to date establish how FLT3-ITD MRD can be used to guide therapy with FLT3 inhibitors. BMT CTN (Blood and Marrow Transplant Clinical Trials Network) 1506 (“MORPHO”; ClinicalTrials.gov identifier: NCT02997202) is a phase 3 randomized double-blinded trial in which participants with FLT3-ITD–mutated AML who had successfully undergone an allogeneic HCT in first remission were randomized to receive 2 years of post-HCT maintenance therapy with the FLT3 inhibitor gilteritinib vs placebo.17 Bone marrow aspirates were collected before HCT and at multiple time points after HCT specifically for FLT3-ITD MRD assessment. A statistically significant difference in relapse-free survival (RFS) between gilteritinib vs placebo as posttransplant maintenance, which was the primary end point, was not detected (P = .0518). However, prespecified subgroup analysis revealed that in those participants with any detectable FLT3-ITD MRD (to a level of 1 × 10−6) in the peri-HCT period (immediately before HCT and/or after HCT but before randomization), maintenance with gilteritinib for 2 years after HCT conferred significantly improved RFS (P = .0065).
Participants randomized to gilteritinib on BMT CTN 1506/MORPHO had a lower incidence of relapse but a higher incidence of myelosuppression, infection, graft-versus-host disease (GVHD), and adverse events leading to discontinuation of study treatment compared with those on the placebo arm. This highlights the importance of understanding how reliable the FLT3-ITD MRD assay is in identifying which patients are at highest risk of relapse, and when the relapse is most likely to occur, so that only patients who are likely to benefit are treated with this therapy. We therefore carried out this post hoc analysis of the BMT CTN 1506/MORPHO MRD data, examining the clinical significance of multiclonal FLT3-ITD mutations detected as MRD as well as the kinetics of FLT3-ITD clonal eradication or relapse. These data demonstrate the remarkable potential for using FLT3-ITD MRD to guide therapeutic decisions for patients with FLT3-ITD–mutated AML.
Methods
Patients
All data herein are derived from participants enrolled on the BMT CTN 1506/MORPHO trial.17 The study population consisted of 356 adult participants aged 18 to 78 years enrolled between 17 August 2017 and 8 July 2020 at 122 centers in 16 different countries. Eligibility criteria for MORPHO required participants to have FLT3-ITD–mutated AML in CR after no more than 1 or 2 courses of intensive chemotherapy, and to proceed to allogeneic HCT within 1 year of achieving remission. Participants with relapsed or refractory disease were excluded. The study was conducted in compliance with the Declaration of Helsinki and approved by institutional review boards at each site. All participants provided informed consent. FLT3 inhibitor use before HCT was not required but was used in 62.6% of participants. At the time of HCT, participants were required to be in ongoing first remission, and for the HCT any type of conditioning, hematopoietic cell source, and GVHD prophylaxis was permitted. Gilteritinib 120 mg/d or placebo was started after engraftment was confirmed and a bone marrow examination showed no morphologic evidence of AML, no sooner than 30 days and no later than 90 days after stem cell infusion. Bone marrow aspirates were collected at defined intervals: (1) before HCT within 30 days before the start of conditioning; (2) after HCT, immediately before randomization; (3) at 3, 6, 12, 18, and 24 months after the start of gilteritinib/placebo; and (4) at the end of treatment. Whenever possible, relapse specimens were centrally analyzed for FLT3 mutations by conventional polymerase chain reaction (PCR) amplification and capillary electrophoresis. Relapses were centrally adjudicated by a blinded committee.
FLT3-ITD MRD assay
In this post hoc study, only patients with a successful MRD result from a marrow aspirate in the peri-HCT period are included. The peri-HCT period is defined as before HCT without any further consolidation (the “pre-HCT bone marrow biopsy”) up until randomization. Before randomization, participants were required to undergo a bone marrow biopsy to confirm the ongoing morphologic absence of leukemia and to collect a sample for MRD. Therefore, there were 2 peri-HCT samples: 1 collected immediately before HCT, and 1 collected after engraftment but before randomization (which was required to take place between 30-90 days after stem cell infusion). The first 2-mL pull of marrow aspirate from any bone marrow sample was designated for MRD analysis, which was performed as described using an amplicon-based approach (Invivoscribe, San Diego, CA).8 Briefly, after genomic DNA extraction, 700 ng DNA was amplified by PCR for 25 cycles using primers flanking exons 14 to 15 of FLT3. NGS was carried out by running purified amplicons from multiple samples simultaneously on an Illumina MiSeq sequencer (Illumina, San Diego, CA) and analyzed using proprietary software (Invivoscribe). The FLT3-ITD variant allele frequency (VAF) was calculated by dividing the number of mutant reads (a minimum of 3) by the number of total reads per sample.
Statistical analysis
Survival times were calculated from the day of randomization. Overall survival, RFS, CR, and relapse were defined by standard criteria.1,18 Survival was estimated using Kaplan-Meier analysis, with differences between groups compared using the 2-sided log-rank test. GraphPad Prism10 was used for graphing and statistical analysis. In the previously published intention-to-treat analysis of this clinical trial,17 participants for whom MRD was not performed before or after HCT were included as MRD negative. In this post hoc study, any participants for whom either or both peri-HCT samples were missing were excluded from analysis.
Results
Marrow aspirates successfully analyzed for peri-HCT FLT3-ITD MRD (eg, both before HCT and after HCT/before randomization) were available from 341 of 356 (95.8%) participants (see CONSORT diagram, supplemental Figure 1, available on the Blood website). In 6 participants the pre-HCT aspirate sample was not sent to the reference laboratory for MRD analysis, and the same was true for 9 completely different participants after HCT/before randomization. These 15 participants were therefore excluded from the present analysis. The baseline characteristics and incidence of relapse for the 15 excluded participants compared with the other 341 participants appear similar (supplemental Table 1). The VAFs or allelic ratios of the FLT3-ITD mutation(s) identified at the original AML diagnosis were not available for most participants, because these were not formal requirements for enrollment. The length of the FLT3-ITD clone identified at diagnosis was voluntarily reported for 124 of 341 (36.4%) participants overall, 65 of whom had MRD detected at some point before or after HCT. Of these 65 participants, the length of an MRD clone matched a reported diagnostic clone in 49 of 65 (75.4%) of cases. Participant characteristics with respect to MRD are displayed in Table 1. Comparing participants with and without MRD, there were no obvious clinical characteristics, including age, sex, karyotype, number of courses of pre-HCT chemotherapy, or conditioning regimen that distinguished MRD-positive from MRD-negative groups. Multiple FLT3-ITD clones were detected in 54 total participants either before HCT, after HCT, or both. Analyzed as a group, the mean, median, and range of VAFs for the individual FLT3-ITD clones were similar between participants with a single clone compared with the 54 participants with multiclonal FLT3-ITD mutations. However, when the VAF for each participant was calculated as the sum of the VAFs of all detectable clones in any given participant, then participants with multiclonal disease had, on average, a ninefold higher VAF than those with only a single clone, before commencing maintenance with gilteritinib or placebo (Table 1).
