CD34 is a cell-surface transmembrane protein expressed specifically at the stem/progenitor stage of lymphohematopoietic development that appears to regulate adhesion. To elucidate intracellular signals modified by CD34, we designed and constructed glutathione-S–transferase (GST)– fusion proteins of the intracellular domain of full-length CD34 (GST-CD34ifull). Precipitation of cell lysates using GST-CD34ifullidentified proteins of molecular mass 39, 36, and 33 kd that constitutively associated with CD34 and a 45-kd protein that associated with CD34 after adhesion. By Western analysis, we identified the 39-kd protein as CrkL. In vivo, CrkL was coimmunoprecipitated with CD34 using CD34 antibodies, confirming the association between CrkL and CD34. CD34 peptide inhibition assays demonstrated that CrkL interacts at a membrane-proximal region of the CD34 tail. To identify the CrkL domain responsible for interaction with CD34, we generated GST-fusion constructs of adapter proteins including GST-CrkL3′ (C-terminal SH3) and GST-CrkL5′ (N-terminal SH2SH3). Of these fusion proteins, only GST-CrkL3′ could precipitate endogenously expressed CD34, suggesting that CD34 binds the C-terminal SH3 domain of CrkL. Interestingly, there appears to be differential specificity between CrkL and CrkII for CD34, because GST-CD34ifull did not precipitate CrkII, a highly homologous Crk family member. Furthermore, GST-CD34ifull did not bind c-Abl, c-Cbl, C3G, or paxillin proteins that are known to associate with CrkL, suggesting that CD34 directly interacts with the CrkL protein. CD34ifull association with Grb or Shc adapter proteins was not detected. Our investigations shed new light on signaling pathways of CD34 by demonstrating that CD34 couples to the hematopoietic adapter protein CrkL.
Introduction
CD34 is a 116-kd type I transmembrane glycophosphoprotein expressed at the primitive state of hematopoietic development as well as on most endothelial cells, some neuronal cells and, rarely, on primitive cells from other tissues.1,2 In the hematopoietic system, the presence of CD34 on the cell surface identifies cells that can, upon cytokine or growth factor stimulation, expand and differentiate into all the lymphohematopoietic lineages. Following hematopoietic maturation, CD34 expression is entirely suppressed. The clinical importance of CD34 is illustrated by its utility as a selective marker for identification and isolation of the lymphohematopoietic stem/progenitor cell population. Patients transplanted with CD34+ stem/progenitor cells demonstrate long-term engraftment. While the functional role of CD34 is not entirely understood, one group reported CD34 knockout mice have significantly decreased hematopoiesis in the yolk sac, marrow, and spleen although mature circulating blood cells appear normal.3
The structure of CD34 is typical of the sialomucin family of adhesion molecules. The highly sialylated O-glycosylated extracellular domain of CD34 is believed to encourage a highly charged rodlike structure that projects from the cell surface, available for intercellular interactions. The extracellular domain of CD34 shares structural similarity with another sialomucin, CD43, a hematopoietic cell-specific protein implicated in cell-cell adhesion, tyrosine kinase activation, and cytoskeletal interactions.4 Like CD43, CD34 is capable of inducing adhesion and signal transduction. The predominant form of CD34 expressed in hematopoietic cells is full-length CD34, having a relatively short intracellular region of 73 amino acids. An alternatively spliced form with a shorter intracellular tail is expressed at lower abundance. Full-length CD34 is highly phosphorylated by protein kinase C (PKC).5 CD34 itself does not have intrinsic kinase activity.
The natural ligand for CD34 expressed on progenitor cells within the bone marrow compartment is unknown. Endothelial CD34 binds to L-selectin within high endothelial venules6; thus, by analogy it may be that hematopoietic CD34 binds a selectin-like protein, although this remains to be demonstrated. CD34 antibodies including My10, 9C5, and QBEND10 have been reported to mimic ligand engagement by inducing homotypic and heterotypic cell adhesion, including adhesion of normal progenitor cells and KG1a cells to bone marrow stroma.7-11 CD34-mediated adhesion appears to be epitope-specific, as demonstrated by the inability of the epitope class III-specific HPCA-2 and 581 CD34 antibodies to induce adhesion. Signals transduced after engagement of CD34 lead to actin polymerization and are dependent on activation of tyrosine kinases and the presence of an intact cytoskeleton. The intracellular tail of CD34 is essential for cytoadhesion signaling but not sufficient for proliferation signaling.8 Engagement of the extracellular region of either full-length or truncated CD34 (with only 16 intracellular amino acids) can induce tyrosine phosphorylation of intracellular proteins.10 Like many adhesion molecules, CD34 may have roles in both adhesion and differentiation. In a myeloid leukemia cell line we demonstrated that forced expression impaired terminal differentiation.12 To address how CD34 transduces signals during adhesion, we sought to identify proteins that physically associated with the intracellular tail of the CD34 protein.
Adapter proteins such as the Crk family, Grb2, Shc, and Nck are known to link surface receptors that do not possess inherent kinase activity to intracellular signaling cascades. These adapters can enable receptors such as integrins to directly or indirectly interact with tyrosine or serine/threonine kinases to transmit signals. First identified by ten Hoeve et al,13 CrkL is a 39-kd adapter protein that consists of 1 SH2 and 2 SH3 domains and is 60% homologous to CrkII. CrkL is most abundantly expressed in hematopoietic cells14 and has been implicated in pathogenesis of chronic myelogenous leukemia. Recent studies indicate CrkL may play a major role in chronic myelogenous leukemia, in which it binds the fusion protein BCR-ABL and becomes phosphorylated at Tyr207.15-17In vitro evidence suggests the formation of CrkL complexes with proteins such as C3G, c-Abl, Sos, and Dock180.18,19 Uemura et al20 reported the involvement of CrkL in adhesion to fibronectin, and recent studies reveal that overexpression of CrkL in 32D cells leads to an increased adhesion to fibronectin through activation of C3G.21 Although a direct signaling role for CrkL has not been defined, strong evidence suggests that CrkL associates with several other proteins that have been implicated in adhesion, such as Cbl, Cas, Hef1, and paxillin.19 Thus, the Crk family of proteins has been found to participate in a variety of signaling events following T-cell stimulation, hematopoietic cytokine activation, and integrin cross-linking.19
A role for CD34 in hematopoietic cell homing has been suggested. It has been postulated that interaction of CD34 with the substratum within the bone marrow compartment leads to signaling events. Because CD34 has no intrinsic kinase activity, and therefore probably requires coupling to intracellular adapter proteins to transduce intracellular signals, we examined the association of CD34 with known adapter proteins (eg, CrkII, CrkL, Grb2, Nck, and Shc). As a first step, we investigated the possibility that CrkL may interact with CD34, based on the reports indicating that CrkL is mainly expressed in hematopoietic cells and involved in signal pathways and adhesion. Our results indicate that CD34 and CrkL interact specifically and preferentially and raise the possibility that CrkL modulates CD34 signaling pathways, coupling pathways controlling differentiation and adhesion.
Materials and methods
Antibodies
The monoclonal antibodies used in this work were as follows: 581 (class III CD34; immunoglobulin G1 (IgG1; Immunotech, Marseille, France); 9C5 (class II CD34; IgG1; Baxter Healthcare, Glendale, CA); MOPC 21 (IgG1 isotype control; Sigma, St Louis, MO); HPCA-1 (My10, class I CD34; IgG1; purified from hybridoma supernatant); QBEND 10 (class II CD34; IgG1; Southern Biotechnology, Birmingham, AL); CrkII (IgG2a; BD Transduction Laboratories, Lexington, KY); Grb2 (IgG1; BD Transduction Laboratories); paxillin (5H11, IgG1; Upstate Biotechnology, Lake Placid, NY); and Abl (8E9, IgG1; BD Pharmingen, San Diego, CA). Rabbit polyclonal antibodies used in this work were as follows: CrkL (C-20), C3G (C-19), and Cbl (C-15), purchased from Santa Cruz Biotechnology (Santa Cruz, CA); and Shc (S14630) purchased from BD Transduction Laboratories. Baxter Healthcare provided the 9C5 peptide, sold as Stem Cell Releasing Peptide (9069N). This peptide had been previously identified and found by Tseng-Law et al22 to release immunomagnetically selected hematopoietic progenitor cells from Isolex 300i beads. Definitions of CD34 epitopes include the following: class I: neuraminidase-sensitive, O-sialoglycoproteinase–resistant; class II: neuraminidase-resistant, O-sialoglycoproteinase–sensitive; and class III: neuraminidase-resistant, O-sialoglycoproteinase–resistant.1 23
CD34 adhesion assay
KG1a cells growing logarithmically were adjusted to 6 × 105 cells/mL and seeded in a 6-well plate (4 mL/well) overnight at 37°C. At the specified time points, 15 μg/mL antibody was added, mixed by gentle stirring, and cells were allowed to settle at 37°C. At the end of the assay, cells were taken for microscopic examination or were harvested for biochemical analyses. Antibody engagement inhibition assays using the 9C5 peptide were performed by addition of 250 μg/mL peptide 30 minutes prior to antibody treatment.
35S metabolic labeling
Prior to metabolic labeling, logarithmically growing KG1a cells were centrifuged (300g for 10 minutes at room temperature) and resuspended in methionine/cysteine-free media (Life Technologies, Grand Island, NY) supplemented with 10% dialyzed fetal calf serum. After starving cells of methionine and cysteine for 1 hour at 37°C in 5% CO2, cells were metabolically labeled overnight with 100 μCi/mL 35S-methionine/cysteine (Trans-35S Label, ICN Pharmaceuticals, Costa Mesa, CA). Subsequently, cells were stimulated with 15 μg/mL antibody or 200 nM 12-O-tetradecanoylphorbol-13-acetate (TPA) (Sigma-Aldrich, St Louis, MO). Control samples received isotype-matched antibody or vehicle-control dimethyl sulfoxide (DMSO), respectively, for 5 minutes at 37°C. The reaction was stopped by the addition of 20 mL ice-cold wash buffer and lysates prepared as described below.
Cell extracts
Following adhesion assays, KG1a cells were immediately transferred to cold wash buffer containing RPMI 1640 with 20 mM HEPES buffer, pH 7.3; 0.1 mM sodium orthovanadate; 0.2 mM sodium metavanadate; 1 mM sodium fluoride; and 1 mM sodium pyrophosphate. Cells were centrifuged at 300g for 10 minutes at 2°C, the supernatant was poured off, and the cells were resuspended in 500 μL cold wash buffer. Cell samples were transferred to 1.5 mL microfuge tubes, briefly centrifuged, and the supernatant pipetted off. While on ice, cells were lysed in 200 μL cold lysis buffer containing 1% Nonidet P-40 (NP-40), 10 mM Tris, pH 7.4, 50 mM NaCl, 50 mM sodium fluoride, 2 mM sodium orthovanadate, 4 mM sodium metavanadate, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 μg/mL aprotinin, and 1 μg/mL leupeptin for 10 minutes and then centrifuged at 16 000g for 10 minutes at 2°C. The supernatant was transferred to new tubes either for precipitation with glutathione-S–transferase (GST)-fusion proteins or for immunoprecipitation with antibodies (see below).
GST–fusion protein construction
TheGST-CD34ifull fusion protein was made by fusion of GST protein encoded in the vector pGEX-2TK (Pharmacia, Piscataway, NJ) to the CD34 sequence encoding the intracellular tail of full-length CD34 (Figure 2). To accomplish this, polymerase chain reaction (PCR) products with flanking EcoRI sites were generated that encoded the entire intracellular region and 14 of 23 amino acids of the transmembrane region of full-length CD34, using as a template the pCD34full plasmid.24 The primers for PCR were as follows: sense, 5′-CTGGAATTCTGCTGGCCATCTTGGGCACC-3′ and antisense, 5′-GAGGAATTCATCACAGTTCTGTGTCAGCCACC-3′. PCR conditions for amplification were 94°C for 3 minutes, then 35 cycles (94°C for 45 seconds, 60°C for 45 seconds, 72°C for 90 seconds), followed by an extension cycle of 72°C for 10 minutes. The PCR products were gel-purified, restricted with EcoRI, and inserted in-frame into the EcoRI site of pGEX-2TK. GST-fusion proteins of CrkL (CrkL3′ and CrkL5′) were generated from reverse transcriptase–PCR products from K562 CrkL complementary DNA in-frame into theBamHI and EcoRI cloning sites of the pGEX-3X vector (Pharmacia) as described in Figure 6A. The primers for PCR were as follows: CrkL: sense, 5′-GCGCAGGATCCTGTCCTCCGCCAGGTTCGAC-3′ and antisense, 5′-GCGCAGAATTCGCAATCACTCGTTTTCATCTGGG-3′. PCR parameters for amplification were 35 cycles at 94°C for 1 minute, 50°C for 1 minute, and 72°C for 1 minute; and 1 extension cycle of 10 minutes at 72°C. The PCR products were gel-purified. The CrkL PCR product was digested at internal restriction sites for EcoRI (nucleotide 1088) and BamHI (nucleotide 1106), resulting in 2 fragments, which were cloned separately into pGEX-3X (Pharmacia). Gst-CrkL 5′ contained amino acids 1 to 192, encompassing regions encoding the N-terminal SH2 and SH3 domains. Gst-CrkL 3′ contained amino acids 199 to 214, encompassing the C-terminal SH3 only. Plasmids were transformed into DH5α Escherichia coli (Life Technologies) according to the manufacturer's instructions. All constructs were verified for correct sequence. Fusion proteins were expressed in BL21 E coli (Pharmacia). Control GST protein was derived from the pGEX-2TK vector. Correct expression of fusion proteins was verified by immunoblotting with antibodies directed against GST, CD34, or CrkL as appropriate.
Fusion protein production
All GST-fusion proteins were produced following the method of Smith and Johnson25 with some modification. Briefly, bacteria were grown overnight at 37°C with shaking at 225 revolutions per minute (rpm). The bacteria were diluted 1:8 and grown at 18°C in a 225-rpm shaker until the OD550 reached 0.6 units (about 3 hours). Fusion protein production was induced with 0.1 mM isopropyl beta-D-thiogalactopyranoside for 3 hours at 18°C while shaking at 225 rpm. Bacteria were centrifuged for harvest, supernatant was discarded, and pellets were solubilized in cold MTBS buffer (16 mM Na2HPO4, 4 mM NaH2PO4, 150 mM NaCl containing 1% wt/vol Triton X-100, and 1 mM PMSF). The bacterial lysate was subjected to alternating 15-second bursts of sonication with 30-second incubations on ice 4 times. Following sonication, the lysate underwent 3 cycles of freeze-thaw (freezing in an alcohol–dry ice slurry followed by thawing in a 37°C water bath) for final lysis. Bacterial debris was pelleted and then discarded. Glutathione Sepharose (1 mL bed volume for 500 mL bacterial culture) was added to the supernatant and allowed to incubate at room temperature for 1 hour while rocking. The protein-Sepharose complex was then centrifuged and washed 4 times with MTBS buffer containing 0.1% Triton X-100. GST-fusion protein–Sepharose complexes were routinely stored at 4°C for less than 6 months.
GST precipitation
Prior to precipitation glutathione-Sepharose and GST-fusion protein–Sepharose complexes were equilibrated with 2 washes of 1% NP-40 lysis buffer (see above). KG1a whole-cell lysates of 4 × 106 cells per point were precleared by incubation with equilibrated 20 μg GST-Sepharose and 20 μL bed volume glutathione-Sepharose beads for 1 hour while rotating at 4°C. GST-fusion proteins (20 μg bound to Sepharose/point) were aliquotted into 1.5 mL microfuge tubes and then were equilibrated with 2 washes of 1% NP-40 lysis buffer. The precleared lysates were then transferred to the equilibrated specific GST-fusion protein and incubated for 3 hours while rotating at 4°C. The complexes were washed 5 times in 1% NP-40 lysis buffer containing 1 mM PMSF. Laemmli sample buffer (3 ×) was added, the samples were boiled for 5 minutes, and the sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE)/Western blot analysis was performed.
Peptide competition assays
To demonstrate the specificity of the CrkL:CD34 interaction, peptides covering sequences of the intracellular domain of CD34 were synthesized. These included the intracellular CD34 sequences peptide No. 1 (KNGTGQATSRNGHSAR) or peptide No. 2 (RRSWSPTGER). Peptides were solubilized in deionized, distilled water and used at 10-fold higher concentration than GST-fusion proteins. Following cell harvest and lysis, lysate samples were precleared of nonspecific interactions as described for GST-fusion protein precipitations. Lysates were then incubated with 200 μg specified peptide for 1 hour at 4°C while rocking. GST-fusion protein precipitation followed as detailed above.
Immunoprecipitation
To verify the interaction of CD34 with CrkL in vivo, coimmunoprecipitation studies were performed. The 9C5 antibody is nonimmunoprecipitating; therefore, to immunoprecipitate CD34 the My10 antibody was used. KG1a cells were lysed as described. Cell lysates were precleared for 1 hour at 4°C while rocking with protein A beads equilibrated in lysis buffer. After preclearing, lysates were centrifuged and the supernatant was transferred to fresh 1.5 mL microfuge tubes. My10 or control isotype-matched antibody was added and allowed to incubate for 2 hours at 4°C while rocking. The lysates were precipitated with equilibrated protein A beads (washed twice with lysis buffer) for an additional 1 hour at 4°C while rocking. The captured antibody protein complexes were washed 5 times with lysis buffer. Laemmli sample buffer (3 ×) was added, and the samples were boiled and then loaded onto SDS-PAGE. Western blot analysis was then performed to detect interacting proteins.
Results
CD34-induced adhesion is epitope specific
Others8-11 have observed CD34-mediated homotypic adhesion following engagement of the CD34 antigen. Several lines of evidence indicate that CD34-activated adhesion is dependent upon epitope binding of anti-CD34 antibodies to the CD34 antigen on the cell surface. Reports indicate that only antibodies of class I and II (binding to epitopes near the N terminus of CD34) cause adhesion, while antibodies of class III (binding to an epitope closer to the plasma membrane) cannot mediate adhesion. We confirmed these reports in studies shown in Figure 1. KG1a human myeloid leukemia cells were treated with CD34 monoclonal antibodies in homotypic adhesion assays. Class I (HPCA-1 [My10]) and class II (9C5 and QBEND10) but not class III (581) antibodies rapidly induced cell-cell adhesion (within 30 minutes). To rule out any possibility that this adhesion involved Fc receptor binding, we performed competition studies using CD34 peptide. For these studies, the 9C5 peptide 9069N,22 which mimics amino acids 14 to 19 of the mature CD34 antigen, was added to KG1a cells 30 minutes prior to addition of the 9C5 antibody. As shown in Figure 1B, at 250 μg/mL blocking peptide cell-cell adhesion was 100% inhibited. At 50 μg/mL, approximately 50% inhibition was observed (data not shown). These results clearly demonstrate that engagement of the extracellular domain of CD34 induced cell-cell adhesion and that CD34 antibody-mediated adhesion was not the result of Fc receptor stimulation. Engagement of CD34 by antibody did not change the rate of growth of KG1a cells, or change the cell cycle profile, compared with cells treated with control isotype-matched immunoglobulin alone (data not shown).
Engagement of the extracellular domain of the CD34 antigen induces homotypic adhesion.
(A) Adhesion is epitope class–specific: Monoclonal antibodies recognizing class I and II, but not class III, CD34 epitopes induce cell-cell adhesion. KG1a cells (0.7 × 106-1.0 × 106 cells/mL) were stimulated with 15 μg/mL IgG1 monoclonal antibody for 30 minutes at 37°C. Engagement of CD34 class I (with HPCA-1 [My10]) or class II epitopes (with QBEND 10) strongly induced homotypic adhesion of KG1a cells. Engagement of class III epitope (with 581) had no effect. (B) Cell-cell adhesion can be competitively inhibited by CD34 peptide. Prior to adhesion assays, cells in the right panel were pretreated for 30 minutes with 250 μg/mL blocking peptide (Stem Cell Releasing Peptide [9069N]). Each cell sample then received 15 μg/mL 9C5 antibody, and adhesion assays were carried out as in panel A. The 9C5 peptide mimics amino acids 14 to19 of the mature CD34 antigen. At this concentration, cell-cell adhesion was entirely inhibited. At 50 μg/mL, approximately 50% inhibition was observed (data not shown).
Engagement of the extracellular domain of the CD34 antigen induces homotypic adhesion.
(A) Adhesion is epitope class–specific: Monoclonal antibodies recognizing class I and II, but not class III, CD34 epitopes induce cell-cell adhesion. KG1a cells (0.7 × 106-1.0 × 106 cells/mL) were stimulated with 15 μg/mL IgG1 monoclonal antibody for 30 minutes at 37°C. Engagement of CD34 class I (with HPCA-1 [My10]) or class II epitopes (with QBEND 10) strongly induced homotypic adhesion of KG1a cells. Engagement of class III epitope (with 581) had no effect. (B) Cell-cell adhesion can be competitively inhibited by CD34 peptide. Prior to adhesion assays, cells in the right panel were pretreated for 30 minutes with 250 μg/mL blocking peptide (Stem Cell Releasing Peptide [9069N]). Each cell sample then received 15 μg/mL 9C5 antibody, and adhesion assays were carried out as in panel A. The 9C5 peptide mimics amino acids 14 to19 of the mature CD34 antigen. At this concentration, cell-cell adhesion was entirely inhibited. At 50 μg/mL, approximately 50% inhibition was observed (data not shown).
Construction of GST-CD34 proteins
To delineate intracellular signals modified by CD34, we designed and constructed GST-fusion proteins of the intracellular domain of full-length CD34. Of interest, the CD34 tail is highly conserved between species (Figure 2). We predict that important molecular interactions occur at the CD34 tail, and we were interested in identifying those interactions. Therefore, we fused the entire intracellular region and half of the transmembrane region of CD34 to the C terminus of GST (Figure 2) and used this GST-CD34 fusion protein as “bait” in pull-down experiments. Because CD34 has no intrinsic kinase activity, we predicted that CD34 required coupling to an intracellular adapter protein to transduce signals. GST-CD34 fusion protein was verified by Western blotting with specific antibodies directed against intracellular CD34 (data not shown).
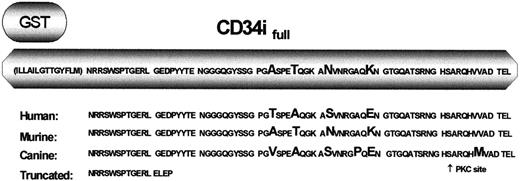
GST-CD34ifull fusion protein.
The native CD34 protein contains 23 transmembrane and 73 intracellular amino acids in addition to the extracellular domain. Depicted is the amino acid sequence of CD34 that was expressed as a GST-fusion protein in studies designed to precipitate CD34 interacting protein(s). This region includes roughly half the transmembrane domain (parentheses) and all of the intracellular domain of full-length CD34. Listed below is a comparison of the predicted amino acid sequences of human, murine, and canine CD34, demonstrating the high degree of conservation between species. Amino acids differing between species are enlarged. Depicted on the bottom line is the corresponding sequence of truncated CD34. Note that the first 12 intracellular amino acids are 100% conserved between species.
GST-CD34ifull fusion protein.
The native CD34 protein contains 23 transmembrane and 73 intracellular amino acids in addition to the extracellular domain. Depicted is the amino acid sequence of CD34 that was expressed as a GST-fusion protein in studies designed to precipitate CD34 interacting protein(s). This region includes roughly half the transmembrane domain (parentheses) and all of the intracellular domain of full-length CD34. Listed below is a comparison of the predicted amino acid sequences of human, murine, and canine CD34, demonstrating the high degree of conservation between species. Amino acids differing between species are enlarged. Depicted on the bottom line is the corresponding sequence of truncated CD34. Note that the first 12 intracellular amino acids are 100% conserved between species.
Specific proteins interact with GST-CD34ifull
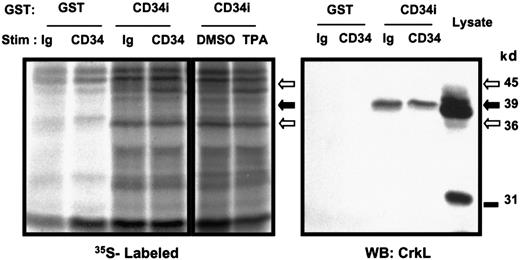
To profile different proteins that might associate with the intracellular tail of CD34, KG1a cells were metabolically labeled and GST-pull-down experiments were performed. Logarithmically growing KG1a cells were starved of methionine and cysteine briefly, and35S-methionine/cysteine was then added to allow radiolabeling of cellular protein. Subsequently, 9C5 or control MOPC antibodies were added for 5 minutes to the KG1a cell suspension to stimulate homotypic adhesion. In parallel experiments, cells were treated 5 minutes either with 200 nM TPA to activate PKC or with the DMSO vehicle control (Figure 3). We have previously shown that CD34 is rapidly and highly phosphorylated by activated PKC.5 Activation of PKC coincides with a change in cellular localization of CD34, translocating CD34 from the cytosol to the cell surface.26 By precipitation of whole-cell lysates using GST protein alone or GST-CD34ifull, we detected several radiolabeled proteins (39, 36, 33, and 45 kd) that associated with GST-CD34ifull fusion protein but not GST protein. Of the proteins identified to bind GST-CD34ifull, the 45-kd protein band showed an increased association with GST-CD34ifull protein following CD34 antibody ligation or stimulation with TPA, with respect to controls (Figure 3). The 36-kd protein showed a strong constitutive signal associated with GST-CD34ifull compared with parental GST protein, as evident in Figure 3. Further studies are in progress to identify the 45- and 36-kd proteins. Thus, we have demonstrated that it is possible to detect proteins that bind CD34 in both an inducible and constitutive manner under the assay conditions used. We proceeded to identify the faint 39-kd band as CrkL protein, as described below.
Detection of GST-CD34ifull–associated proteins.
To screen for potential CD34-associated proteins, logarithmically growing KG1a cells were adjusted to 6 × 105/mL and loaded with 35S-methionine/cysteine for 16 hours to label intracellular proteins. Cells were stimulated with 15 μg/mL CD34 antibody (“CD34”; 9C5), Ig control (“Ig”; MOPC 21), 200 nM TPA, or 0.1% DMSO control. After 30 minutes at 37°C, cells were washed, lysed, and 35S-labeled proteins were pulled down with 20 μg GST-CD34ifull fusion protein (“CD34i”) or control GST protein (“GST”). Shown is an autoradiograph indicating inducible precipitation of an unidentified 45-kd band with GST-CD34ifull fusion protein after stimulation of KG1a cells with anti-CD34 or TPA. Also shown is a 36-kd protein band constitutively associated with GST-CD34ifull. A faint 39-kd protein band constitutively associated with GST-CD34ifullis visible below the 45-kd band. Results demonstrate that under these assay conditions, both inducible and constitutive protein interactions with GST-CD34 can be detected. To specifically identify any known signaling intermediates present in material precipitated with GST-CD34ifull, in a parallel experiment nitrocellulose filters were immunoblotted with anti-CrkL antibody (results shown here) or with antibodies corresponding to certain other adapter proteins (Figure 7). The data indicate that CrkL protein was precipitated with the GST-CD34ifull fusion protein, and not with GST control, and indicate interaction (direct or indirect) between CD34 and CrkL proteins in vitro. WB indicates Western immunoblot analysis.
Detection of GST-CD34ifull–associated proteins.
To screen for potential CD34-associated proteins, logarithmically growing KG1a cells were adjusted to 6 × 105/mL and loaded with 35S-methionine/cysteine for 16 hours to label intracellular proteins. Cells were stimulated with 15 μg/mL CD34 antibody (“CD34”; 9C5), Ig control (“Ig”; MOPC 21), 200 nM TPA, or 0.1% DMSO control. After 30 minutes at 37°C, cells were washed, lysed, and 35S-labeled proteins were pulled down with 20 μg GST-CD34ifull fusion protein (“CD34i”) or control GST protein (“GST”). Shown is an autoradiograph indicating inducible precipitation of an unidentified 45-kd band with GST-CD34ifull fusion protein after stimulation of KG1a cells with anti-CD34 or TPA. Also shown is a 36-kd protein band constitutively associated with GST-CD34ifull. A faint 39-kd protein band constitutively associated with GST-CD34ifullis visible below the 45-kd band. Results demonstrate that under these assay conditions, both inducible and constitutive protein interactions with GST-CD34 can be detected. To specifically identify any known signaling intermediates present in material precipitated with GST-CD34ifull, in a parallel experiment nitrocellulose filters were immunoblotted with anti-CrkL antibody (results shown here) or with antibodies corresponding to certain other adapter proteins (Figure 7). The data indicate that CrkL protein was precipitated with the GST-CD34ifull fusion protein, and not with GST control, and indicate interaction (direct or indirect) between CD34 and CrkL proteins in vitro. WB indicates Western immunoblot analysis.
CD34ifull binds CrkL
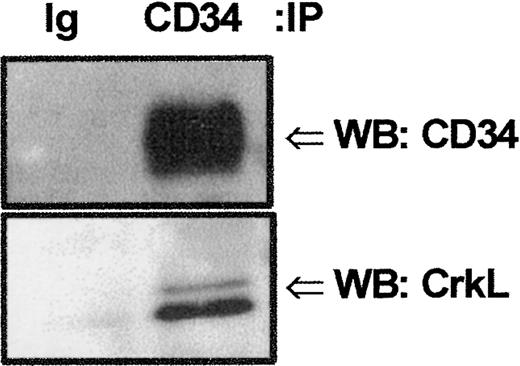
We screened by Western blot analysis against a panel of signaling intermediates known to transduce signals from surface receptors to downstream intracellular signaling events. Because the adapter family of proteins functions as bridges within protein complexes to link certain cell-surface receptors without intrinsic kinase activity to tyrosine or serine/threonine kinases or phosphatases,13,14 21 we initially focused our attention on whether any of these proteins interacted with CD34. The 39-kd CrkL protein has been shown to be preferentially expressed in hematopoietic cells. Therefore, we performed SDS-PAGE/Western blot analyses with anti-CrkL following GST-fusion protein precipitation of KG1a whole-cell lysates to determine if CrkL associated with GST-CD34ifull. These studies indicated the adapter protein CrkL bound to GST-CD34ifull (Figure 3) and not to GST alone. Engagement of the extracellular region of CD34 antigen with antibody did not detectably change the intensity of CrkL binding as detected by Western analyses; therefore, CrkL association with CD34 appeared to be constitutive. To demonstrate that endogenously expressed CD34 can bind to CrkL in vivo, we investigated the ability of monoclonal anti-CD34 (My10) antibody to coimmunoprecipitate CrkL from KG1a cells. Without prior antibody engagement of the CD34 antigen, KG1a cells were harvested, lysed, and immunoprecipitated with My10 (Figure4). CD34 and CrkL coimmunprecipitated with the My10 antibody, but not to control IgG, as detected by immunoblotting. Our studies indicate endogenous CD34 and CrkL associate in vivo.
Detection of potential CrkL:CD34 interactions.
Endogenous CrkL and CD34 proteins associate in vivo. Coimmunoprecipitation (IP) studies were performed to confirm CrkL:CD34 interaction. KG1a cell lysates (8 × 106cells/point) were precleared with protein A–Sepharose, and proteins tightly associated with CD34 protein were coimmunoprecipitated with CD34 antibody (HPCA-1; 5 μg). Precipitated proteins were separated by 10% SDS-PAGE and transferred to nitrocellulose. Filters were cut according to protein size and immunoblotted (WB) either with rabbit anti-CrkL antibody or with mouse anti-CD34 antibody. CrkL and CD34 proteins were detected in the same sample coimmunoprecipitated with anti-CD34 but not in the sample precipitated with isotype-matched control immunoglobulin (MOPC 21). These results demonstrate that endogenous CrkL and CD34 proteins are associated, either directly or indirectly within a protein complex.
Detection of potential CrkL:CD34 interactions.
Endogenous CrkL and CD34 proteins associate in vivo. Coimmunoprecipitation (IP) studies were performed to confirm CrkL:CD34 interaction. KG1a cell lysates (8 × 106cells/point) were precleared with protein A–Sepharose, and proteins tightly associated with CD34 protein were coimmunoprecipitated with CD34 antibody (HPCA-1; 5 μg). Precipitated proteins were separated by 10% SDS-PAGE and transferred to nitrocellulose. Filters were cut according to protein size and immunoblotted (WB) either with rabbit anti-CrkL antibody or with mouse anti-CD34 antibody. CrkL and CD34 proteins were detected in the same sample coimmunoprecipitated with anti-CD34 but not in the sample precipitated with isotype-matched control immunoglobulin (MOPC 21). These results demonstrate that endogenous CrkL and CD34 proteins are associated, either directly or indirectly within a protein complex.
CD34ifull interacts with CrkL at a membrane-proximal intracellular CD34 domain
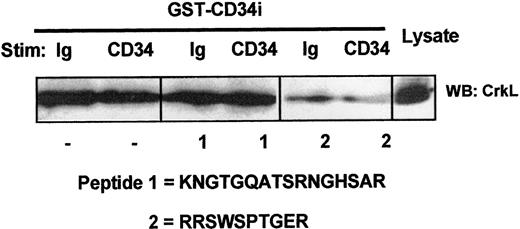
We sought to confirm the specificity of CD34:CrkL interaction as well as to identify the domain of GST-CD34ifull associating with the endogenous CrkL protein. Using synthesized peptides of the intracellular region of CD34, including peptide No. 1 (KNGTGQATSRNGHSAR) and peptide No. 2 (RRSWSPTGER), we performed competition assays. As shown in Figure 5, our investigations demonstrate that only CD34 peptide No. 2 competitively inhibited GST-CD34ifull from precipitating CrkL from whole-cell lysates of KG1a cells. Thus, the association of CD34 with CrkL appears to be dependent on a 10–amino acid intracellular sequence (Figure 5) near the transmembrane region of CD34 (Figure 2). This highly conserved domain includes an RRSWS sequence and a proline, which may be critical to CrkL binding.
Localization of the CrkL:CD34ifullinteraction domain on CD34.
Competitive inhibition of CrkL:CD34ifull association by intracellular CD34 peptide. To localize the putative CD34 domain that serves to interact with CrkL, competitive inhibition studies were performed using synthetic peptides corresponding to highly conserved domains within the intracellular tail of CD34. KG1a cell lysates (4 × 106 cells/point) were precleared with GST-Sepharose. Prior to pull-down with GST-CD34ifull fusion protein, certain samples were incubated 30 minutes with 200 μg/mL CD34 peptide No. 1 or No. 2, as indicated. Following SDS-PAGE and transfer to nitrocellulose membrane, proteins present in GST-CD34ifull–precipitated material were immunoblotted (WB) with rabbit anti-CrkL antibody. Binding of CrkL was competed with peptide No. 2 (RRSWSPTGER), thereby demonstrating that CrkL specifically interacts with CD34 (directly or indirectly) at a CD34 domain corresponding to this sequence. The location of this intracellular sequence corresponds to a region near the transmembrane domain that is encoded in both full-length and truncated CD34 proteins and that is entirely conserved between human, murine, and canine species. Peptide No. 1 was not able to inhibit CrkL, suggesting that interactions with CrkL do not occur at the corresponding CD34 domain that is nearer to the carboxyl tail of CD34.
Localization of the CrkL:CD34ifullinteraction domain on CD34.
Competitive inhibition of CrkL:CD34ifull association by intracellular CD34 peptide. To localize the putative CD34 domain that serves to interact with CrkL, competitive inhibition studies were performed using synthetic peptides corresponding to highly conserved domains within the intracellular tail of CD34. KG1a cell lysates (4 × 106 cells/point) were precleared with GST-Sepharose. Prior to pull-down with GST-CD34ifull fusion protein, certain samples were incubated 30 minutes with 200 μg/mL CD34 peptide No. 1 or No. 2, as indicated. Following SDS-PAGE and transfer to nitrocellulose membrane, proteins present in GST-CD34ifull–precipitated material were immunoblotted (WB) with rabbit anti-CrkL antibody. Binding of CrkL was competed with peptide No. 2 (RRSWSPTGER), thereby demonstrating that CrkL specifically interacts with CD34 (directly or indirectly) at a CD34 domain corresponding to this sequence. The location of this intracellular sequence corresponds to a region near the transmembrane domain that is encoded in both full-length and truncated CD34 proteins and that is entirely conserved between human, murine, and canine species. Peptide No. 1 was not able to inhibit CrkL, suggesting that interactions with CrkL do not occur at the corresponding CD34 domain that is nearer to the carboxyl tail of CD34.
The CrkL C-terminal SH3 domain interacts with endogenous CD34
To further confirm the interaction between CD34 and CrkL and to identify the CrkL domain associating with endogenous CD34, we constructed GST-fusion constructs of certain domains of CrkL, including GST-CrkL3′ (C-terminal SH3) and GST-CrkL5′ (N-terminal SH2SH3) (Figure 6A). Interestingly, only GST-CrkL3′ (C-terminal SH3) demonstrated the ability to precipitate endogenously expressed CD34 from whole-cell lysates of KG1a cells (Figure 6B). By contrast, GST-CrkL5′ fusion protein precipitated the c-abl protein, but not CD34, presumably by association between c-abl and the CrkL N-terminal SH3 domain, as has been previously reported27 (Figure7B). Thus, these data indicate CrkL specifically interacts with CD34 through its C-terminal SH3 domain.
Localization of the CrkL:CD34ifullinteraction domain on CrkL.
To localize the putative CrkL domain that serves to interact with CD34, GST-CrkL fusion proteins containing partial CrkL sequences were constructed and then used to precipitate endogenous CD34 protein. (A) GST-CrkL fusion proteins. The native CrkL protein contains 1 SH2 and 2 SH3 domains, as shown. Depicted are the CrkL constructs used in studies designed to precipitate interacting proteins, including CD34. Two GST-CrkL fusion proteins were constructed: GST-5′CrkL encompassed CrkL amino acids 1 to 194, covering the SH2 and N-terminal SH3 domains (but not the C-terminal SH3); GST-3′CrkL encompassed CrkL amino acids 197 to 303, covering the C-terminal SH3 only. (B) GST-CrkL precipitation studies. KG1a cells were stimulated 5 minutes with 9C5 antibody (“CD34”) to engage the extracellular domain of CD34 in adhesion assays (or with control immunoglobulin MOPC 21 [“Ig”]). Lysates from 4 × 106 cells per point were precleared with GST-Sepharose, and then proteins were pulled down with 20 μg GST, 20 μg GST-CD34ifull (“CD34i”), 2 μg GST-3′CrkL C-terminal SH3 domain (“CrkL 3′SH3”), or 2 μg 5′CrkL SH2–N-terminal SH3 domain (“CrkL 5′SH2-SH3”) bound to GST-Sepharose. Precipitated proteins were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted (WB) with QBEND10 CD34 antibody. Results demonstrate constitutive association between endogenous CD34 and the C′SH3 domain of GST-CrkL protein but not to the SH2-N′SH3 CrkL domain. In contrast, the SH2-N′SH3 CrkL domain, but not the C′SH3 domain, precipitated C-Abl (Figure 7B).
Localization of the CrkL:CD34ifullinteraction domain on CrkL.
To localize the putative CrkL domain that serves to interact with CD34, GST-CrkL fusion proteins containing partial CrkL sequences were constructed and then used to precipitate endogenous CD34 protein. (A) GST-CrkL fusion proteins. The native CrkL protein contains 1 SH2 and 2 SH3 domains, as shown. Depicted are the CrkL constructs used in studies designed to precipitate interacting proteins, including CD34. Two GST-CrkL fusion proteins were constructed: GST-5′CrkL encompassed CrkL amino acids 1 to 194, covering the SH2 and N-terminal SH3 domains (but not the C-terminal SH3); GST-3′CrkL encompassed CrkL amino acids 197 to 303, covering the C-terminal SH3 only. (B) GST-CrkL precipitation studies. KG1a cells were stimulated 5 minutes with 9C5 antibody (“CD34”) to engage the extracellular domain of CD34 in adhesion assays (or with control immunoglobulin MOPC 21 [“Ig”]). Lysates from 4 × 106 cells per point were precleared with GST-Sepharose, and then proteins were pulled down with 20 μg GST, 20 μg GST-CD34ifull (“CD34i”), 2 μg GST-3′CrkL C-terminal SH3 domain (“CrkL 3′SH3”), or 2 μg 5′CrkL SH2–N-terminal SH3 domain (“CrkL 5′SH2-SH3”) bound to GST-Sepharose. Precipitated proteins were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted (WB) with QBEND10 CD34 antibody. Results demonstrate constitutive association between endogenous CD34 and the C′SH3 domain of GST-CrkL protein but not to the SH2-N′SH3 CrkL domain. In contrast, the SH2-N′SH3 CrkL domain, but not the C′SH3 domain, precipitated C-Abl (Figure 7B).
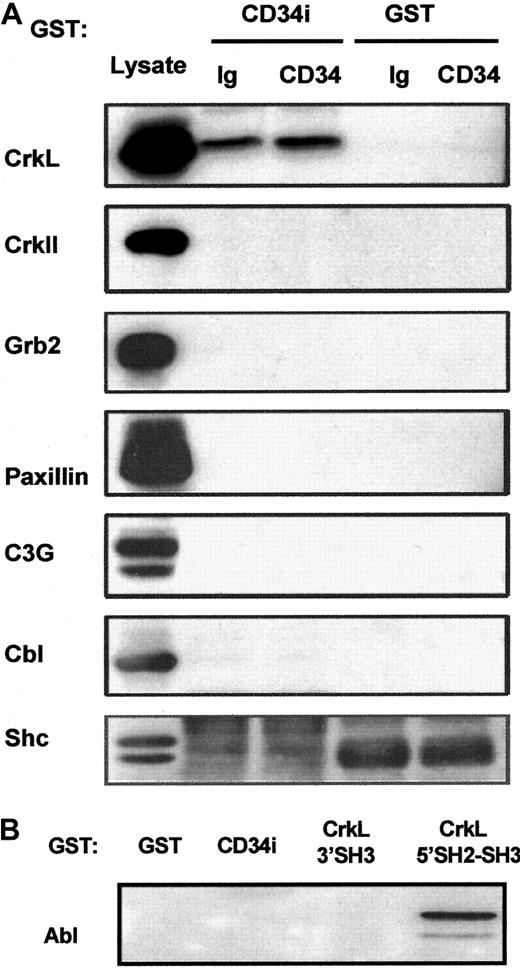
Potential CD34 interactions with CrkII, Grb2, Shc, C3G, Cbl, paxillin, or Abl.
(A) GST-CD34ifull fusion protein pull-down studies. In an effort to detect associations between CD34 and other signaling intermediates in addition to CrkL, GST-CD34ifullprecipitation studies were performed, and precipitated proteins were immunoblotted. KG1a cells were stimulated briefly (30 minutes) with 9C5 anti-CD34 (“CD34”) or MOPC 21 isotype control (“Ig”) in adhesion assays, and cells then were washed and lysed. Lysates from 4 × 106 cells per point were precleared with GST-Sepharose, and proteins were precipitated with 20 μg GST-CD34ifull (“CD34i”) or 20 μg GST (“GST”) protein, as indicated. Proteins were separated by SDS-PAGE, transferred to nitrocellulose membrane, and filters were cut according to protein size. Membranes were then immunoblotted with antibodies, as indicated, to determine whether CrkII, Grb2, paxillin, C3G, Cbl, or Shc proteins were present in the protein complex precipitated by GST-CD34ifull. As a positive control for immunoblotting, KG1a lysate was loaded in neighboring lanes, shown at left. Results demonstrate that CrkL protein was selectively precipitated with the GST-CD34ifull fusion protein and that CrkII, Grb2, Shc C3G, Cbl, and paxillin proteins could not be detected in precipitated material. Thus, GST-CD34ifull apparently does not tightly associate with these proteins under conditions employed in our assays. (B) GST-CD34ifull and GST-CrkL pull-down studies. In experiments similar to those described in Figure 5, GST-fusion proteins were used to precipitate c-Abl from KG1a lysates. Abl was identified by immunoblotting with anti-Abl antibody. GST-CD34ifull did not detectably precipitate c-Abl protein; in contrast, Abl was precipitated with GST-CrkL5′SH2-SH3, an important positive control for our studies.
Potential CD34 interactions with CrkII, Grb2, Shc, C3G, Cbl, paxillin, or Abl.
(A) GST-CD34ifull fusion protein pull-down studies. In an effort to detect associations between CD34 and other signaling intermediates in addition to CrkL, GST-CD34ifullprecipitation studies were performed, and precipitated proteins were immunoblotted. KG1a cells were stimulated briefly (30 minutes) with 9C5 anti-CD34 (“CD34”) or MOPC 21 isotype control (“Ig”) in adhesion assays, and cells then were washed and lysed. Lysates from 4 × 106 cells per point were precleared with GST-Sepharose, and proteins were precipitated with 20 μg GST-CD34ifull (“CD34i”) or 20 μg GST (“GST”) protein, as indicated. Proteins were separated by SDS-PAGE, transferred to nitrocellulose membrane, and filters were cut according to protein size. Membranes were then immunoblotted with antibodies, as indicated, to determine whether CrkII, Grb2, paxillin, C3G, Cbl, or Shc proteins were present in the protein complex precipitated by GST-CD34ifull. As a positive control for immunoblotting, KG1a lysate was loaded in neighboring lanes, shown at left. Results demonstrate that CrkL protein was selectively precipitated with the GST-CD34ifull fusion protein and that CrkII, Grb2, Shc C3G, Cbl, and paxillin proteins could not be detected in precipitated material. Thus, GST-CD34ifull apparently does not tightly associate with these proteins under conditions employed in our assays. (B) GST-CD34ifull and GST-CrkL pull-down studies. In experiments similar to those described in Figure 5, GST-fusion proteins were used to precipitate c-Abl from KG1a lysates. Abl was identified by immunoblotting with anti-Abl antibody. GST-CD34ifull did not detectably precipitate c-Abl protein; in contrast, Abl was precipitated with GST-CrkL5′SH2-SH3, an important positive control for our studies.
Lack of CD34ifull association with certain other adapters (CrkII, Grb2, or Shc) and signaling intermediates (C3G, c-Cbl, paxillin, c-Abl)
In an attempt to determine whether certain other adapter and signaling intermediates associate with CD34, we probed nitrocellulose filters containing SDS-PAGE–separated GST-CD34ifull–precipitated proteins with antibodies against other adapter proteins (Figure 7). We did not demonstrate binding between CD34ifull and CrkII, Grb2, or Shc adapter protein (although shc studies were complicated by high background with 2 independent anti-shc antibodies). This indicates that association between CD34 and the CrkL adapter is specific. Importantly, because GST-CD34ifull did not precipitate CrkII, it appears that CrkL and CrkII do not have overlapping specificity with regard to CD34. Nitrocellulose membranes were also probed with antibodies against CrkL binding proteins C3G, Cbl, paxillin, and Abl, which could potentially physically link CD34 to CrkL. We did not detect their presence in GST-CD34ifull–precipitated material either with or without engagement of the CD34 antigen with anti-CD34. We also did not detect these proteins in coimmunoprecipitated complexes with CD34, indicating that neither C3G, Cbl, paxillin, nor Abl can interact with high affinity with CD34 in vivo. Together the data suggest that CD34 directly associates with CrkL.
Discussion
In this study, we provide evidence that the sialomucin CD34 can associate with the 39-kd adapter protein CrkL in a constitutive manner in hematopoietic progenitor cells. We also show the likelihood that during adhesion CD34 associates with a 45-kd protein, and studies are ongoing to identify this protein. To our knowledge, this is the first report identifying CD34-associating proteins.
In accordance with previous findings,7-11 we demonstrated that class I and II, but not class III, monoclonal antibodies can induce adhesion mediated by CD34 (Figure 1A). We conclusively show by CD34 peptide adhesion inhibition studies that adhesion is mediated by engagement of CD34 rather than mediated by Fc receptor stimulation (Figure 1B). The peptide competition assays illustrate the epitope specificity of CD34 engagement and its consequently induced homotypic cellular adhesion. Taken together, this evidence supports that engagement of the CD34 antigen via specific epitopes leads to homotypic adhesion.
A 45-kd protein was detected that associated with the intracellular tail of full-length CD34 immediately after engagement of CD34 with antibody (Figure 3). Because this protein also associated with the CD34 tail after TPA stimulation, it is interesting to speculate that this protein may provide a link between pathways activated by PKC and CD34 adhesion. In support of this notion, we found that certain inhibitors of PKC can suppress CD34-mediated adhesion (data not shown). Previously, we determined that CD34 is potentially involved in signal transduction in myeloid progenitor cells through its involvement with PKC. CD34 is highly phosphorylated by PKC5,26,28 and also stably phosphorylated by other unidentified serine kinases.29 Studies are in progress to identify the 45-kd protein detected in this study in order to provide important insights in the adhesion signaling pathway of CD34.
The viral Crk oncogene (v-Crk) is known to induce sarcomas in chicken, and its cellular homologs c-Crk I, c-Crk II, and Crk-like (CrkL) have been implicated in many signal transduction events, including cell differentiation and migration.30 Although the precise function of CrkL is not known, evidence suggests that CrkL links signals generated from extracellular stimuli to multiple intracellular pathways. For example, signals generated from ligation of β1 integrin receptor, M-CSF receptor, c-kit, interleukin receptors, and the insulin receptor can be transduced by CrkL and can alter cytoskeletal reorganization, migration, proliferation, and differentiation. Multiple protein-protein interactions with Crk proteins have been observed. Virtually all CrkL SH2 domains analyzed bind to specific surface motifs of proteins that contain a phosphorylated tyrosine residue essential for high-affinity binding. The CrkL SH2-dependent interactions conform to pY-X-X-P–containing epitopes expressed on c-Cbl, paxillin, and p130 Cas. CrkL N-terminal SH3-dependent interactions tend to conform to the proline-rich consensus sequence P-X-X-P-X-R/K, and interactions with Abl, Bcr-Abl, C3G, SOS, EPS15, and DOCK180 have been demonstrated. In general, the specificities of CrkII and CrkL overlap. However, Platanias et al31 recently reported that in vivo C3G interacts with CrkL but not CrkII in hematopoietic cells, although the SH3 domains of both CrkL and CrkII interacted in vitro with C3G.32 33These data demonstrate that interactions among Crk family members are not entirely overlapping. Similarly, we show that CD34 interacts with CrkL, but we have repeatedly been unable to demonstrate CD34 interaction with CrkII in vitro (discussed below).
Our studies definitively demonstrate that CD34 and CrkL can associate with each other. In vitro, CD34 fusion protein precipitated endogenous CrkL from KG1a lysates (Figure 3), and CrkL fusion protein precipitated endogenous CD34 (Figure 6B). In vivo, the presence of a complex containing both endogenous CrkL and CD34 was found (Figure 4). In contrast, associations between CD34 and Abl, C3G, Cbl, paxillin, Shc, and Grb2 were not detectable (Figure 7), indicating that the CD34:CrkL interaction was highly specific. CD34 peptide competition studies supported this conclusion (Figure 5). It is unclear whether CD34 and CrkL interact directly. However, certain known CrkL-binding proteins Abl, C3G, Cbl, and paxillin (discussed below) did not bind CD34 in vitro (Figure 7) or in vivo (data not shown) in our studies. This suggests that the association between CrkL and CD34 may be direct or that some unidentified third partner is involved. If directly binding, CrkL may constitutively associate with CD34, and then certain of these CrkL-binding proteins may transduce signals from CrkL, which convey the downstream effects of CD34 (eg, that lead to actin polymerization during CD34-mediated adhesion). Thus, our inability to detect association between CD34 and Abl, C3G, Cbl, paxillin, Shc, or Grb2 proteins suggests that these proteins probably do not serve to link CrkL to CD34, but the results leave open the possibility that these proteins could link downstream CD34 signals to the cytoskeleton during CD34-mediated adhesion.
Our studies have localized potential sites of interaction between CD34 and CrkL. For CD34 we identified the sequence RRSWSPTGER that is present on both full-length and truncated CD34, which prevented CrkL precipitation with intracellular CD34 in vitro in a competition assay (Figure 5). This region is identical in full-length and truncated CD34 and is 100% conserved between species (Figure 2). Thus, it appears from the evolutionary conservation that this is a region critical to function of the CD34 molecule. We delineated the motif of CrkL interaction with CD34 by constructing GST-fusion proteins covering different domains of the adapter protein CrkL (Figure 6). We determined that the CrkL C-terminal SH3 domain is the putative CD34-interacting domain. These results were intriguing because CrkL C-terminal SH3-associating proteins have not been previously reported to our knowledge. While our studies failed to detect association between CD34 and CrkL SH2–N-terminal SH3, we found as expected that the endogenous Abl protein associated with this CrkL fusion protein, an important control for our studies (Figure 7B). Thus, CD34 may be the first binding protein to be reported that associates with the CrkL C-terminal SH3 domain. Interestingly, CD34 does not contain the known proline consensus sequence PXXPXR/K important for N-terminal SH3 binding. Shi et al proposed that CrkL might also interact at other less well characterized motifs, such as are present in GCKR.34 Future studies aimed at characterizing the amino acids (within the NRRSWSPTGERL sequence) of CD34 that interact with this SH3 domain of CrkL should enhance our understanding of CrkL protein interactions. Collectively these and other studies have indicated that CD34 is a hematopoietic stem/progenitor cell adhesion-regulatory molecule. CD34 engagement may help to localize cells to the appropriate hematopoietic compartment by promoting adhesive interactions occurring at the bone marrow compartment (as proposed by Majdic et al9). For example, CD34 could anchor stem/progenitor cells to extracellular matrix components expressed on bone marrow stroma, thus enhancing their retention within this developmental niche. Down-regulation of CD34 would release cells from this niche at the time of terminal differentiation. CD34 could also contribute to tissue-specific homing of stem/progenitors to the marrow. Engagement of CD34 could enhance efficient stem/progenitor cell homing/engraftment and therefore may be therapeutically useful.
Our results report the first interactions identified between CD34 and other signaling intermediates. Significantly, we show that CrkL is implicated as a binding partner of CD34. Furthermore, we report the first interaction of the C-terminal SH3 domain of CrkL, which may reveal a clue to the role of CrkL in hematopoiesis. The functional relevance of this association is still under investigation.
Supported in part by National Institutes of Health grants DK-48374 and CA-70970.
The Johns Hopkins University holds patents on CD34 monoclonal antibodies and related inventions. C.I.C. is entitled to a share of the sales royalty received by the University under licensing agreements between the University, Becton Dickinson, and Baxter Healthcare. The terms of these arrangements have been reviewed and approved by the University in accordance with its conflict of interest policies.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Mary Jo Fackler Johns Hopkins Oncology Center, Johns Hopkins University School of Medicine, Bunting-Blaustein Cancer Research Bldg, 1650 Orleans St, Baltimore, MD; e-mail:mfackler@jhmi.edu.

![Fig. 1. Engagement of the extracellular domain of the CD34 antigen induces homotypic adhesion. / (A) Adhesion is epitope class–specific: Monoclonal antibodies recognizing class I and II, but not class III, CD34 epitopes induce cell-cell adhesion. KG1a cells (0.7 × 106-1.0 × 106 cells/mL) were stimulated with 15 μg/mL IgG1 monoclonal antibody for 30 minutes at 37°C. Engagement of CD34 class I (with HPCA-1 [My10]) or class II epitopes (with QBEND 10) strongly induced homotypic adhesion of KG1a cells. Engagement of class III epitope (with 581) had no effect. (B) Cell-cell adhesion can be competitively inhibited by CD34 peptide. Prior to adhesion assays, cells in the right panel were pretreated for 30 minutes with 250 μg/mL blocking peptide (Stem Cell Releasing Peptide [9069N]). Each cell sample then received 15 μg/mL 9C5 antibody, and adhesion assays were carried out as in panel A. The 9C5 peptide mimics amino acids 14 to19 of the mature CD34 antigen. At this concentration, cell-cell adhesion was entirely inhibited. At 50 μg/mL, approximately 50% inhibition was observed (data not shown).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/97/12/10.1182_blood.v97.12.3768/6/m_h81211161001.jpeg?Expires=1769321140&Signature=Ucvtp0KHN0kEjgGSp0ksAqN-K69UhvZm5CfrQhRqKsYcqrfP9gMzTd9SyLVWYavr4fzvgui0TVveN90KaKZ~29XTyOAKeypEbOunXL1ml15gKBNXf1~9fyK8gYcBlwMwHWha-LO4KCAg5h98M5o9detaNgHyaeBP7UNvuGYQh4e--NIYueRIjHTfnbFoOjMcTrtBrLSVcyUPC6~nq3ej4BsYjysXR-PrlxmxcV704bBkq~8dFvXxIacxVT3uHLHzpg5iDg7uwMHSDapG8ysIIbPKHOS55KMZKJ3i2GcKA-p7Nxh6-z5GKvjhJsqfDq593xeqt0RncTVI~SF3Zy5ULQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 6. Localization of the CrkL:CD34ifullinteraction domain on CrkL. / To localize the putative CrkL domain that serves to interact with CD34, GST-CrkL fusion proteins containing partial CrkL sequences were constructed and then used to precipitate endogenous CD34 protein. (A) GST-CrkL fusion proteins. The native CrkL protein contains 1 SH2 and 2 SH3 domains, as shown. Depicted are the CrkL constructs used in studies designed to precipitate interacting proteins, including CD34. Two GST-CrkL fusion proteins were constructed: GST-5′CrkL encompassed CrkL amino acids 1 to 194, covering the SH2 and N-terminal SH3 domains (but not the C-terminal SH3); GST-3′CrkL encompassed CrkL amino acids 197 to 303, covering the C-terminal SH3 only. (B) GST-CrkL precipitation studies. KG1a cells were stimulated 5 minutes with 9C5 antibody (“CD34”) to engage the extracellular domain of CD34 in adhesion assays (or with control immunoglobulin MOPC 21 [“Ig”]). Lysates from 4 × 106 cells per point were precleared with GST-Sepharose, and then proteins were pulled down with 20 μg GST, 20 μg GST-CD34ifull (“CD34i”), 2 μg GST-3′CrkL C-terminal SH3 domain (“CrkL 3′SH3”), or 2 μg 5′CrkL SH2–N-terminal SH3 domain (“CrkL 5′SH2-SH3”) bound to GST-Sepharose. Precipitated proteins were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted (WB) with QBEND10 CD34 antibody. Results demonstrate constitutive association between endogenous CD34 and the C′SH3 domain of GST-CrkL protein but not to the SH2-N′SH3 CrkL domain. In contrast, the SH2-N′SH3 CrkL domain, but not the C′SH3 domain, precipitated C-Abl (Figure 7B).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/97/12/10.1182_blood.v97.12.3768/6/m_h81211161006.jpeg?Expires=1769321140&Signature=xCfMrFpTMo50hEyYhh0iz~5H8Xo42s347zXBuAerJFeHr7eglye2NeWhLLOmNvI5Nby4JGnWEhVV86m6B~SXsR3I2mXTy8o~Xa8X0HFB5E2sbJ9UIwbrtkBs9XAihFBb0WQiq7HrYIm8xzBlUVzMV9-xn5Osp5ni4o73UOIzKKB69OOzaoOkoAp0qfAe~7jfcW2dJe3qxmcrXveDEUkyYD3zEbz~mrmIRReYmxGVVJjh4gdpWmbZiUtcb0OkF60dslHI-CdfXCJVeIwLsE57Ivu0NA8yAzj-Zz6bt1bfxoCtq7ZQ32SwcRmkcvmDD3dgns~37LWlP6KrhEB6zqasww__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal