Abstract
We used stochastic modeling and computer simulation to study the replication, apoptosis, and differentiation of murine hemopoietic stem cells (HSCs) in vivo. This approach allows description of the behavior of an unobserved population (ie, HSCs) on the basis of the behavior of observed progeny cells (ie, granulocytes and lymphocytes). The results of previous limiting-dilution, competitive-repopulation studies in 44 mice were compared with the results of simulated transplantation studies to identify parameters that led to comparable outcomes. Using this approach, we estimated that murine HSCs replicate (on average) once every 2.5 weeks and that the frequency of murine HSCs is 8 per 105 nucleated marrow cells. If it is assumed that short-term repopulating cells are distinct from HSCs, that they contribute to hemopoiesis early after transplantation, and that they are independently regulated, a frequency of 4 HSCs per 105nucleated marrow cells also allows simulations that best approximate the observed data. When stochastic modeling and computer simulation were applied to limiting-dilution, autologous-transplantation studies in cats heterozygous for glucose-6-phosphate-dehydrogenase, different estimates of HSC replication rate (1 per 8.3-10 weeks) and frequency (6 per 107 cells) were derived. Therefore, it appears that these parameters vary inversely with increased longevity, size, or both. An implication of these data is that human HSCs may be less frequent and replicate more slowly. These findings on cell kinetics have several implications.
Introduction
Hemopoietic stem cells (HSCs) give rise to all types of mature blood cells, including granulocytes, monocytes, lymphocytes, red cells, and platelets. They are the parent cells that are essential for maintaining homeostasis in the blood system. Because mammalian HSCs are infrequent and are defined functionally, information about their in vivo kinetics must be inferred from analyses of progenitors and differentiated blood cells.
Stochastic modeling is an excellent method for analyzing the behavior of an unobserved cell population (ie, HSCs) on the basis of observations of the behavior of derivative cells. This approach does not require information about the outcome of individual cell divisions; rather, it allows characterization of the behavior of a population of cells on the basis of the assumption that the fate of an individual member is probabilistic. The fate (replication, differentiation, or apoptosis) of an individual HSC depends on the chance that it interacts with specific accessory cells in the marrow microenvironment, the presence or quantity of cytokines it uniquely encounters, its cell-surface expression of receptors, and the integrity of its signal-transduction pathways. A complex, yet regulated, process such as hemopoiesis is especially amenable to stochastic analysis.
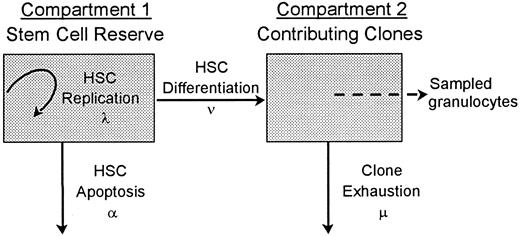
A stochastic model of HSC differentiation is diagrammed in Figure1. A hemopoietic stem cell can replicate (self-renew; the intensity or average rate of HSC replication is λ), differentiate (the average rate of HSC differentiation is ν), or die (the average rate of HSC apoptosis is α). Once an HSC commits to a differentiation pathway, it gives rise to a clone that contributes to hemopoiesis until exhaustion (the average rate of exhaustion is μ). With this approach, there are no obligatory connections or feedback loops. For example, differentiation (or apoptosis) does not require a replication division and the differentiation of one HSC does not trigger the replication of another. This broad conceptualization of hemopoiesis can describe a hemopoietic system in which self-renewal does not occur (λ = 0). It also can describe a system in which the 2 daughter cells generated by HSC replication have separate fates (eg, apoptosis vs further replication vs differentiation) or undergo second events at disparate points in time and thus allows asymmetric outcomes from an initial HSC division. The model has 3 assumptions: (1) HSCs act independently; (2) all events related to HSCs (ie, replication, apoptosis, and differentiation) happen stochastically; and (3) at all points in time, those clones that contribute to hemopoiesis contribute equally.
A 2-compartment stochastic model of HSC behavior.
Each HSC in the hemopoietic stem-cell reserve may by chance replicate (resulting in a second, identical HSC in this compartment), undergo apoptosis, or differentiate. These fates are probabilistic (stochastic) and occur at mean rates of λ, α, and ν, respectively. An HSC that commits to a differentiation and maturation program initiates a clone that contributes to hemopoiesis until exhaustion (the mean rate of clone exhaustion is μ). R is the number of HSCs in the stem-cell reserve. R0 is the number of HSCs in the stem-cell reserve at time zero, ie, the number of transplanted HSCs. C is the number of contributing clones andC0 is the number of contributing clones immediately after transplantation. Although individual HSC decisions are modeled as random or stochastic and are independent in terms of probability theory, the rates at which the different decisions happen in the HSC compartment are density dependent.1
A 2-compartment stochastic model of HSC behavior.