Participant characteristics with regards to MRD
| . | MRD negative before and after HCT (n = 167) . | MRD positive before and after HCT (n = 174) . | MRD positive after HCT (n = 71) . | Multiclonal FLT3-ITD mutations before and after HCT (n = 54) . |
|---|---|---|---|---|
| Age, median, y | 53 | 53 | 52 | 52.5 |
| Female, n (%) | 89 (53.3) | 75 (43.1) | 33 (46.5) | 23 (42.6) |
| Karyotype, n (%) | ||||
| Favorable | 3 (1.8) | 9 (5.2) | 6 (8.5) | 2 (3.7) |
| Intermediate | 159 (95.2) | 157 (5.2) | 63 (88.7) | 49 (90.7) |
| Adverse | 5 (3.0) | 8 (4.6) | 2 (2.8) | 3 (5.6) |
| NPM1mut, n (%) | 66 (39.5) | 51 (29.3) | 24 (33.8) | 14 (25.9) |
| Median ITD length bp (range) | — | 45 (6-235) | 45 (15-183) | 48 (15-165) |
| VAF, median (VAF/clone) | — | 2.47 × 10–5 | 1.37 × 10–5 | 2.35 × 10–5 |
| VAF, median VAF/pt | — | 4.7 × 10–5 | 1.73 × 10–5 | 1.2 × 10–4 |
| VAF, range | — | 1.09 × 10–6 to 3 × 10–1 | 2.16 × 10–6 to 3 × 10–1 | 2.16 × 10–6 to 3 × 10–1 |
| Conditioning, n (%) | ||||
| MAC | 96 (57.5) | 107 (61.5) | 44 (62) | 31 (57.4) |
| RIC | 71 (42.5) | 67 (38.5) | 27 (38) | 23 (42.6) |
| Courses chemotherapy before HCT, mean; median | 2.9; 3 | 2.5; 2 | 2.5; 2 | 2.3; 2 |
| Days from AML diagnosis to HCT, mean; median | 157; 143 | 137; 127 | 135; 121 | 125; 118 |
| FLT3 inhibitor used before HCT, n (%) | 97 (58.1) | 106 (60.9) | 36 (50.7) | 29 (53.7) |
| . | MRD negative before and after HCT (n = 167) . | MRD positive before and after HCT (n = 174) . | MRD positive after HCT (n = 71) . | Multiclonal FLT3-ITD mutations before and after HCT (n = 54) . |
|---|---|---|---|---|
| Age, median, y | 53 | 53 | 52 | 52.5 |
| Female, n (%) | 89 (53.3) | 75 (43.1) | 33 (46.5) | 23 (42.6) |
| Karyotype, n (%) | ||||
| Favorable | 3 (1.8) | 9 (5.2) | 6 (8.5) | 2 (3.7) |
| Intermediate | 159 (95.2) | 157 (5.2) | 63 (88.7) | 49 (90.7) |
| Adverse | 5 (3.0) | 8 (4.6) | 2 (2.8) | 3 (5.6) |
| NPM1mut, n (%) | 66 (39.5) | 51 (29.3) | 24 (33.8) | 14 (25.9) |
| Median ITD length bp (range) | — | 45 (6-235) | 45 (15-183) | 48 (15-165) |
| VAF, median (VAF/clone) | — | 2.47 × 10–5 | 1.37 × 10–5 | 2.35 × 10–5 |
| VAF, median VAF/pt | — | 4.7 × 10–5 | 1.73 × 10–5 | 1.2 × 10–4 |
| VAF, range | — | 1.09 × 10–6 to 3 × 10–1 | 2.16 × 10–6 to 3 × 10–1 | 2.16 × 10–6 to 3 × 10–1 |
| Conditioning, n (%) | ||||
| MAC | 96 (57.5) | 107 (61.5) | 44 (62) | 31 (57.4) |
| RIC | 71 (42.5) | 67 (38.5) | 27 (38) | 23 (42.6) |
| Courses chemotherapy before HCT, mean; median | 2.9; 3 | 2.5; 2 | 2.5; 2 | 2.3; 2 |
| Days from AML diagnosis to HCT, mean; median | 157; 143 | 137; 127 | 135; 121 | 125; 118 |
| FLT3 inhibitor used before HCT, n (%) | 97 (58.1) | 106 (60.9) | 36 (50.7) | 29 (53.7) |
bp, base pair; MAC, myeloablative conditioning; NPM1mut, NPM1 mutation; pt, participant; RIC, reduced-intensity conditioning.
FLT3 mutation status at relapse
Of 341 participants for whom MRD analysis was performed both before and after HCT, 66 relapsed: 46 (69.7%) in the placebo arm and 20 (30.3%) in the gilteritinib arm. Relapse samples were available for FLT3 mutation analysis for 44 (67.7%) of 66 patients: 12 on the gilteritinib arm and 32 on the placebo arm. The remaining 22 participants without a relapse sample available for analysis differed somewhat in baseline characteristics from the other 44 in that they were older and relapsed later in follow-up (supplemental Table 2). Among the 44 participants for whom the mutation status was analyzed at relapse, of 12 patients on the gilteritinib arm, 8 relapsed with wild-type FLT3 and 4 relapsed with FLT3-ITD mutations. Of 31 patients on the placebo arm, 8 relapsed with wild-type FLT3, 22 with a FLT3-ITD mutation, 1 participant relapsed with a FLT3-D835 mutation, and 1 relapsed with a FLT3 juxtamembrane deletion. The mutation findings for the participants who relapsed are summarized in supplemental Table 3. There were 21 participants from whom an ITD length was available at relapse and in whom an MRD clone was detected either before or after HCT. In all 21 cases, the length of the relapse clone matched that of a detected MRD clone.
Correlation between MRD and relapse
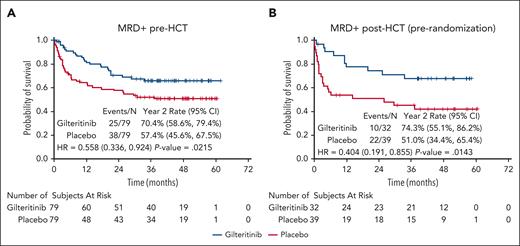
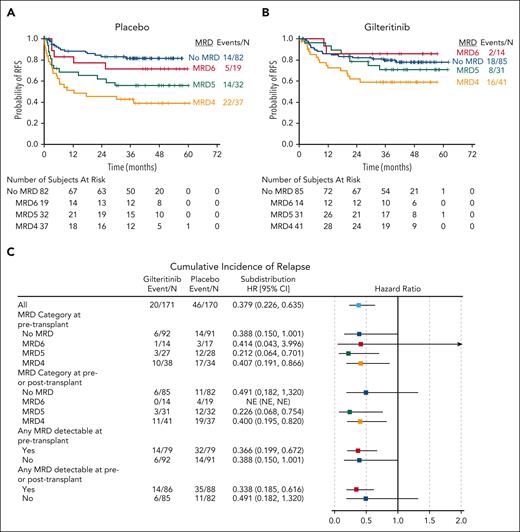
In the primary analysis of BMT CTN 1506/MORPHO,17 participants with peri-HCT MRD (ie, detectable MRD either immediately before, or immediately after HCT and before randomization) had a reduced rate of relapse if they were on the gilteritinib arm compared with those on the placebo arm. A reduced risk of relapse was observed in the gilteritinib arm regardless of whether the MRD was detected before HCT or after HCT, and using either 1 × 10−6 or 1 × 10−4 as a cutoff (Figure 1; supplemental Figures 2-5). Individual participant results are provided in supplemental Table 4. Because of this effect of gilteritinib on the risk of relapse, we used results from the placebo arm to examine the association between MRD and relapse after HCT. Of 88 participants on the placebo arm with detectable peri-HCT MRD, 7 (8.0%) died in remission and 34 (42.0%) of the remaining 81 relapsed. The mutation allelic burden (ie, the VAF) of peri-HCT MRD correlated closely with RFS in the participants on the placebo arm, with higher VAFs associated with decreased RFS (Figure 2A) and increased cumulative incidence of relapse (supplemental Figures 6 and 7). At all levels of MRD, use of maintenance gilteritinib appears to have improved RFS and decreased the incidence of relapse (Figures 2B-C; supplemental Figure 8; supplemental Tables 5-7). However, the absence of detectable peri-HCT MRD did not guarantee against relapse. Of 82 participants in the placebo arm who had no detectable MRD in the peri-HCT period, 11 relapsed. Three of these relapsed with AML lacking any FLT3 mutation. Because FLT3 inhibitor use during induction and consolidation therapy is now known to induce deeper remissions,10,16 we examined whether FLT3 inhibitor treatment before HCT was associated with a negative peri-HCT MRD result. Of 82 MRD-negative placebo arm participants, 47 had been treated before HCT with intensive chemotherapy combined with an FLT3 inhibitor, and only 2 of 47 (4.3%) of these relapsed with a FLT3-ITD mutation (2 others relapsed with wild-type FLT3 AML). The remaining 35 placebo arm participants received induction and consolidation without an FLT3 inhibitor before HCT, and 6 of 35 (17.1%) of these relapsed with a FLT3-ITD mutation.
RFS according to FLT3-ITD MRD detected before or after HCT. (A) RFS by randomization arm for all participants with FLT3-ITD MRD detected immediately before beginning pre-HCT conditioning. (B) RFS by randomization arm for all participants with FLT3-ITD MRD detected after engraftment was confirmed and before randomization to either gilteritinib or placebo. 95% CI, 95% confidence interval; HR, hazard ratio.
RFS according to FLT3-ITD MRD detected before or after HCT. (A) RFS by randomization arm for all participants with FLT3-ITD MRD detected immediately before beginning pre-HCT conditioning. (B) RFS by randomization arm for all participants with FLT3-ITD MRD detected after engraftment was confirmed and before randomization to either gilteritinib or placebo. 95% CI, 95% confidence interval; HR, hazard ratio.
Relapse risk according to MRD level. RFS for participants with no detectable FLT3-ITD MRD is compared with RFS for participants with increasing levels of MRD. No MRD refers to no detectable MRD before or after HCT; MRD6 indicates mutant reads per total reads of ≥10−6 but <10−5; MRD5 indicates mutant reads per total reads of ≥10−5 but <10–4; and MRD4 indicates mutant reads per total reads of ≥10−4. (A) Placebo arm. (B) Gilteritinib arm. (C) Forest plot of cumulative incidence of relapse for gilteritinib vs placebo by level of MRD. 95% CI, 95% confidence interval; HR, hazard ratio; NE, not estimable.
Relapse risk according to MRD level. RFS for participants with no detectable FLT3-ITD MRD is compared with RFS for participants with increasing levels of MRD. No MRD refers to no detectable MRD before or after HCT; MRD6 indicates mutant reads per total reads of ≥10−6 but <10−5; MRD5 indicates mutant reads per total reads of ≥10−5 but <10–4; and MRD4 indicates mutant reads per total reads of ≥10−4. (A) Placebo arm. (B) Gilteritinib arm. (C) Forest plot of cumulative incidence of relapse for gilteritinib vs placebo by level of MRD. 95% CI, 95% confidence interval; HR, hazard ratio; NE, not estimable.
Kinetics of clonal relapse or eradication
There were 71 participants with detectable MRD after HCT (ie, immediately after HCT and before randomization), and we analyzed these cases to better understand the kinetics of clonal relapse and clonal eradication in the presence and absence of gilteritinib. Clinical relapse in this trial was an adjudicated end point defined according to the revised International Working Group criteria.1 We defined clonal relapse as clinical relapse in combination with detection of a FLT3-ITD mutation using the conventional PCR capillary electrophoresis technique. The mutation had to be of identical length as a clone detected using the MRD assay during follow-up after HCT. Clonal eradication was defined as an MRD clone becoming undetectable in follow-up and with the participant neither relapsing nor dying. There were 37 participants with post-HCT MRD randomized to placebo, 20 of whom relapsed, and there were 29 participants with post-HCT MRD randomized to gilteritinib, 7 of whom relapsed. The details of relapse are summarized in supplemental Table 8.
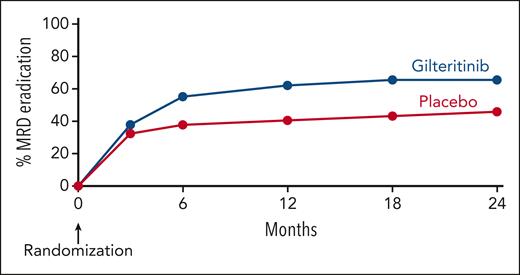
Figure 3 displays the kinetics of clonal relapse and eradication of individual FLT3-ITD clones. Most relapses on the placebo arm occurred quickly, 64.3% (9/14) within 8 weeks of randomization and 93.9% (13/14) by 16 weeks after randomization. Likewise, clonal eradication on the placebo arm, presumably because of the effects of allogeneic HCT, was typically noted on the first MRD assessment (3 months after randomization, 12-14 weeks). In contrast, on the gilteritinib arm, clones were eradicated over a broader range of time points. Figure 4 summarizes the course of clonal eradication for both arms. Post-HCT MRD at any level appeared to affect the relative risk of relapse (supplemental Figure 9), with the impact diminishing as the level of MRD decreased. However, when the data are broken down into these MRD groups, the resultant small numbers preclude being able to draw statistically robust conclusions.
Kinetics of clonal relapse and eradication in participants with post-HCT MRD. Each line on the plots depicts a single FLT3-ITD clone detected with the MRD assay, starting with randomization. In a limited number of cases, a sample was available at relapse, analyzed with conventional capillary electrophoresis PCR, and the length of the FLT3-ITD clone was identical to the clone detected in the MRD assay. These clones are depicted with red lines. The blue lines depict cases in which the relapse specimen was not available for analysis and so the length could not be confirmed. (A) Placebo arm, from randomization to 2 years of follow-up. (B) Placebo arm, depicting only the first 24 weeks of follow-up after randomization. (C) Gilteritinib arm, from randomization to 2 years of follow-up. (D) Gilteritinib arm, depicting only the first 24 weeks of follow-up after randomization.
Kinetics of clonal relapse and eradication in participants with post-HCT MRD. Each line on the plots depicts a single FLT3-ITD clone detected with the MRD assay, starting with randomization. In a limited number of cases, a sample was available at relapse, analyzed with conventional capillary electrophoresis PCR, and the length of the FLT3-ITD clone was identical to the clone detected in the MRD assay. These clones are depicted with red lines. The blue lines depict cases in which the relapse specimen was not available for analysis and so the length could not be confirmed. (A) Placebo arm, from randomization to 2 years of follow-up. (B) Placebo arm, depicting only the first 24 weeks of follow-up after randomization. (C) Gilteritinib arm, from randomization to 2 years of follow-up. (D) Gilteritinib arm, depicting only the first 24 weeks of follow-up after randomization.
Time course of clonal eradication by treatment arm.FLT3-ITD clones detected after HCT (ie, immediately after HCT, before randomization). Clonal eradication is defined as a previously detected FLT3-ITD clone becoming undetectable and not recurring again in a participant who was alive and disease free at completion of follow-up.
Time course of clonal eradication by treatment arm.FLT3-ITD clones detected after HCT (ie, immediately after HCT, before randomization). Clonal eradication is defined as a previously detected FLT3-ITD clone becoming undetectable and not recurring again in a participant who was alive and disease free at completion of follow-up.
Although relapses were expected to occur on the placebo arm, when they occurred on the gilteritinib arm they presumably represented a failure of the maintenance therapy. There were only 21 reported relapse events on the gilteritinib arm (of 178 total randomized to gilteritinib), so it is difficult to draw any statistically robust conclusions as to the specific causes. However, to at least illustrate in anecdotal fashion some of the different circumstances under which relapse occurred while on gilteritinib, 6 cases are displayed in Figure 5. Participants on the gilteritinib who discontinued the study treatment early (for any reason) had a higher incidence of relapse than those who completed the 24 months of maintenance (supplemental Figure 10). Gilteritinib was most commonly interrupted or stopped altogether for reasons of toxicity (eg, count suppression, liver enzyme elevation, etc), and in the cases displayed in Figure 5, this was usually in a setting in which there was a persistent MRD clone. As noted above, there were also relapses with wild-type FLT3. Participants with multiple clones tended to relapse with the clone present at the highest VAF at initial detection (a “dominant” clone), although the relatively few number of relapses overall limits the strength of this conclusion.
Case examples from gilteritinib arm. MRD, relapse, and drug exposures are displayed for 6 participants randomized to gilteritinib. Note that all investigators and participants were blinded to both study drug and MRD status. The colored thin lines represent the FLT3-ITD MRD clones and the thick black bars depict the periods when the participant was being treated with gilteritinib. Pt (participant) 1: 3 clones (21, 30, 45 base pairs [bp]) detected before and after HCT. Started gilteritinib on post-HCT day +41, maintained continuously on 120 mg/d for 24 months. The 30-bp clone was not detected at 2 time points, then rose steadily until 24 months after HCT. Gilteritinib was stopped per protocol, with the pt still in morphologic remission, and the pt relapsed 6 weeks later. Pt 2: single 30-bp clone detected before and after HCT. Started gilteritinib on post-HCT day 43. Clone eradicated, but pt relapsed 8 months after HCT with AML confirmed by PCR/capillary electrophoresis to be wild-type FLT3. Pt 3: single 54-bp clone detected both before and after HCT. Started gilteritinib on post-HCT day 35, with concomitant posaconazole. Lactate dehydrogenase was elevated to grade 3 and gilteritinib was decreased briefly to 80 mg/d, and then discontinued permanently on day +51. Relapsed on post-HCT day 134. Pt 4: single 42-bp clone detected before and after HCT. Started gilteritinib on post-HCT day 86, maintained continuously for 24 months, then stopped per protocol. The 42-bp clone was detected throughout, slowly decreasing in level. Pt alive and relapse free 49 months after HCT. Pt 5: 5 clones detected before HCT, all negative immediately after HCT. A new 159-bp clone appeared after HCT, before randomization. Started gilteritinib with concomitant itraconazole, but the gilteritinib had to be held twice for thrombocytopenia and resumed at 80 mg/d, with a resultant gilteritinib trough level of only 77.5 ng/mL (which is approximately one-third the median trough level for all other participants17). The 159-bp clone became undetectable, but the 21-bp clone that had been detected before HCT reappeared on post-HCT day 142. Pt relapsed with the 21-bp and the 57-bp clone while on 80 mg/d gilteritinib. Pt 6: 2 clones detected before HCT, and a 63-bp clone persisted after HCT. Started gilteritinib on post-HCT day 50 and posaconazole started prophylactically on day post-HCT day 70. The absolute neutrophil count dropped to 100/mm3 on post-HCT day 105 and gilteritinib was stopped. Gilteritinib was resumed on post-HCT day 131 (again with concomitant posaconazole), but the neutrophil count decreased again and the study drug was permanently stopped on post-HCT day 138. Pt relapsed 2 months later with the 63-bp clone.
Case examples from gilteritinib arm. MRD, relapse, and drug exposures are displayed for 6 participants randomized to gilteritinib. Note that all investigators and participants were blinded to both study drug and MRD status. The colored thin lines represent the FLT3-ITD MRD clones and the thick black bars depict the periods when the participant was being treated with gilteritinib. Pt (participant) 1: 3 clones (21, 30, 45 base pairs [bp]) detected before and after HCT. Started gilteritinib on post-HCT day +41, maintained continuously on 120 mg/d for 24 months. The 30-bp clone was not detected at 2 time points, then rose steadily until 24 months after HCT. Gilteritinib was stopped per protocol, with the pt still in morphologic remission, and the pt relapsed 6 weeks later. Pt 2: single 30-bp clone detected before and after HCT. Started gilteritinib on post-HCT day 43. Clone eradicated, but pt relapsed 8 months after HCT with AML confirmed by PCR/capillary electrophoresis to be wild-type FLT3. Pt 3: single 54-bp clone detected both before and after HCT. Started gilteritinib on post-HCT day 35, with concomitant posaconazole. Lactate dehydrogenase was elevated to grade 3 and gilteritinib was decreased briefly to 80 mg/d, and then discontinued permanently on day +51. Relapsed on post-HCT day 134. Pt 4: single 42-bp clone detected before and after HCT. Started gilteritinib on post-HCT day 86, maintained continuously for 24 months, then stopped per protocol. The 42-bp clone was detected throughout, slowly decreasing in level. Pt alive and relapse free 49 months after HCT. Pt 5: 5 clones detected before HCT, all negative immediately after HCT. A new 159-bp clone appeared after HCT, before randomization. Started gilteritinib with concomitant itraconazole, but the gilteritinib had to be held twice for thrombocytopenia and resumed at 80 mg/d, with a resultant gilteritinib trough level of only 77.5 ng/mL (which is approximately one-third the median trough level for all other participants17). The 159-bp clone became undetectable, but the 21-bp clone that had been detected before HCT reappeared on post-HCT day 142. Pt relapsed with the 21-bp and the 57-bp clone while on 80 mg/d gilteritinib. Pt 6: 2 clones detected before HCT, and a 63-bp clone persisted after HCT. Started gilteritinib on post-HCT day 50 and posaconazole started prophylactically on day post-HCT day 70. The absolute neutrophil count dropped to 100/mm3 on post-HCT day 105 and gilteritinib was stopped. Gilteritinib was resumed on post-HCT day 131 (again with concomitant posaconazole), but the neutrophil count decreased again and the study drug was permanently stopped on post-HCT day 138. Pt relapsed 2 months later with the 63-bp clone.
Multiclonal MRD
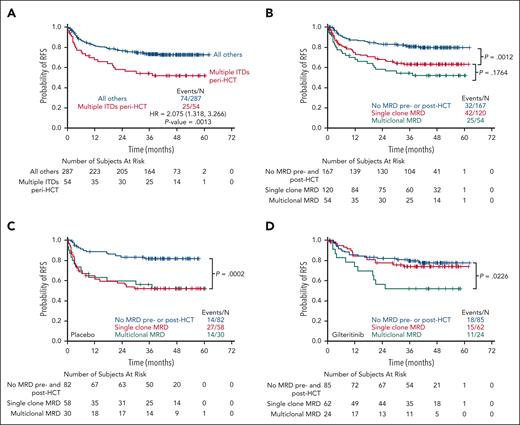
In virtually all prior studies using the PCR-NGS FLT3-ITD MRD platform, multiple low VAF FLT3-ITD clones have been detected in patients in remission. The clinical significance of this multiclonality has been unclear. In this study, 54 of 174 (31%) of participants with peri-HCT MRD had ≥2 FLT3-ITD mutations, and the presence of this multiclonality was associated with a worse RFS irrespective of treatment arm when compared with all other participants on the trial and when compared with participants with MRD consisting of just a single FLT3-ITD clone, although that difference did not reach statistical significance (Figure 6; supplemental Figures 11 and 12). There were no obvious differences in clinical characteristics between participants with multiclonal mutations and those without (Table 1). The only apparent difference was that when the individual VAFs for each participant were summed, the total allelic burden was greater in participants with multiclonality. However, on the placebo arm, participants with single-clone peri-HCT FLT3-ITD MRD had a similar outcome compared with those with multiclonal disease (Figure 6C). In contrast, participants on the gilteritinib arm with single-clonal FLT3-ITD MRD closely resembled those without any peri-HCT MRD, whereas those with multiclonal MRD had a worse RFS (Figure 6D).
Multiclonal FLT3-ITD mutations and RFS. (A) 54 participants were found to have multiple FLT3-ITD clones using the MRD assay peri-HCT. The Kaplan-Meier curve depicts RFS, irrespective of treatment arm, for these 54 participants compared with all other participants. (B) RFS for these same 54 participants compared with those with no MRD before and after HCT or those with only a single FLT3-ITD clone detected either before and after HCT. (C) RFS for participants on the placebo arm according to peri-HCT FLT3-ITD MRD status. The P value refers to the comparison between the “no MRD” and “single-clone MRD.” The P value comparing the single-clone and multiclonal MRD curves is .022 (not shown). (D) RFS for participants on the gilteritinib arm according to peri-HCT FLT3-ITD MRD status. The P value refers to the comparison between the “no MRD” and “multiclonal MRD.” The P value comparing “no MRD” and “single-clonal MRD” curves is .044 (not shown). HR, hazard ratio.
Multiclonal FLT3-ITD mutations and RFS. (A) 54 participants were found to have multiple FLT3-ITD clones using the MRD assay peri-HCT. The Kaplan-Meier curve depicts RFS, irrespective of treatment arm, for these 54 participants compared with all other participants. (B) RFS for these same 54 participants compared with those with no MRD before and after HCT or those with only a single FLT3-ITD clone detected either before and after HCT. (C) RFS for participants on the placebo arm according to peri-HCT FLT3-ITD MRD status. The P value refers to the comparison between the “no MRD” and “single-clone MRD.” The P value comparing the single-clone and multiclonal MRD curves is .022 (not shown). (D) RFS for participants on the gilteritinib arm according to peri-HCT FLT3-ITD MRD status. The P value refers to the comparison between the “no MRD” and “multiclonal MRD.” The P value comparing “no MRD” and “single-clonal MRD” curves is .044 (not shown). HR, hazard ratio.
Discussion
These prospective results, derived from prespecified MRD data collection, clearly establish the potential of using FLT3-ITD MRD to guide therapeutic decisions for patients with AML. They confirm the findings of recent retrospective studies using banked DNA samples collected before HCT, namely that pre-HCT FLT3-ITD MRD, detected using NGS-based methods, predicts for worse outcomes after HCT.11,13,14 At this time, we suggest that regular monitoring of patients for FLT3-ITD MRD using the PCR-NGS approach should be a new standard of care, certainly for patients undergoing allogeneic transplantation, but likely for any patient with FLT3-ITD-mutated AML. Furthermore, the ability of gilteritinib to eradicate MRD provides additional evidence that post-HCT gilteritinib should be standard therapy for any patient with any level of peri-HCT MRD detectable with PCR-NGS. There are multiple FLT3 inhibitors available to intervene if and when MRD is detectable, and similar findings on the prognostic impact of FLT3-ITD MRD detected by PCR-NGS are emerging from studies of other FLT3 inhibitors.16,19
The quantifiable nature of the PCR-NGS assay is important, and with these new data correlating the risk of relapse with level of MRD, we can conclude that there appears to be no level of detectable MRD that is “safe.” These findings are consistent with a recent study using the same MRD assay with banked samples from patients with AML with FLT3-ITD AML undergoing HCT.14 The amount of MRD correlates tightly with relapse risk, whether as a single mutation or multiple mutations. The assay is commercially available worldwide with a methodology that is well described and 1 that can be performed in any laboratory with familiarity with NGS assays.8,10
An important issue is the sensitivity and specificity of the FLT3-ITD MRD assay. There were 21 cases in which a patient with MRD relapsed and the relapse sample was available for FLT3 mutation analysis, and in all 21 cases, the ITD length at relapse matched the ITD length identified as MRD. There was less concordance between the ITD length reported at diagnosis and detected either at MRD or relapse, although what was reported at diagnosis may have been less reliable because it was essentially additional information volunteered by the study sites. When a FLT3-ITD clone was detected in the peritransplant period, the relapse incidence in the placebo arm was 42.2%, which would seem to justify using gilteritinib after HCT when MRD is detectable. In contrast, any association between an MRD result and the potential for relapse should probably be viewed in the context of the patient’s pre-HCT therapy. It is well established that for patients with AML, any MRD detected, regardless of method, before or after HCT adversely impacts post-HCT outcome,20 and it has been more recently established that the use of FLT3 inhibitors with induction and consolidation therapy results in deeper remission with respect to MRD.10,16 For MRD-negative participants on the placebo arm of BMT CTN 1506/MORPHO who had received a FLT3 inhibitor before HCT, only 2 of 47 (4.3%) relapsed with a FLT3-ITD mutation, which probably justifies not starting gilteritinib in this circumstance, particularly if there are active post-HCT complications such as infection or GVHD unless MRD recurs.
Ever since the discovery of FLT3-ITD mutations,21 patients with multiple clones detected by conventional PCR have been recognized.22 The clinical impact of multiclonality has not been clear, but our data here suggest that the presence of multiple FLT3-ITD peri-HCT MRD clones may blunt the beneficial impact of gilteritinib. In the case examples illustrated in Figure 5, participants 1, 5, and 6 had multiclonal disease, which was persisting despite gilteritinib therapy, and stopping or dose-reducing gilteritinib was followed by relapse. These findings lead us to speculate that there may be a different biology for multiclonal FLT3-ITD disease. Regardless, these admittedly limited data suggest that dose interruption or reduction should be avoided at all costs in patients with peri-HCT multiclonal MRD on gilteritinib maintenance. Further investigation into the biology and impact of FLT3-ITD multiclonality in other clinical settings is clearly warranted.
Based on these data, combined with the data from the primary analysis and other published data,23 we offer guidance on both the general use of gilteritinib after HCT, as well as in the setting in which FLT3-ITD MRD is detectable before and/or after HCT. Clinicians should be aware that high steady-state trough levels of gilteritinib can cause myelosuppression, particularly when the drug is administered followed intensive chemotherapy or HCT. They should also be aware that the drug has a half-life of almost a week, and, as with most CYP3A4-metabolized drugs, has trough levels with high interpatient variability.24 Concomitant use of gilteritinib with strong CYP3A4 inhibitors such as posaconazole, voriconazole, or itraconazole can often, but not always, lead to higher steady-state trough levels.17 All of this must be balanced against the relative benefit that gilteritinib may confer on the patient, based in a large part on the presence or absence of detectable FLT3-ITD MRD.
If there is detectable MRD (at any level) immediately before HCT, every effort should be made to use gilteritinib after HCT, and to minimize disruptions of this treatment. Given the rapid kinetics of relapse we observed, gilteritinib should be started as early as possible after engraftment is confirmed. When there are medical issues making this problematic, MRD analysis from a bone marrow biopsy obtained relatively soon after HCT can establish whether relapse is imminent or whether gilteritinib can be delayed. Once gilteritinib is started in a patient with MRD before or after HCT, it should be continued for a 2-year period, and only discontinued if a repeat MRD assessment after that time is negative. When multiclonal disease is detected, every effort should be made to avoid interruptions in gilteritinib and, again, it should not be discontinued until it can be confirmed that there is no detectable MRD. Monitoring MRD during this period may be performed, but we do not have any data on what might be an effective intervention in the case of a rising clone. Certainly, donor lymphocyte infusions, gilteritinib dose increases, or switching to an alternative FLT3 inhibitor can be considered as potential interventions in these cases.
Because 4.5% of participants on the trial only had post-HCT MRD and because many relapses occur early, for patients who are MRD negative before HCT, we recommend starting gilteritinib as soon as possible after engraftment and continuing until an MRD assessment can be obtained to confirm the absence of post-HCT MRD. For patients with no peri-HCT MRD, gilteritinib therapy can be stopped, although ongoing MRD monitoring at frequent intervals can be considered, with the knowledge that gilteritinib can be added as an intervention should detectable MRD appear.
In summary, this post hoc study of the BMT CTN 1506/MORPHO data set provides clinicians with more granular details regarding the use of FLT3-ITD MRD and gilteritinib in the setting of allogeneic transplantation. The FLT3-ITD MRD assay is likely to become incorporated into standard-of-care practice at all phases of the disease, and in the context of therapy with other FLT3 inhibitors.
Acknowledgments
Support for this study was provided by grants to the Blood and Marrow Transplant Clinical Trials Network from the National Heart, Lung, and Blood Institute, National Institutes of Health (U10HL069294) and the National Cancer Institute, National Institutes of Health (U24HL138660), as well as funding from Astellas Pharma Global Development Inc.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the above-mentioned parties.
Authorship
Contribution: M.J.L. contributed to study design, analyzed the data, and wrote the manuscript; and all other authors contributed to study design, helped analyze the data, and edited the manuscript.
Conflict-of-interest disclosure: M.J.L. reports a consulting or advisory role with Daiichi Sankyo, Amgen, Astellas Pharma, Bristol Myers Squibb (BMS), AbbVie/Genentech, GlaxoSmithKline, Syndax, Takeda; reports expert testimony with Novartis; reports research funding from Astellas Pharma (to institution); and received travel, accommodations, and expenses from Astellas Pharma. M.H. reports honoraria from Celgene; reports consulting or advisory role with Incyte, ADC Therapeutics, Puma Biotechnology, Verastem, Kite/Gilead, MorphoSys, Omeros, Novartis, Gamida Cell, Seagen, Genmab, Myeloid Therapeutics, BeiGene, AstraZeneca, Sanofi, BMS/Celgene, CRISPR Therapeutics, Caribou Biosciences, AbbVie, and Genentech; reports speakers bureau participation with Genzyme, AstraZeneca, BeiGene, ADC Therapeutics, and Kite/Gilead; and reports research funding from Takeda, Spectrum Pharmaceuticals, Otsuka, Astellas Pharma, and Genzyme. M.R.L. reports honoraria from BeiGene Shanghai and Amgen; reports speakers bureau participation with BeiGene Shanghai and Amgen; report research funding from Amgen, Astellas Pharma, Actinium Pharmaceuticals, Syndax; received travel, accommodations, and expenses from BeiGene Shanghai and Amgen; and reports other relationship with Biosight. J.R.W. reports consulting or advisory role with Shire, Celgene, Cidara Therapeutics, F2G, and ORCA Therapeutics. E.B.P. reports employment with Biogen, Exelixis, Regulus Therapeutics, Graviton Bioscience Corp, and EpiKast; reports leadership with Biogen, Exelixis, and Regulus Therapeutics; reports stock and other ownership interests with Biogen, Exelixis, Regulus Therapeutics, Apellis Pharmaceuticals, Leap Therapeutics, and Actio Biosciences Inc; reports consulting or advisory role with Actio Biosciences; received research funding from AbbVie; and received travel, accommodations, and expenses from Biogen, Exelixis, and Regulus Therapeutics. A.E.P. received honoraria from Astellas Pharma, and Daiichi Sankyo; served in a consulting or advisory role with Astellas Pharma, Actinium Pharmaceuticals, Daiichi Sankyo, AbbVie, Forma Therapeutics, Sumitomo Dainippon, Celgene/BMS, Syndax, Genentech, BerGenBio, Immunogen, Foghorn Therapeutics, Rigel, and Curis; received research funding from Astellas Pharma (to institution), Bayer (to institution), Daiichi Sankyo (to institution), Fujifilm (to institution), AbbVie (to institution), and Syndax (to institution); and received travel, accommodations, and expenses from Daiichi Sankyo. R.J.S. reports leadership role with Kiadis Pharma, Be The Match/National Marrow Donor Program; served in a consulting or advisory role with Juno Therapeutics, Gilead Sciences, Rheos Medicines, Cugene, Jazz Pharmaceuticals, Precision Biosciences, Takeda, Jasper Therapeutics, Alexion Pharmaceuticals, Neovii, Vor Biopharma, Smart Immune, BlueSphere Bio; provided expert testimony for Pfizer; and received travel, accommodations, and expenses from Gilead Sciences. C.U. reports employment with Takeda and Blueprint Medicines; received honoraria from Novartis and Blueprint Medicines; and reports speakers bureau participation with Novartis. G.L.U. served in a consulting or advisory role with Jazz Pharmaceuticals. E.K.W. reports leadership role with Cambium Medical Technologies and Cambium Oncology; reports stock and other ownership interests in Cambium Medical Technologies, Cambium Oncology, Cerus, and Chimerix; received honoraria from Novartis, Verastem, Kite (a Gilead company), Pharmacyclics, Karyopharm Therapeutics, Sanofi, and Janssen Oncology; served in a consulting or advisory role with Novartis, Verastem, Pharmacyclics, Karyopharm Therapeutics, Partners Healthcare, Kite (a Gilead company), Cambium Medical Technologies, Alimera Sciences, and Sanofi; received research funding from Novartis, Amgen, Juno Therapeutics, Verastem, Partners Healthcare, and Sanofi; receives royalties from patent on preparing platelet lysate that has been licensed to Cambium Medical Technologies; and received travel, accommodations, and expenses from Janssen Oncology. S.V. served in a consulting or advisory role with Omeros and Johnson and Johnson; and received research funding from Sanofi (to institution). M. Solh reports speakers bureau participation with BMS, Amgen, Seagen, and GlaxoSmithKline; and received research funding from Partner Therapeutics. A. Mishra received research funding from Novartis. L.S.M. reports stock and other ownership interests in Corvus Pharmaceuticals; received honoraria from UpToDate; served in a consulting or advisory role with Amgen, Medexus Pharmaceuticals, Astellas Pharma, Kite (a Gilead company), and CTI BioPharma Corp; and received research funding from Adaptive Biotechnologies, Astellas Pharma, Jasper Therapeutics, Kite (a Gilead company), and BMS. H.-J.K. received honoraria from AbbVie, AML Hub, BMS, Hando, Novartis, Aston Sci, Amgen, Takeda, Green Cross, AIM ImmunoTech, Astellas Pharma, Jazz Pharmaceuticals, Janssen, LG Chemical, Pfizer, ViGen Cell, Ingenium, Sanofi, Meiji Pharm, and Merck Sharp & Dohme (MSD); served in a consulting or advisory role with Jazz Pharmaceuticals, Novartis, AbbVie, Astellas Pharma, MSD, BMS, Takeda, Sanofi, Handok, and AML Hub; and reports speakers bureau participation with Jazz Pharmaceuticals, Takeda, and Novartis. M. Stelljes served in a consulting or advisory role with Pfizer, MSD, BMS, Incyte, Takeda, Astellas, and Amgen; reports speakers bureau participation with Pfizer, Medac, MSD, Astellas, Jazz Pharmaceuticals, Amgen, Novartis, Gilead, Celgene, BMS, AbbVie, and Incyte; received research funding from Pfizer; and received travel support from Medac and Pfizer. Y.N. served in a consulting or advisory role with Daiichi Sankyo/UCB Pharma Japan, and Astellas Pharma; and reports speakers bureau participation with Astellas Pharma, Daiichi Sankyo/UCB Pharma Japan, AbbVie, Amgen, BMS Japan, Chugai Pharma, CSL Behring, Janssen Pharma, Kyowa, Nippon Shinyaku, Novartis, Otsuka, Sumitomo Pharma Oncology, Takeda, MSD, and JCR Pharmaceuticals. M.O. received honoraria from Astellas Pharma; and reports speakers bureau participation with Astellas Pharma, Daiichi Sankyo, Otsuka, and Novartis. A.H.W. received honoraria from Amgen, Servier, Novartis, Celgene, AbbVie/Genentech, Pfizer, Janssen Oncology, Astellas Pharma, MacroGenics, AstraZeneca, Gilead/Forty Seven, Stemline Therapeutics, and BeiGene; served in a consulting or advisory role with Servier, Novartis, Amgen, AbbVie/Genentech, Celgene, MacroGenics, Pfizer, Astellas Pharma, AstraZeneca, Janssen, Stemline Therapeutics, and BeiGene; reports speakers bureau participation with AbbVie/Genentech, Novartis, Celgene/BMS, Astex Pharmaceuticals, and Servier; received research funding from Novartis (to institution), Celgene (to institution), AbbVie (to institution), AstraZeneca (to institution), Servier (to institution), Amgen (to institution), and Roche (to institution); is a current employee of the Walter and Eliza Hall Institute (WEHI) which receives milestone and royalty payments related to venetoclax, and is eligible for benefits related to these payments; and receives payments from WEHI related to venetoclax. G.M. reports stock and other ownership interests in Ostentus Therapeutics, Inc; reports honoraria from Novartis and AbbVie; and reports speakers bureau participation with Novartis and AbbVie. C.C., N.H., M.R., J.H., S.C.G., and R.N. report employment with Astellas Pharma. S.M.D. reports leadership role with National Marrow Donor Program. M.M.H. served in a consulting or advisory role with Medac (to institution); and received research funding from Jazz Pharmaceuticals (to institution), Novartis (to institution), Sanofi (to institution), Astellas Pharma (to institution), Xenikos (to institution), and Gamida Cell (to institution). Y.-B.C. reports leadership role with ImmunoFree; reports stock and other ownership interests in ImmunoFree; and served in a consulting or advisory role with Magenta Therapeutics, Incyte, Novo Nordisk, Editas Medicine, Alexion Pharmaceuticals, Astellas Pharma, Takeda, Pharmacosmos, and Vor Biopharma. The remaining authors declare no competing financial interests.
Correspondence: Mark J. Levis, The Sidney Kimmel Comprehensive Cancer Center, Johns Hopkins University, 1650 Orleans St Room 2M44, Baltimore, MD 21287; email: levisma@jhmi.edu.
References
Author notes
Presented in abstract form at the 65th annual meeting of the American Society of Hematology, San Diego, CA, 11 December 2023.
Researchers may request access to anonymized participant-level data, trial-level data, and protocols from Astellas-sponsored clinical trials at www.clinicalstudydatarequest.com.
For the Astellas criteria on data sharing see: https://clinicalstudydatarequest.com/Study-Sponsors/Study-Sponsors-Astellas.aspx.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.


![Case examples from gilteritinib arm. MRD, relapse, and drug exposures are displayed for 6 participants randomized to gilteritinib. Note that all investigators and participants were blinded to both study drug and MRD status. The colored thin lines represent the FLT3-ITD MRD clones and the thick black bars depict the periods when the participant was being treated with gilteritinib. Pt (participant) 1: 3 clones (21, 30, 45 base pairs [bp]) detected before and after HCT. Started gilteritinib on post-HCT day +41, maintained continuously on 120 mg/d for 24 months. The 30-bp clone was not detected at 2 time points, then rose steadily until 24 months after HCT. Gilteritinib was stopped per protocol, with the pt still in morphologic remission, and the pt relapsed 6 weeks later. Pt 2: single 30-bp clone detected before and after HCT. Started gilteritinib on post-HCT day 43. Clone eradicated, but pt relapsed 8 months after HCT with AML confirmed by PCR/capillary electrophoresis to be wild-type FLT3. Pt 3: single 54-bp clone detected both before and after HCT. Started gilteritinib on post-HCT day 35, with concomitant posaconazole. Lactate dehydrogenase was elevated to grade 3 and gilteritinib was decreased briefly to 80 mg/d, and then discontinued permanently on day +51. Relapsed on post-HCT day 134. Pt 4: single 42-bp clone detected before and after HCT. Started gilteritinib on post-HCT day 86, maintained continuously for 24 months, then stopped per protocol. The 42-bp clone was detected throughout, slowly decreasing in level. Pt alive and relapse free 49 months after HCT. Pt 5: 5 clones detected before HCT, all negative immediately after HCT. A new 159-bp clone appeared after HCT, before randomization. Started gilteritinib with concomitant itraconazole, but the gilteritinib had to be held twice for thrombocytopenia and resumed at 80 mg/d, with a resultant gilteritinib trough level of only 77.5 ng/mL (which is approximately one-third the median trough level for all other participants17). The 159-bp clone became undetectable, but the 21-bp clone that had been detected before HCT reappeared on post-HCT day 142. Pt relapsed with the 21-bp and the 57-bp clone while on 80 mg/d gilteritinib. Pt 6: 2 clones detected before HCT, and a 63-bp clone persisted after HCT. Started gilteritinib on post-HCT day 50 and posaconazole started prophylactically on day post-HCT day 70. The absolute neutrophil count dropped to 100/mm3 on post-HCT day 105 and gilteritinib was stopped. Gilteritinib was resumed on post-HCT day 131 (again with concomitant posaconazole), but the neutrophil count decreased again and the study drug was permanently stopped on post-HCT day 138. Pt relapsed 2 months later with the 63-bp clone.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/145/19/10.1182_blood.2024025154/1/m_blood_bld-2024-025154-gr5.jpeg?Expires=1769079661&Signature=Ii1GaV4gYPmAr1EPMVHMZG5-8otu2mBN97DUuLRAtbaH1ISd5sl2vIWPdhqQIXgo7A47K5unINcHpYPLjB9m8BvWTs73oyzGKqFShW4zMLYgSfa3~cimVs8b2IEd31F~cuXczEslDkkErC4ABDqoW949xsnXu8kUQeqUbe4WNo9QkKx8-Pz38SDYFDXBLfNW4Dx~C9dmZJ5DalYRopC3AFC1tgVStpqriYqGSOds~2baQHBQexaQnWwNe8zhKZwYdLf~BJHhWW5ZOS5C2790IbfknyFs~PJKwRAWdmo2a0ljohmVV6iGLVD-j~6euFq846B0-Ou434cScW91OE~EMw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal