Abstract
Patients with cytogenetically normal acute myeloid leukemia (CN-AML) show heterogeneous treatment outcomes. We used gene-expression profiling to develop a gene signature that predicts overall survival (OS) in CN-AML. Based on data from 163 patients treated in the German AMLCG 1999 trial and analyzed on oligonucleotide microarrays, we used supervised principal component analysis to identify 86 probe sets (representing 66 different genes), which correlated with OS, and defined a prognostic score based on this signature. When applied to an independent cohort of 79 CN-AML patients, this continuous score remained a significant predictor for OS (hazard ratio [HR], 1.85; P = .002), event-free survival (HR = 1.73; P = .001), and relapse-free survival (HR = 1.76; P = .025). It kept its prognostic value in multivariate analyses adjusting for age, FLT3 ITD, and NPM1 status. In a validation cohort of 64 CN-AML patients treated on CALGB study 9621, the score also predicted OS (HR = 4.11; P < .001), event-free survival (HR = 2.90; P < .001), and relapse-free survival (HR = 3.14, P < .001) and retained its significance in a multivariate model for OS. In summary, we present a novel gene-expression signature that offers additional prognostic information for patients with CN-AML.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous disorder in terms of its genetic basis, its pathophysiology, and its prognosis. In the past, the karyotype of the AML blasts has emerged as the single most important prognostic factor, and patients can be classified into favorable, intermediate, and unfavorable prognostic groups according to the presence of specific chromosomal aberrations.1,2 However, almost half of all adult AML patients present with a normal karyotype at diagnosis. These patients are usually assigned to the intermediate prognostic group. More recently, mutations in specific genes that allow the identification of prognostic subgroups in cytogenetically normal (CN) AML have been described.3 Internal tandem duplications (ITD) of the fms-like tyrosine kinase 3 (FLT3) gene are strong predictors of inferior outcome. Mutations in the nucleophosmin 1 (NPM1) gene lead to cytoplasmic localization of the NPM protein4 and are a favorable prognostic factor, especially in patients lacking FLT3 ITD.5-9 Other prognostically relevant alterations include mutations in the MLL and CEBPA genes as well as overexpression of BAALC, ERG, and EVI1.3 However, in approximately 24% of CN-AML cases, none of the aforementioned mutations can be detected.6 These findings illustrate that CN-AML itself constitutes a heterogeneous disease and that there probably are many genetic alterations in CN-AML to be discovered that affect response to treatment and survival
More recently, microarray technology has made it possible to study deregulated gene expression in AML. Several groups have used unsupervised clustering algorithms to identify subgroups of AML with different prognoses.10-12 Although such clustering results may offer insights into the biology of AML, they cannot be directly used to estimate the prognosis of newly diagnosed individual patients. Bullinger et al used cDNA microarrays and a semisupervised clustering approach to develop an outcome classifier for AML patients,13 using a dataset that contained samples with both normal and aberrant karyotypes. The prognostic utility of this signature was recently validated in an independent set of 64 CN-AML patients, using Affymetrix oligonucleotide microarrays (Affymetrix, Santa Clara, CA).14 Chromosomal abnormalities are a major prognostic factor in AML, and they strongly affect gene-expression profiles.10,15 Thus, some of the genes identified in these studies probably are surrogate markers for cytogenetic aberrations, and they may be of little value for outcome prediction in CN-AML. We therefore investigated a cohort of CN-AML patients and present a gene signature, based on the expression of 66 genes, that was specifically developed to predict treatment outcome in patients with a normal karyotype.
Methods
Patients
In the current analysis, 3 independent cohorts of patients were studied. The training cohort consisted of 163 adult patients with CN-AML. A total of 156 (95.7%) of these patients were enrolled in the multicenter AMLCG-1999 trial of the German AML Cooperative Group between 1999 and 2003.16 All patients in the training cohort received intensive double-induction and consolidation chemotherapy. The independent test cohort consisted of 79 adult patients who were diagnosed with CN-AML in 2004. Sixty-two of them were treated according to the AMLCG-1999 protocol, whereas 17 received intensive chemotherapy outside the study. For both cohorts, pretreatment diagnostic evaluation was performed centrally at the Laboratory for Leukemia Diagnostics, University of Munich, and included standard cytomorphology, cytogenetics, fluorescence in situ hybridization, polymerase chain reaction-based testing for NPM1 mutations and FLT3 ITD,5 and gene-expression profiling. FLT3 ITD to wild-type (FLT3 ITD/wt) ratio was quantified using the Genescan technique.17 All patients had a normal karyotype, based on the analysis of at least 20 metaphases.
The third independent cohort (called the “validation cohort”) comprised 64 CN-AML patients that were treated in the Cancer and Leukemia Group B (CALGB) 9621 study. Details regarding the treatment protocol and patient characteristics have been published elsewhere.18 Karyotypes were reviewed centrally, and evaluation of FLT3 ITD and NPM1 status and gene-expression profiling were performed centrally at the Ohio State University Comprehensive Cancer Center.14 The AMLCG and CALGB clinical trials were approved by the local institutional review boards of all participating centers, and informed consent was obtained from all patients in accordance with the Declaration of Helsinki. Both the AMLCG and CALGB trials have been registered at www.clinicaltrials.gov (NCT00266136 and NCT00002925).
Gene-expression profiling and data preprocessing
Gene-expression profiling in the training cohort was performed using Affymetrix HG-U133 A&B microarrays, and samples from the test cohort were analyzed using Affymetrix HG-U133 plus 2.0 microarrays. Details regarding sample preparation, hybridization, and image acquisition have been described previously.15,19 Briefly, mononuclear cells from bone marrow (BM) or peripheral blood (PB) were enriched by Ficoll gradient centrifugation. Total RNA was isolated using the RNeasy Mini Kit (QIAGEN, Hilden, Germany). cDNA preparation, in vitro transcription, labeling, hybridization, washing, and staining steps were performed according to standard Affymetrix protocols. Quality control consisted of visual inspection of the array image for artifacts, assessment of RNA degradation plots, and inspection of rank versus residual plots after normalization and probe set summarization. All microarray data has been submitted to Gene Expression Omnibus and can be found under the accession number GSE12417.
Initially, 200 HG-U133 A&B chip pairs and 102 HG-U133 plus 2.0 microarrays were evaluated. Microarrays from 8 patients were excluded because of poor quality. The remaining sets of 194 and 100 arrays were separately normalized using the variance stabilizing normalization algorithm,20 and probe set expression values were calculated by the median polish method. Finally, 163 HG-U133 A&B chip pairs (161 BM, 2 PB samples) and 79 HG-U133 plus 2.0 chips (74 BM, 5 PB samples) were selected for further analysis, based on the availability of follow-up data and information on NPM1 and FLT3 ITD status. The HG-U133 A&B microarrays formed the training set, whereas the HG-U133 plus 2.0 data served as the test set.
Pretreatment PB samples from the 64 patients in the validation cohort were analyzed using Affymetrix HG-U133 plus 2.0 microarrays. Details on sample preparation and array hybridization have been published elsewhere.21 Briefly, PB mononuclear cells were enriched by Ficoll-Hypaque gradient, total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA), and the samples were then prepared according to standard Affymetrix protocols. Invariant-set normalization and calculation of log2-model based expression index values were performed using the dChip software package (Harvard University, Cambridge, MA).14
Development of a prognostic signature
To identify a prognostic gene signature for CN-AML, we applied the “supervised principal components” approach suggested by Bair and Tibshirani.22 Univariate Cox scores, which measure the correlation between gene-expression levels and overall survival of the 163 patients in the training cohort, were computed for each of the 44 754 probe sets common to both microarray types. In all subsequent analyses, only the 86 probe sets with absolute Cox scores of greater than 2.9 were used (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). The decision regarding this threshold was reached using a 10-fold cross-validation procedure that evaluated the prognostic significance of the resulting model within the training data.22
To construct a prognostic score, the expression values for each of the 86 probe sets were first centered to a mean of 0. Next, a principal component analysis (PCA) was performed on the training data, using the expression values of only those 86 probe sets. We then used the correlation of each gene with the first principal component as weights to construct a continuous outcome predictor. The predictor for each patient can be calculated as a linear combination of the centered expression values of the 86 probe sets, each multiplied with the weight obtained by PCA (Figure 1; Table S1).
Heatmaps visualizing the expression levels of the 86 probe sets used in the prognostic gene signature. The heatmaps show the relative levels of expression for the 86 probe sets in 163 patients in the training cohort (A), 79 patients in the test cohort (B), and in the 64 patients in the validation cohort (C). The probe sets are arranged according to their univariate Cox score for correlation with overall survival, with those having the highest negative score shown on top and those with the highest positive score shown at the bottom. Green spots indicate below-average expression; red spots, above-average expression. Below each heatmap, the continuous score calculated from these 86 expression values is visualized. NPM1 and FLT3 status are color-coded: A yellow mark indicates the presence of an NPM1 mutation or FLT3 ITD; blue mark, the absence of the mutation. In panel A, the 2 patient groups generated by 2-means clustering of the continuous score values are depicted in green and red. The mean of the 2 cluster centers was subsequently used as a cutoff threshold in the test and validation cohorts to allow dichotomization of the continuous risk score, as indicated in green and red in panels B and C.
Heatmaps visualizing the expression levels of the 86 probe sets used in the prognostic gene signature. The heatmaps show the relative levels of expression for the 86 probe sets in 163 patients in the training cohort (A), 79 patients in the test cohort (B), and in the 64 patients in the validation cohort (C). The probe sets are arranged according to their univariate Cox score for correlation with overall survival, with those having the highest negative score shown on top and those with the highest positive score shown at the bottom. Green spots indicate below-average expression; red spots, above-average expression. Below each heatmap, the continuous score calculated from these 86 expression values is visualized. NPM1 and FLT3 status are color-coded: A yellow mark indicates the presence of an NPM1 mutation or FLT3 ITD; blue mark, the absence of the mutation. In panel A, the 2 patient groups generated by 2-means clustering of the continuous score values are depicted in green and red. The mean of the 2 cluster centers was subsequently used as a cutoff threshold in the test and validation cohorts to allow dichotomization of the continuous risk score, as indicated in green and red in panels B and C.
Validation of the prognostic gene signature
In contrast to the training population, the 79 samples in the test cohort were analyzed with a newer version of the Affymetrix microarray. Probe set-wise comparison of the expression values between the 2 cohorts showed that in the test cohort (hybridized on HG-U133 plus 2.0 arrays), most probe sets tended to have lower mean signal levels and higher SDs than in the training dataset (analyzed on HG-U133 A&B chips; Figure S1). To achieve a similar distribution of expression values, we adjusted the mean expression value of each probe set in the test dataset to 0 and then scaled the SD of each probe set to the values observed in the training cohort. The 64 patients in the validation cohort were also analyzed on HG-U133 plus 2.0 microarrays. Rescaling of the expression values in the validation cohort was performed as in the test cohort. A continuous risk score was calculated for each patient in the test and validation group on the basis of the 86 selected probe sets and their respective weights, as determined in the training dataset.
Statistical analyses
Overall survival (OS) was defined as time from study entry until death from any cause, and event-free survival (EFS) was defined as time from study entry until removal from study because of failure to achieve complete remission, relapse, or death from any cause. Relapse-free survival (RFS) was defined as time from the date of complete remission (CR) until relapse or death, regardless of cause.23 Patients alive without an event were censored at the time of their last follow-up. Clinical variables were compared between the study cohorts using Fisher exact test for categorical variables, and the Mann-Whitney U test for continuous variables.
One-way analysis of variance was used to assess the association of baseline clinical and molecular characteristics with the continuous microarray risk score, and Cox proportional hazards models were used to test the prognostic value of the gene-expression score. Hazard ratios (HRs) were computed for a difference in the score equal to its interquartile range. Therefore, the HRs for the microarray-based score represent the increase in risk associated with a change in score equal to the difference between the 25th and 75th percentiles of the score values.
To assess the prognostic impact of our microarray-based score after adjusting for the influence of other clinical and molecular risk factors, multivariate Cox regression models were constructed. The following variables, in addition to the gene-expression risk score, were included in the multivariate model based on their established prognostic relevance in CN-AML patients and the availability of complete data: age (as a continuous variable), NPM1 mutation status, and FLT3 ITD. In the test cohort, Genescan analysis of the FLT3 ITD/wt allelic ratio was available for 78 of the 79 patients, and patients were thus divided into 3 groups: those with no detectable FLT3 ITD, those with detectable FLT3 ITD and a low FLT3 ITD/wt ratio (ie, a value of ≤ 0.8), and those with a high FLT3 ITD/wt ratio (ie, a value of > 0.8).17 Information on FLT3 ITD/wt ratio was not available for the validation cohort; therefore, FLT3 ITD status was used as a binary variable. Because we sought to assess the prognostic value of the gene-expression score after adjustment for possible confounders, no variable selection algorithm was used, and all 4 variables were retained in the Cox regression model. The proportional hazards assumption was checked by examining Schoenfeld residuals for each variable individually. To account for possibly important unobserved covariates, we also considered models including an additional random effect (a so-called frailty term). The results were very similar to those of the models without a frailty term (data not shown).
For the purpose of visualization, we also derived a dichotomized version of the predictor. We performed 2-means clustering (using a k-means clustering algorithm) on the values of the continuous predictor for the 163 patients in the training set. This method of dichotomization does not make use of information about survival times. The mean of the 2 cluster centers (ie, a score value of 0.1703) was then chosen as the cutoff threshold, which was subsequently used to classify patients in the test and validation cohorts into predicted “good” and “poor” outcome groups (Figure 1). The Kaplan-Meier method was used to generate survival curves for patient subgroups identified by dichotomization of the gene-expression score. All statistical analyses were performed using the R 2.7.0 software package24 and routines from the biostatistics software repository Bioconductor.25
Results
Characteristics of the patient cohorts
The clinical characteristics of the 3 patient cohorts at the time of the initial diagnostic evaluation are shown in Table 1. The patients in the training cohort were younger and presented with higher leukocyte counts and lower hemoglobin levels than those in the test cohort. The incidence of FLT3 ITD was higher in the training cohort than in the test cohort (P = .005), whereas the proportion of patients with NPM1 mutations was similar in both groups. The prognostically favorable combination of an NPM1 mutation in the absence of FLT3 ITD (FLT3 ITD−/NPM1+) was present in 23% of the patients in the training cohort and in 34% of subjects in the test cohort (P = .09). Because of the inclusion criteria of the CALGB 9621 study (with an upper age limit of 59 years), the 64 patients in the validation cohort were significantly younger than those in the training and test cohorts. A total of 71% of the patients had an NPM1 mutation, a significantly higher percentage than in the training cohort (P = .01). A total of 35% of the patients in the validation cohort were FLT3 ITD−/NPM1+, similar to the other 2 cohorts.
Identification of prognostic genes
Using univariate Cox regression, we identified 86 probe sets on the Affymetrix microarrays that were significantly associated with OS in the training cohort, with an absolute Cox score of more than 2.9 (Figure 1; Table S1). These 86 probe sets corresponded to 60 annotated genes and 6 expressed sequence tag clones, and 13 genes were represented by more than one probe set each. We then used principal component analysis to assign a weight to each of these probe sets, and a linear combination of the weighted expression values of the 86 probe sets was tested for its ability to serve as a “risk score.”
Association of the gene-expression score with baseline patient characteristics and clinical outcomes
Our test cohort consisted of 79 patients treated on the same therapeutic protocol as the patients in the training cohort. A continuous risk score was calculated for each patient using the probe sets and weights identified in the training cohort. The presence of a FLT3 ITD was associated with higher values of the gene-expression–based continuous risk score (P < .001), and the score also showed a positive association with higher leukocyte counts (P = .001; Table S2). In an analysis of treatment outcomes, a higher continuous risk score was a significant negative predictor of both OS (HR = 1.85; 95% CI, 1.25-2.74; P = .002) and EFS (HR = 1.73; 95% CI, 1.24-2.42; P = .001). Information on the response to induction treatment was available for 77 of the 79 patients, and 50 of them (65%) had a CR. In a logistic regression model, a higher gene-expression score was associated with a lower chance of reaching CR (P = .045). Among patients in CR, a higher gene-expression score predicted shorter RFS (HR = 1.76; 95% CI, 1.07-2.89; P = .025). For visualization of these results, survival curves generated by dichotomization of the continuous risk score are shown in Figure 2.
Outcomes in the test dataset according to the prognostic gene-expression score. The prognostic signature was developed exclusively in the training data and then applied to the test data. For visualization of patient survival according to the gene-expression score, a cutoff value was defined in the training cohort using a k-means clustering algorithm. This threshold was then used for dichotomization of score values in the test cohort. Kaplan-Meier plots of (A) OS and (B) EFS were generated for patients with high versus low gene-expression scores. Data on EFS were available for 77 of the 79 patients in the test cohort. (C) RFS for the 50 patients in the test cohort who reached CR.
Outcomes in the test dataset according to the prognostic gene-expression score. The prognostic signature was developed exclusively in the training data and then applied to the test data. For visualization of patient survival according to the gene-expression score, a cutoff value was defined in the training cohort using a k-means clustering algorithm. This threshold was then used for dichotomization of score values in the test cohort. Kaplan-Meier plots of (A) OS and (B) EFS were generated for patients with high versus low gene-expression scores. Data on EFS were available for 77 of the 79 patients in the test cohort. (C) RFS for the 50 patients in the test cohort who reached CR.
To further corroborate our results, we assessed our prognostic signature in an entirely independent validation cohort of patients from the CALGB study group. For these 64 patients, we used the same probe sets and weights as in our original training and test cohorts to calculate the microarray-based risk score. Similar to the test cohort, we found a positive association of the continuous risk score with the presence of FLT3 ITD (P < .001). No significant association of the gene-expression score and other clinical variables was observed. In the validation cohort, the continuous risk score showed a significant negative correlation with both OS (HR = 4.11; 95% CI, 2.10-8.03; P < .001) and EFS (HR = 2.90; 95% CI, 1.63-5.18; P < .001). Fifty-four patients (84%) had a CR after induction chemotherapy, but the continuous risk score did not predict the chance of reaching CR (P = .21). However, among patients who had a CR after induction therapy, a higher gene-expression score predicted a shorter RFS (HR = 3.14; 95% CI, 1.62-6.07, P < .001). Survival curves generated by dichotomization of the continuous risk score are displayed in Figure 3.
Outcome prediction in the independent validation cohort. The gene-expression risk score was calculated using the same probe set weights as in the original training and test cohorts, and the cutoff value defined in the training cohort was used for dichotomization. (A) OS and (B) EFS for the entire cohort of 64 patients. (C) RFS for the 54 patients who reached CR.
Outcome prediction in the independent validation cohort. The gene-expression risk score was calculated using the same probe set weights as in the original training and test cohorts, and the cutoff value defined in the training cohort was used for dichotomization. (A) OS and (B) EFS for the entire cohort of 64 patients. (C) RFS for the 54 patients who reached CR.
Performance of the gene-expression score in the context of FLT3 ITD, NPM1 mutations, and age
Because we observed an association of the continuous gene-expression score with FLT3 ITD status, Cox proportional hazards models were used to determine whether our gene-expression signature provided additional prognostic information after adjustment for FLT3 and other known risk factors. In multivariate analyses, after adjusting for age, FLT3 ITD/wt allelic ratio and NPM1 mutation status, the gene-expression score remained a significant predictor for OS in the test cohort (P = .037; Table 2). Along with the continuous risk score, higher age was a negative prognostic factor for OS, and the presence of an NPM1 mutation was of borderline significance. A higher gene-expression score also was significantly associated with shorter EFS (P = .024) in the test cohort after adjusting for age, FLT3 ITD/wt allelic ratio, and NPM1 status (Table 2). In the validation cohort, where only patients younger than 60 years were included, a higher gene-expression score (P = .007) and wild-type NPM1 predicted a worse OS, whereas age was not significant. In a multivariate analysis of EFS, the gene-expression score was of borderline significance in the smaller validation cohort (P = 0.09; Table 3).
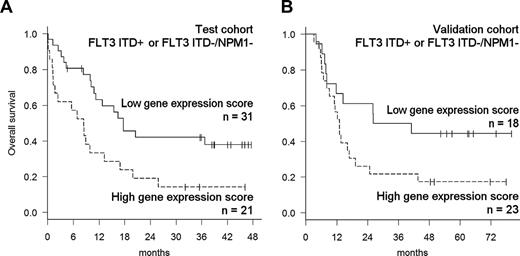
Patients with an isolated NPM1 mutation in the absence of FLT3 ITD (FLT3 ITD−/NPM1+) constitute a subgroup of CN-AML with favorable treatment outcomes. On the other hand, the presence of FLT3 ITD or the FLT3 ITD−/NPM1− genotype are commonly regarded as high-risk molecular features in CN-AML.5-9 We therefore tested whether microarray profiling provided additional prognostic information for the latter subgroup of patients. A higher value of the gene-expression score was associated with inferior OS of such “molecular high risk” patients in both the test cohort (HR = 1.71; 95% CI, 1.14-2.58; P = .009) and the validation cohort (HR = 2.07; 95% CI, 1.21-3.53; P = .008; Figure 4).
Prognostic value of the gene-expression signature in patients with high-risk molecular features. To visualize the prognostic value of the gene-expression predictor in patients who either carry a FLT3 ITD or are FLT3 ITD−/NPM1−, score values were dichotomized based on the cutoff value defined in the training cohort. (A) Kaplan-Meier plots of OS of molecular high-risk patients in the test cohort. (B) OS of molecular high-risk patients in the validation cohort.
Prognostic value of the gene-expression signature in patients with high-risk molecular features. To visualize the prognostic value of the gene-expression predictor in patients who either carry a FLT3 ITD or are FLT3 ITD−/NPM1−, score values were dichotomized based on the cutoff value defined in the training cohort. (A) Kaplan-Meier plots of OS of molecular high-risk patients in the test cohort. (B) OS of molecular high-risk patients in the validation cohort.
Discussion
We present a prognostic gene signature, based on the expression levels of 66 genes and expressed sequence tags represented by 86 oligonucleotide microarray probe sets, which is able to predict OS, EFS, and RFS in AML patients with a normal karyotype. Because generating a prognostic score based on thousands of expression values per patient involves the risk of “overfitting,” assessing the performance of a prognostic gene signature on independent datasets is of critical relevance.26 In this respect, it is important to note that we verified the prognostic value of our signature in 2 independent and diverse patient cohorts from different study groups.
Our initial test cohort consisted of patients from a German multicenter treatment study (AMLCG-1999). The clinical characteristics of these patients were generally similar to the training cohort, which included patients from the same trial. To corroborate our findings, we also tested our predictor in a second group of 64 patients from a US multicenter trial (CALGB 9621). There were substantial differences between the CALGB validation cohort and the German patients in the test and training datasets: The German AMLCG trial compared 2 double-induction chemotherapy regimens using either one or 2 cycles of high-dose cytarabine, followed by consolidation chemotherapy and either autologous stem cell transplantation or prolonged monthly maintenance chemotherapy.16 In contrast, the CALGB trial tested induction therapy with variable doses of daunorubicin and etoposide along with conventional-dose cytarabine, with or without multidrug resistance modulation, followed by autologous stem cell transplantation or an alternative intensification regimen.18 Furthermore, the CALGB trial did not include patients with secondary or treatment-related AML. The median age in the validation cohort was much lower than in the training and test cohorts because the CALGB study only enrolled patients younger than 60 years; thus, the baseline risk of this population was lower. Moreover, the frequency of important molecular markers, namely, FLT3 ITD and NPM1 mutations, varied among the cohorts. Despite these differences, the gene-expression score was a significant predictor for OS, EFS, and RFS in both populations. We used multivariate models to adjust for the differences in age and the frequencies of FLT3 ITD and NPM1 mutations, and the results of these analyses are remarkably consistent between the test and the validation cohort. In summary, our results indicate that the performance of our gene-expression predictor is robust in patient populations with different baseline characteristics.
Another important difference between the study cohorts is the predominant use of BM specimens for gene-expression profiling in the training and test cohorts, whereas PB samples were analyzed in the validation cohort. The performance of our predictor did not appear to be compromised by the different starting materials. Similarly, in the study by Radmacher et al,14 PB specimens were used to reproduce the gene signature by Bullinger et al13 that had been developed from a mixed PB/BM dataset. Other studies of gene-expression in acute leukemia have included both BM and PB samples, and in unsupervised analyses, no clustering of samples according to starting material was reported.10,11,13 There are very few studies that directly compared expression patterns in paired PB and BM samples in AML, but the available reports indicate a high degree of correlation between both sources.13,27
Despite the important differences between the patient populations and the analytical methods, our gene-expression score was a significant predictor of survival in both the test and validation cohorts. Assessing the generalizability of a prognostic marker requires testing both its reproducibility in an independent patient sample as well as its transportability to patient cohorts with different geographic and methodologic background.28 In this respect, the fact that we tested our predictor in 2 patient cohorts from different study groups from Europe and the United States strongly supports the validity of our gene signature.
The prognostic value of our signature was conserved after adjustment for age and NPM1 and FLT3 ITD mutations, and it also allowed prognostic stratification of the high-risk subgroup of patients who were either FLT3 ITD+ or FLT3 ITD−/NPM1−. Patients with these genotypes have similar survival according to many,5-9 but not all,29 studies; therefore, additional prognostic factors in addition to NPM1 and FLT3 would be useful. Moreover, the analyses of RFS, although limited by the smaller patient numbers, suggest that the gene-expression signature may identify patients at increased risk for relapse after CR. This would be helpful for guiding risk-adapted postremission therapy. Taken together, these findings underscore the value of our signature, which apparently works well in diverse populations of patients with CN-AML.
The 64 patients in the validation cohort have been used previously to confirm the prognostic value of a gene signature published by Bullinger at al.13 That signature was originally developed in a cohort of patients with both normal and abnormal karyotypes using cDNA microarrays and comprises 133 genes. Subsequently, Radmacher et al constructed a prognostic classifier using 81 of the genes from the original signature (represented by 157 Affymetrix probe sets) and tested it in a group of 64 CN-AML patients.14 Interestingly, the Radmacher classifier also showed a strong correlation with patients' FLT3 ITD status. It predicted OS and disease-free survival in univariate analyses, but it did not provide additional prognostic information in multivariate models that adjusted for FLT3 status. Of note, only 7 probe sets in that classifier, corresponding to 5 genes, overlap with our prognostic gene signature.
Similar to the study of Radmacher et al,14 there was a strong correlation between the risk score derived from our prognostic gene signature and FLT3 ITD status. This finding is not surprising, considering that the presence of a FLT3 length mutation is a strong negative risk factor, and that the genes in our signature were selected according to their correlation with OS in the training cohort, without adjustment for FLT3 status. However, our results suggest that our gene signature is not merely a surrogate marker for FLT3 ITD because in multivariate analyses the gene-expression score turned out to be the stronger prognostic factor. We therefore hypothesize that the genes in our signature do not simply mirror the presence of a FLT3 ITD but are also influenced by other (and maybe unknown) genetic alterations associated with poor survival in CN-AML. In this respect, our results are similar to a recent report on an expression signature developed to predict FLT3 ITD in CN-AML.30 Although this signature misclassified approximately 20% of cases in an independent test set, the gene signature in fact was a stronger predictor of survival than FLT3 status itself. Six of the 20 genes in the FLT3 ITD prediction signature overlap with our prognostic signature. Neben et al examined differential gene expression in CN-AML patients with and without FLT3 ITD, and published a list containing the 11 top-ranking genes.31 Of note, there is no overlap between the genes represented in our survival predictor and the FLT3 target genes described in that study.
It will be interesting to explore how the genes in our risk signature are linked to cellular pathways that are involved in the pathogenesis of the disease and to investigate how these pathways influence a patient's risk of death. We therefore compared our results with other reports on deregulated gene expression in AML. A recent study examined the gene-expression profiles of cytarabine-sensitive and cytarabine-resistant murine cell lines.32 The homolog of a gene that was strongly up-regulated in chemo-resistant cell lines, Wbp5, is also part of our predictor. Another gene from our prognostic signature, TCF4, is part of a gene set that defines chemo-resistance in childhood acute lymphoblastic leukemia.33 Finally, Heuser et al34 analyzed samples from adult AML patients with resistance to induction chemotherapy. Three genes contained in our signature (FHL1, CD109, and SPARC) were associated with a poor response to chemotherapy in that study.34 Such overlap between studies performed in various patient populations and in animal models might point toward genes that are functionally relevant.
Besides NPM1 mutations and FLT3 ITD, several other molecular prognostic markers have been described in CN-AML, including CEBPA mutations, FLT3 TKD mutations, MLL PTD, and overexpression of ERG, BAALC, and EVI1.3 Currently, little information is available on the interactions and relative importance of these risk factors. In our present study, we were unable to assess the performance of our gene-expression score in the context of these novel markers due to missing data and the relatively small sample sizes in our study, which make it difficult to capture the effects of low-frequency markers, such as CEBPA mutations. Therefore, our microarray-based signature needs to be tested together with other new risk markers in a sufficiently large patient cohort before definitive conclusions about their relative importance can be drawn.
Gene-expression profiling currently is not part of the standard diagnostic workup of AML. However, previous studies have proven that microarrays can serve as a comprehensive tool for the diagnosis and classification of leukemia.15 Based on these results, diagnostic microarray platforms for routine clinical use are being developed. In this context, a gene signature that offers prognostic information for a large subgroup of AML cases (ie, CN-AML) would be very useful. Another potential advantage of gene-expression profiling is its ability to capture the effects of multiple genetic alterations simultaneously; thus, a gene-expression signature may summarize the prognostic implications of multiple “conventional” risk markers into one single score. Further prospective studies are necessary to determine whether the signature identified herein is clinically useful to stratify patients to risk-adapted remission induction or postremission treatments beyond the current capacities of conventional single molecular marker screening. Such studies could investigate, for example, whether or not this signature identifies patients with high FLT3 ITD/wt allelic ratio who do not relapse, or patients with a favorable NPM1/FLT3 profile who do ultimately relapse. Moreover, although the current study focused on deriving a signature from CN-AML and assessing its prognostic capability within this cytogenetic subset, we do not know whether this signature might have clinical utility for patients with AML and aberrant karyotypes. Further investigations should address this possibility.
In conclusion, we present a prognostic gene signature that was developed to predict outcome in patients with CN-AML. A predictor based on this signature was associated with OS, EFS, and RFS in 2 different and independent patient cohorts. Although our gene-expression score was strongly influenced by the presence of a FLT3 ITD, it provided prognostic information beyond the effects of FLT3 ITD and NPM1 mutation status in multivariate analyses. We think that, given the heterogeneity of CN-AML, a predictor that allows for refined risk assessment would be of value to identify patients who may or may not benefit from intensive therapeutic strategies, such as allogeneic stem cell transplantation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Eric Bair for helpful comments on the supervised principal components algorithm; Gudrun Mellert, Evelin Zellmeier, and Marlene Seibl for expert technical assistance; and all the patients and local investigators who participated in the AMLCG and CALGB trials and thereby made this work possible.
This work was supported by grants from the Bundesministerium für Bildung und Forschung (German National Genome Research Network; grant 01 GR 0459, M.H., U.M.; and grant 01 GS 0448, C.B., S.K.B.) and the Deutsche Forschungsgemeinschaft (DFG-German Research Council) SFB 684 projects A6 and A8 (S.K.B. and C.B.), by the National Institutes of Health (grants CA101140, CA114725, CA077658, and CA016058), and the Coleman Leukemia Research Foundation (C.D.B.).
Authorship
Contribution: K.H.M. designed research, analyzed and interpreted data, and drafted the manuscript; M.H. performed statistical analyses and cowrote the manuscript; K.S., J.B., G.M., S.P.W., P.P., R.A.L, W.E.B., T.B., and B.W. provided data and were involved in patient care; M.-C.S., A.H., M.R., and K.M. performed statistical analyses; U.M. supervised statistical analysis and provided funding; C.D.B., W.H., S.K.B., and C.B. supervised research, provided funding; and all coauthors reviewed and discussed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christian Buske, Department of Internal Medicine III, University Hospital Groβhadern, Ludwig-Maximilians Universität, Campus Groβhadern, Marchioninistr 15, 81377 München, Germany; e-mail: christian.buske@med.uni-muenchen.de; or Stefan K. Bohlander, Department of Internal Medicine III, University Hospital Groβhadern, Ludwig-Maximilians Universität, Campus Groβhadern, Marchioninistr 15, 81377 München, Germany; e-mail: stefan.bohlander@med.uni-muenchen.de.