Each HSC in the hemopoietic stem-cell reserve may by chance replicate (resulting in a second, identical HSC in this compartment), undergo apoptosis, or differentiate. These fates are probabilistic (stochastic) and occur at mean rates of λ, α, and ν, respectively. An HSC that commits to a differentiation and maturation program initiates a clone that contributes to hemopoiesis until exhaustion (the mean rate of clone exhaustion is μ). R is the number of HSCs in the stem-cell reserve. R0 is the number of HSCs in the stem-cell reserve at time zero, ie, the number of transplanted HSCs. C is the number of contributing clones andC0 is the number of contributing clones immediately after transplantation. Although individual HSC decisions are modeled as random or stochastic and are independent in terms of probability theory, the rates at which the different decisions happen in the HSC compartment are density dependent.1
In previous studies, we used this stochastic model and computer simulation to determine the in vivo kinetics of HSCs in female cats heterozygous for the x-chromosome–linked enzyme, glucose-6-phosphate-dehydrogenase (G6PD) after transplantation of small numbers of autologous cells (1-2 × 107 nucleated marrow cells/kg).1 Because only a small number of HSCs were present in the marrow inoculum, there was competition to repopulate and maintain hemopoiesis between HSCs expressing the G6PD phenotype of the domestic parent and HSCs expressing the G6PD phenotype of the Geoffroy parent.
The parameter values yielding simulations that best fit the experimental observations were λ, 1 per 10 weeks; ν, 1 per 12.5 weeks; α, 0; μ, 1 per 6.7 weeks; and R0 (the number of HSCs in the stem-cell reserve at time 0, ie, the number of transplanted HSCs), 30 HSCs, implying that 30 HSCs were present per 5 × 107 nucleated marrow cells (the average number of nucleated marrow cells that were transplanted) and that the frequency of feline HSCs was 6 per 107 nucleated marrow cells.1 A similar fit was obtained when λ was 1 per 8.3 weeks, α was 1 per 50 weeks, and other parameters were unchanged (unpublished data). Because C0 (the number of contributing clones at time 0) values from 0 to 2000 yielded equivalent results, the number (or existence) of short-term repopulating cells (STRCs) could not be established from the feline data.1
The aim of this study was to apply these methods to the analysis of data generated from experiments in which small numbers of congenic donor (Gpi-1a) and competitor (Gpi-1b) marrow cells were transplanted in mice.2 By analyzing studies with a comparable experimental design with use of a similar analysis, we hoped to gain insight into the in vivo behavior of murine HSCs and to define the evolutionary adaptations of HSCs to increasing animal size and lifespan. In its lifetime (2 years), a mouse (25 g) makes the same number of red blood cells as does a human (70 kg) in 1 day or a cat (4 kg) in 8 days.3 Similar discrepancies exist in the requirements for other blood-cell lineages. We wanted to determine whether feline (and human) HSCs are more numerous than murine HSCs, have higher proliferative potentials per cell, or are regulated by different kinetics to satisfy this increased demand.
Methods
Data from congenic murine transplantations
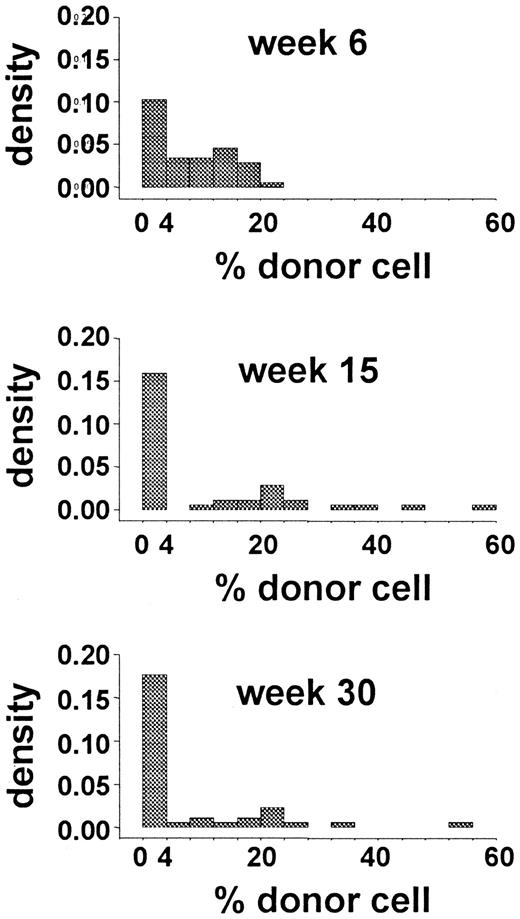
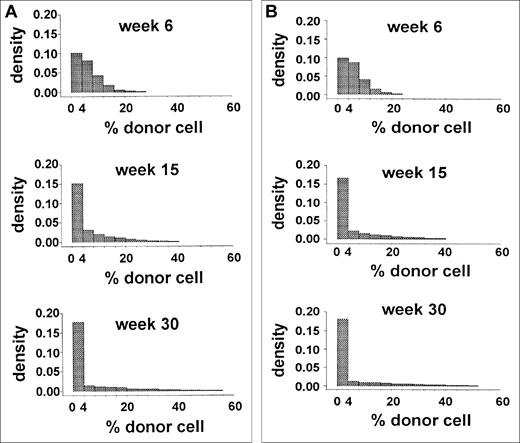
The data from a limiting-dilution, competitive-repopulation assay of murine marrow cells were reported previously.2Forty-four mice were lethally irradiated and then given transplants of 0.25 × 105 Gpi-1a cells and 4.0 × 105 Gpi-1b cells (ie, 1.7 × 107 nucleated marrow cells/kg). Blood samples were obtained 6, 15, and 30 weeks after transplantation, and the proportions of Gpi-1a–type granulocytes, as well as platelets, T cells, and B cells were recorded. The distribution of the proportion of Gpi-1a–type granulocytes at each observation time is shown in Figure 2. Early after transplantation (6 weeks), this distribution reflected the proportion of Gpi-1a cells transplanted (0.25 × 105Gpi-1a cells divided by 4.25 × 105 total marrow cells equals 6%). The excess of 0 values suggests that no reconstituting cell was present among the 0.25 × 105Gpi-1a cells transplanted in several animals. Variance in this proportion at 15 and 30 weeks after transplantation (Figure 2) was higher, and a large percentage of animals had exclusively Gpi-1b granulocytes.
Data from limiting-dilution transplantation studies in mice.
The histogram shows the distribution of the percentage of donor (Gpi-1a) granulocytes at 6, 15, and 30 weeks after transplantation.
Data from limiting-dilution transplantation studies in mice.
The histogram shows the distribution of the percentage of donor (Gpi-1a) granulocytes at 6, 15, and 30 weeks after transplantation.
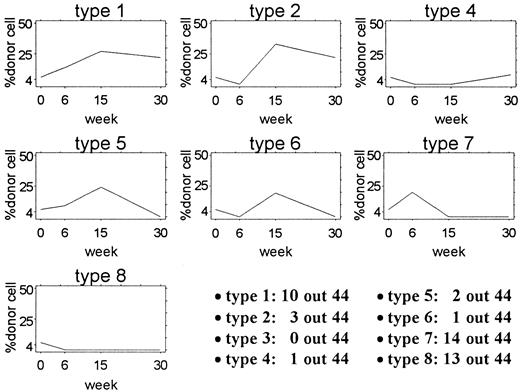
Each of the 44 mice given transplants were placed into 1 of 8 categories defined by the pattern of the percentage of Gpi-1a cells 6, 15, and 30 weeks after transplantation. Representative studies are diagrammed in Figure3. For category type 1, the proportion of Gpi-1a cells was 4% or higher at 6 weeks (w6), 15 weeks (w15), and 30 weeks (w30). For type 2, it was less than 4% at w6 and 4% or higher at w15 and w30. For type 3, it was 4% or higher at w6 and w30 and less than 4% at w15. For category type 4, the proportion of Gpi-1a cells was less than 4% at w6 and w15 and 4% or higher at w30. For type 5, it was 4% or higher at w6 and w15 and less than 4% at w30. For type 6, it was less than 4% at w6 and w30 and 4 % or higher at w15. For type 7, it was 4% or higher at w6 and less than 4% at w15 and w30. Finally, for category type 8, the proportion of Gpi-1acells was less than 4% at all 3 assessment times. Ten of the 44 mice had a reconstitution pattern similar to that of type 1, whereas 3 of the 44 mice had a type 2 pattern. The distribution of other pattern types is shown in Figure 3.
Patterns of contribution in 44 mice given transplants.
Each plot shows the actual pattern of one example mouse from the 8 categories used for analysis. The number of mice with each pattern is shown in the lower right corner. No mouse in this experiment had a type 3 pattern.
Patterns of contribution in 44 mice given transplants.
Each plot shows the actual pattern of one example mouse from the 8 categories used for analysis. The number of mice with each pattern is shown in the lower right corner. No mouse in this experiment had a type 3 pattern.
Statistical methods for the analysis of murine studies
To determine whether specific values for the stochastic model parameters λ, ν, α, μ, and R0 (the total number of HSCs at time 0, ie, the number of transplanted Gpi-1a HSCs plus Gpi-1b HSCs) could lead to the experimental observations, we performed simulation studies. All simulations of murine transplantations mimicked the actual experiment. For example, “granulocytes” were evaluated at 6, 15, and 30 weeks after transplantation, as in the transplantation data. TheR0 values for HSCs were selected randomly from a pool containing Gpi-1a HSCs and Gpi-1b HSCs (ratio 1:16) so that when R0 was small, Gpi-1a HSCs might be absent from the transplanted cell inoculum. Also, each simulation set consisted of independent studies of 44 mice.
The 3 observations pertaining to each mouse (ie, the percentage of “granulocytes” that expressed Gpi-1a at 6, 15, and 30 weeks after transplantation) can be seen as a random draw from a 3-dimensional distribution with a continuous range [0-100].3 Therefore, the entire data set represents a simple random sample of size 44 from such a distribution.
We next discretized the simulated data to better understand the empiric distribution and to simplify the comparison with experimental data. Our discretization used a cutoff point of 4% (ie, values ≤ 4% were recorded as 0) for consistency with the experimental data, since recipient mice were scored negative for donor granulocytes if less than 4% of their granulocytes had a GpI-1aphenotype.2 This categorization of the data has several advantages. First, it maintains the 3-dimensional structure of the data and therefore allows examination of the joint behavior of the 3 components (at 6, 15, and 30 weeks) instead of only an analysis of these compartments marginally. Second, every mouse can be classified as having 1 of the 8 possible patterns shown in Figure 3, thereby allowing an easily interpreted picture of the data. Third, the data set can be seen as an observation drawn from an 8-dimensional multinomial distribution with a parameter of 44 and unknown cell probabilities, thus providing the opportunity to use standard statistical techniques to compare the experimental data with the simulated data.
Given a combination of parameter values for λ, ν, α, μ, andR0, the underlying distribution of the adopted stochastic model with observations obtained at 6, 15, and 30 weeks does not have a known form. However, since observations may be simulated from the model, generating a large number of simulations provides a reference distribution or a Monte Carlo estimate of the underlying distribution that can be used to compare the experimental and simulated data. Therefore, for each combination of the parameters tested, we simulated 150 data sets, each of which contained 44 transplantations. We next applied the discretization procedure described above to each of the 150 data sets and generated a reference distribution. We then tested the null hypothesis that the experimental results were drawn from the reference distribution. This last task was performed by using 8 Monte Carlo tests,4 1 for each of the dimensions of the multinomial distribution.
To determine the ranges of values for the parameters that best reproduced the experimental data, we performed a coarse lattice search over the plausible range of values; 100 different combinations of parameters were considered. We determined the appropriateness of each parameter set by means of 2 criteria. The first criterion (criterion 1) was based on the comparison between the distributions of the proportion of Gpi-1a–type cells by week of the experimental and simulated data. In this comparison, unique features of the experimental data (ie, the high density of 0% Gpi-1a–positive cells at each evaluation time and the broad distribution of observed outcomes, including outcomes where > 30% Gpi-1a–positive cells occurred at 15 and 30 weeks; Figure 2), needed to be maintained. Criterion 1 was satisfied if the distributions of the simulated data were similar to those of the experimental data. The second criterion (criterion 2) was based on the 8 Monte Carlo tests.
A combination of parameters was considered acceptable if (1) criterion 1 was satisfied and at least 6 of the 8 Monte Carlo tests yielded nonsignificant results simultaneously (P > .1); or (2) criterion 1 was not satisfied but at least 7 of the 8 Monte Carlo tests yielded nonsignificant results simultaneously. A parameterization was considered a best fit if criterion 1 was satisfied and all 8 Monte Carlo tests yielded nonsignificant results simultaneously. When a best fit was found, the robustness of the parameterization was confirmed through sensitivity analysis, ie, slightly changing some of the parameter values, the random seed, or both and assuring that these modifications also satisfied best-fit criteria.
Results
Parameters generated from studies of hemopoiesis in cats could not explain the murine data
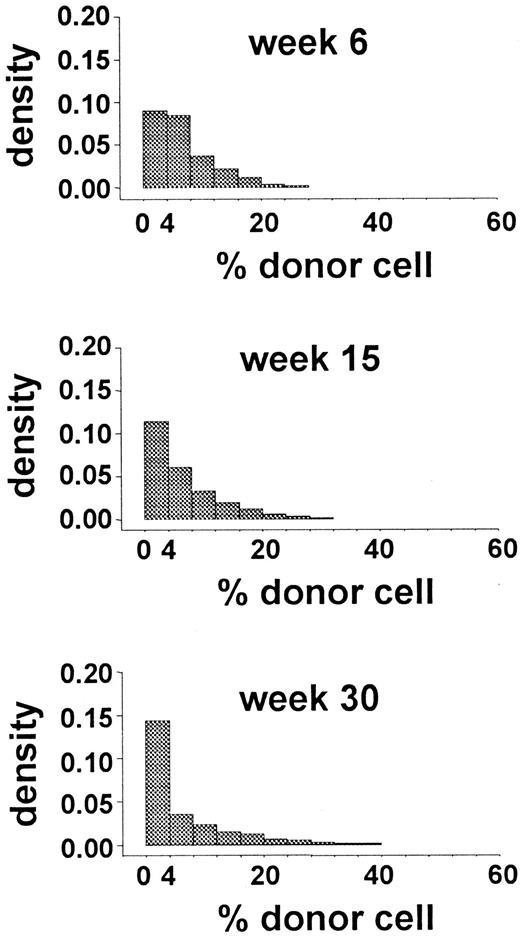
Initially, we simulated murine transplantation studies by using the parameters generated from the analysis of autologous transplantations in G6PD heterozygous cats. Values for λ, ν, α, and μ were 1 per 10 weeks, 1 per 12.5 weeks, 0, and 1 per 6.7 weeks, respectively. Figure 4 shows the results that were most comparable to the experimental data (obtained withR0 = 35). Because these results failed to meet both criterion 1 (Figure 4) and criterion 2 (only 4 of the 8 Monte Carlo tests had P values > .1), the in vivo kinetics of murine HSCs must be different from that of feline HSCs. When λ was 1 per 8.3 weeks and α was 1 per 50 weeks, all simulations also failed to meet both criterion 1 and criterion 2.
Distributions of simulated data obtained by using the parameter values estimated for hemopoiesis in cats.
The data are from 150 studies with 44 independently simulated murine transplantations per study. The distribution of the percentage of donor granulocytes at 6, 15, and 30 weeks after this hypothetical transplantation is shown. These distributions are not similar to the distributions shown in Figure 2 and thus fail to approximate the observed data. Both the densities of 0% donor-cell values at 6 and 15 weeks and the densities of donor-cell values above 30% at 15 and 30 weeks differ from the experimental results.
Distributions of simulated data obtained by using the parameter values estimated for hemopoiesis in cats.
The data are from 150 studies with 44 independently simulated murine transplantations per study. The distribution of the percentage of donor granulocytes at 6, 15, and 30 weeks after this hypothetical transplantation is shown. These distributions are not similar to the distributions shown in Figure 2 and thus fail to approximate the observed data. Both the densities of 0% donor-cell values at 6 and 15 weeks and the densities of donor-cell values above 30% at 15 and 30 weeks differ from the experimental results.
Estimation of parameters for murine HSCs
To define the kinetic parameters of murine HSCs, a coarse lattice search was performed over the plausible range of parameter values of λ, ν, α, μ, and R0. For each combination of parameters (n = 100), 150 data sets, each of which contained 44 transplantations, were generated. These were compared with the experimental results (Figures 2 and 3). A combination of parameters was considered acceptable if criterion 1 and criterion 2 were met. An acceptable combination of parameters was considered a best fit if all 8 Monte Carlo tests yielded nonsignificant results simultaneously (P value > .1) and sensitivity analyses were satisfied. The values of 1 per 2.5 weeks for λ, 1 per 3.4 weeks for ν, 1 per 20 weeks for α, 1 per 6.9 weeks for μ, and 35 HSCs forR0 satisfied these requirements. The histogram describing the percentage of Gpi-1a–positive cells in simulated outcomes obtained with these parameters is shown in Figure5A. If 35 HSCs were present per 4.25 × 105 total nucleated cells transplanted per mouse, then the frequency of HSCs was 8 HSCs per 105 nucleated marrow cells. The ranges of acceptable parameters are shown in Table1. HSC number and λ were well defined by this approach, whereas ν, α, and μ had broader ranges of acceptable values and thus could not be as precisely estimated. Although ranges are provided, only certain relations between λ, ν, and α were permissible according to the criteria. For example, λ needed to be larger than ν + α.
Distributions of simulated data using the best parameter values.
The results derive from 150 data sets of 44 mice given transplants (6600 simulated murine transplantations). The distribution of the percentage of donor granulocytes at 6, 15, and 30 weeks after these simulated transplantations is shown. In (A), parameter values are 1 per 2.5 weeks for λ, 1 per 3.4 weeks for ν, 1 per 20 weeks for α, 1 per 6.9 weeks for μ, 35 HSCs for R0, and 50 clones for C0. For the simulations of (B), R0:C0 was arbitrarily set equal to 1:4. To achieve these results,R0 was 18 HSCs and thus HSC frequency was 4 per 105 nucleated marrow cells.
Distributions of simulated data using the best parameter values.
The results derive from 150 data sets of 44 mice given transplants (6600 simulated murine transplantations). The distribution of the percentage of donor granulocytes at 6, 15, and 30 weeks after these simulated transplantations is shown. In (A), parameter values are 1 per 2.5 weeks for λ, 1 per 3.4 weeks for ν, 1 per 20 weeks for α, 1 per 6.9 weeks for μ, 35 HSCs for R0, and 50 clones for C0. For the simulations of (B), R0:C0 was arbitrarily set equal to 1:4. To achieve these results,R0 was 18 HSCs and thus HSC frequency was 4 per 105 nucleated marrow cells.
Parameter values in studies of kinetics of murine and feline HSCs
| Parameter . | Murine . | Feline best value . | |
|---|---|---|---|
| Acceptable range . | Best fit . | ||
| HSC replication rate (λ) | 1 per 1.8-6.6 wk | 1 per 2.5 wk | 1 per 8.3-10 wk |
| HSC differentiation rate (ν) | 1 per 1.9-13 wk | 1 per 3.4 wk | 1 per 12.5 wk |
| HSC apoptosis rate (α) | 0-1 per 2.0 wk | 1 per 20 wk | 0-1 per 50 wk |
| HSC frequency in nucleated marrow cells | 4-22/105 | 8/105 | 6/107 |
| Parameter . | Murine . | Feline best value . | |
|---|---|---|---|
| Acceptable range . | Best fit . | ||
| HSC replication rate (λ) | 1 per 1.8-6.6 wk | 1 per 2.5 wk | 1 per 8.3-10 wk |
| HSC differentiation rate (ν) | 1 per 1.9-13 wk | 1 per 3.4 wk | 1 per 12.5 wk |
| HSC apoptosis rate (α) | 0-1 per 2.0 wk | 1 per 20 wk | 0-1 per 50 wk |
| HSC frequency in nucleated marrow cells | 4-22/105 | 8/105 | 6/107 |
If the ratio R0:C0is arbitrarily decreased to 1:4, a murine HSC frequency of 4/105 nucleated marrow cells also leads to simulated results that satisfy the criteria for best fit. The acceptable range for μ, the average rate of extinction of a differentiating clone, is 1 per 0.4 to 29 weeks, and the best-fit value is 1 per 6.9 weeks. Values for feline HSCs are from Abkowitz et al.1
Assumptions regarding the existence of STRCs affect estimates of HSC frequency but not other parameters
An STRC can reconstitute hemopoiesis immediately after transplantation and support blood-cell production for a short period, which is operationally defined as less than 6 to 12 weeks. These characteristics are in contrast to those of a long-term repopulating cell (LTRC), which contributes to hemopoiesis from 3 to 4 months after transplantation and throughout a mouse's lifetime. Whether an STRC is a distinct cell type or a functional description of a subset of HSCs is controversial.2,5-13 With the initial analyses (Figure5A), we assumed that C0 could be derived from the R/C mean at steady state (λ − α − ν + μ)/ν, when R0 = 35 and C0 = 50, and that only a few clones (those transplanted from compartment 2; Figure 1) contributed to blood-cell production immediately after transplantation. HSCs that by chance differentiate quickly would also be considered to be STRCs, whereas those that by chance undergo replication divisions (doubling clone size and proliferative potential) would be considered to be LTRCs according to the traditional definitions. Alternatively, if STRCs exist as a distinct, independently regulated cell type, this approximation might underestimate their numbers. The ratio of LTRCs to STRCs has been estimated as 1:2 to 1:10.2,6,10 13 Therefore, to consider the potential effects of contributions from STRCs at 6 weeks after transplantation, we performed additional simulations in which values were designated for this ratio (and not derived). IfR0:C0 was 1:2, no parameter was affected. Values ofR0:C0 ranging from 1:4 to 1:10 affected estimates of HSC frequency but not estimates of average HSC replication rate (λ) or average HSC apoptosis rate (α). For example, when the ratioR0:C0 was set equal to 1:4, HSC frequencies between 4 and 8 per 105 nucleated marrow cells best fit the data. Figure 5B shows the results for the combination of parameters with an R0 of 18 (HSC frequency = 4/105 nucleated marrow cells; all 8P values were > .1 on Monte Carlo analyses). For values of R0:C0 equal to 1:7, acceptable but not optimal (best fit) parameterizations were achieved. For values of R0:C0under 1:10, the simulated outcomes were not similar to the observed data (criterion 1 was not satisfied and P values were < .1 in 3 or more of the 8 categories analyzed).
The estimated parameter values allow excellent simulations of other experimental data
Data on B lymphocytes in the 44 mice given transplants were also available. There was a strong correlation between B-cell and granulocyte values at 15 and 30 weeks (but not at 6 weeks) after transplantation.2 Similarly, parameters derived from the granulocyte analysis allowed an acceptable fit of the B-lymphocyte data. To assess the validity of the estimations further, experimental data generated by transplantation of different numbers of Gpi-1a and Gpi-1b marrow cells (D.E.H., unpublished data, 1996-1999) were studied by using a comparable approach. These data were less complete (10-26 mice/experimental approach) and predominantly contained measurements of long-lived cells (red cells and total lymphocytes [eg, T and B cells]) and thus were not sufficiently powerful for independent parameter estimates or Monte Carlo analysis. Therefore, for each circumstance, 150 comparable data sets were simulated and compared with the experimental observations by using only criterion 1. An analysis was performed for lymphocytes (1, 5, and 9 months) and red cells (5 and 9 months) after 26 mice were given transplants of 1.3 × 105 Gpi-1a and 4 × 105 Gpi-1b marrow cells (0.25 ratio of donor to total cells) and for lymphocytes and granulocytes 1, 4, and 7 months after 14 mice were given transplants of 2 × 105Gpi-1a and 2 × 105 Gpi-1b marrow cells (0.5 ratio of donor to total cells). All histograms of the distribution of simulated data were similar to the histogram of the experimental observations, confirming that the initial estimations were reasonable.
Discussion
We studied the in vivo behavior of HSCs by analyzing data generated from the transplantation of small numbers of HSCs labeled by GpI-1a or GpI-1b phenotype into congenic mice. Through computer simulation and the assumption that all HSC decisions are stochastic, we estimated the average rates of HSC replication (self-renewal), differentiation, and apoptosis. Our data suggest that murine HSCs are not quiescent; rather, they replicate an average of once every 2.5 weeks. Thus, the median time to replication (ie, the time when 50% of HSCs have divided) is 1.7 weeks. Similarly, we determined that the frequency of HSCs in normal, steady-state marrow is 4 to 8 per 105 nucleated cells.
These results are remarkably consistent with estimates using other approaches. For example, incorporation of bromodeoxyuridine was used to determine the rate at which murine HSCs, phenotypically defined as rhodamine 123lo and Hoechst 33342locells14 or c-kitbrightThy1.1lo Sca-1+ lineage-negative cells15 enter the cell cycle over time. In these 2 studies, approximately 4.3% and 7.8%, respectively, of these cells were found to enter the cell cycle each day, apparently in random fashion. Because the cell cycle was estimated to last 3 days,15 50% of HSCs should complete their cell cycle (ie, replicate) in 9 to 19 days (1.3-2.75 weeks). This approach might overestimate the replication rate of HSCs if cells that were more differentiated and less quiescent than HSCs were included among cells termed HSCs as a result of metabolic or immunologic-phenotype analyses. In the study by Bradford et al,14 for example, 1 of 12 cells termed HSCs supported long-term hematopoiesis in transplantation studies. The comparability of results achieved with 2 different experimental methods reinforces the validity of the findings and also suggests that stem-cell kinetics after transplantation are similar to that during steady-state hemopoiesis.
Our estimate of murine HSC frequency—4 to 8 per 105nucleated marrow cells—is also consistent with the data of others,2,7,12-20 who estimated this frequency to be 1 to 50 per 105 nucleated marrow cells. The large range of reported values likely represents the assumptions implicit in different experimental designs. For example, when HSCs are estimated with respect to functional endpoints, ie, their ability to reconstitute blood-cell production and contribute to hematopoiesis after transplantation, HSC frequency generally ranges from 1 to 2.5 per 105 nucleated marrow cells.17,18 Similarly, when the data from our studies were analyzed by using Poisson calculations, HSC frequency was estimated at 1 per 105 nucleated marrow cells,2 a value lower than obtained by the stochastic modeling approach. When these analytic methods are used, an HSC must contribute to blood-cell production in order to be counted. HSCs that replicate but remain in the hemopoietic stem-cell reserve (compartment 1; Figure 1) during the observation time, and those that die, are not counted, thus leading to an underestimation of actual frequency. Also, to be counted by either analytic approach, a cell must seed efficiently, ie, home to marrow and land in an environment that permits its survival and differentiation. In some studies, HSCs were defined exclusively by phenotype rather than functional characteristics. Because not all cells defined in this manner are indeed HSCs (according to a transplantation definition), this approach could overestimate the numbers of HSCs. Of note, most recent estimates of murine HSC frequency range from 3 to 8 per 105 nucleated marrow cells.12-14 20
At an average replication rate of 1 per 2.5 weeks, the stem-cell reserve (compartment 1; Figure 1) will not be reconstituted to steady-state values until 134 weeks (2.6 years) after limiting-dilution transplantation (R0 = 35), which exceeds the lifespan of mice. Reconstitution will not occur until 94 weeks (1.8 years) after transplantation if more typical numbers of nucleated marrow cells (R0 = 400; 5 × 106cells/mouse) are transplanted (assuming the total number of nucleated marrow cells in a normal mouse is 2.8 × 108(ref21) [results obtained with simulation studies and Markov analyses]). These kinetics findings are consistent with the observation that only a few1,2 serial transplantations can be done before exhaustion of the hemopoietic reserve but that 3 to 5 transplantations can be accomplished if they are performed at intervals of more than 6 months.5 13
Table 1 shows the estimated replication rate and frequency of murine and feline HSCs. Although no studies have been done to confirm these values in other large animals, Wang et al22 transplanted subpopulations of human marrow cells into nonobese diabetic–severe combined immunodeficient mice as an indirect method for enumerating human HSCs. There is controversy about whether xenotropic transplantation assays indeed measure HSC activity or only correlate with this activity, but the value obtained for HSC frequency (3/107 nucleated marrow cells) in their study was less than estimated HSC frequencies in mice and cats.
These data provide insight into the evolutionary adaptation to increased size and longevity. They suggest that in larger animals, HSCs are less frequent and divide more slowly. Therefore, the proliferative potential of each cell (ie, differentiated progeny per clone lifetime) is high. Thus, either murine and feline (and human) HSCs are biologically different in their intrinsic capacities or, if they are biologically similar, there is excess capacity in murine cells that is not needed to support hemopoiesis throughout a mouse's lifetime. An extension of this idea would be that cells that are not technically stem cells (ie, cells that derive from a differentiating stem-cell clone that has undergone 1 or 2 divisions [STRCs]) could exist in mice but perhaps not in larger species.
If feline HSCs had a replicative rate equivalent to that of HSCs in mice (1 per 2.5 weeks), these cells would undergo enough replications (312 vs 42) in the longer lifetime of cats (15 years vs 2 years in mice) that they would vastly exceed cell senescence, which was approximated (but likely underestimated) as 50 divisions in diploid fibroblast cells maintained in vitro.23 Such an outcome could lead to high levels of aplastic anemia or myelodysplasia, but this has not been observed in either cats or humans. The estimated value in cats (one HSC replication every 10 weeks) would predict 78 cell divisions in a 15-year lifespan, a result that is more compatible with the hypotheses of Hayflick and Moorhead.23
Lastly, these data provide insights into the concept of clonal succession.24,25 This term was previously defined in 2 different ways. To some, clonal succession implies that the hemopoietic reserve is composed of a large number of stem cells but that only one or a few are active at any time. The experimental data do not support this idea. A second interpretation of clonal succession is that hemopoiesis is supported by a succession of clones. When transplantation is done with a few hemopoietic stem cells, only a few HSC initially support blood-cell production. However, in the steady state or when transplantation uses a large inoculum of HSCs, clones derived from many HSCs support hemopoiesis.26 Still, each clone has a discrete lifetime and is then succeeded by another.27 This interpretation of clonal succession is consistent with all murine and feline data and the stochastic model diagrammed in Figure 1. Indeed, if either the feline or the murine data are modeled with the assumption that μ is 0 (ie, that differentiating clones never become exhausted), no values for other parameters provide simulations that mimic the observed data (unpublished data).
Aside from the teleologic implications of these data, there are practical consequences. In larger animals, stem cells are quite infrequent and thus more difficult to purify and manipulate. They may be more likely to lose “stem cellness” as measured by repopulating ability after in vitro manipulation, since a vast proliferative potential is required for a cell to be counted in a transplantation assay. In addition, the slow kinetics of the stem-cell cycle explains why it is difficult to label feline, canine, and primate (including human) hemopoietic stem cells with retroviral vectors that require cell division for proviral integration. Although murine cells can be labeled readily at rates (30%-50%) comparable with clinical efficacy, this is not the case in gene-transfer studies in larger animals (rates of 0.01%-10%),28-32 despite pharmacologic stimulation (ie, with granulocyte colony-stimulating factor, kit ligand, or flt3 ligand) in vivo before marrow or peripheral blood stem-cell collection or in vitro. An alternative approach, such as using lentiviral vectors, may be required. The in vivo kinetics of hemopoietic stem cells, not just their number, has major biologic implications.
Acknowledgments
We thank Allan Dimaunahan and Zeny Sisk for help with preparation of the manuscript.
Funded by a grant (R01 HL46598) from the National Institutes of Health. J.L.A. is the recipient of a Faculty Research Award from the American Cancer Society.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Janis L. Abkowitz, Division of Hematology, University of Washington, Box 357710, Seattle, WA 98195-7710.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal