Key Points
CN/AML patients have a high frequency of CSF3R and RUNX1 mutations.
CSF3R and RUNX1 mutations induce elevated proliferation of CD34+ cells.
Abstract
Severe congenital neutropenia (CN) is a preleukemic bone marrow failure syndrome with a 20% risk of evolving into leukemia or myelodysplastic syndrome (MDS). Patterns of acquisition of leukemia-associated mutations were investigated using next-generation deep-sequencing in 31 CN patients who developed leukemia or MDS. Twenty (64.5%) of the 31 patients had mutations in RUNX1. A majority of patients with RUNX1 mutations (80.5%) also had acquired CSF3R mutations. In contrast to their high frequency in CN patients who developed leukemia or MDS, RUNX1 mutations were found in only 9 of 307 (2.9%) patients with de novo pediatric acute myeloid leukemia. A sequential analysis at stages prior to overt leukemia revealed RUNX1 mutations to be late events in leukemic transformation. Single-cell analyses in 2 patients showed that RUNX1 and CSF3R mutations were present in the same malignant clone. Functional studies demonstrated elevated granulocyte colony-stimulating factor (G-CSF)–induced proliferation with diminished myeloid differentiation of hematopoietic CD34+ cells coexpressing mutated forms of RUNX1 and CSF3R. The high frequency of cooperating RUNX1 and CSF3R mutations in CN patients suggests a novel molecular pathway of leukemogenesis: mutations in the hematopoietic cytokine receptor (G-CSFR) in combination with the second mutations in the downstream hematopoietic transcription fator (RUNX1). The detection of both RUNX1 and CSF3R mutations could be used as a marker for identifying CN patients with a high risk of progressing to leukemia or MDS.
Introduction
Congenital neutropenia (CN) is a heterogeneous bone marrow failure syndrome characterized by severe neutropenia (blood neutrophil counts <0.5 × 109/l) and maturation arrest of myelopoiesis at the level of the promyelocytes/myelocytes.1 Autosomal-dominant and sporadic CN cases are predominantly attributable to mutations in ELANE, the gene encoding neutrophil elastase.2 Several other genetic mutations, including those in HAX1 (HCLS1-associated protein X-1), G6PC3 (glucose 6 phosphatase, catalytic, 3), GFI1 (growth factor independent 1 transcription repressor), and WAS (Wiskott-Aldrich syndrome gene), have been described in patients with CN.3-6 The majority of CN patients benefit from treatment with granulocyte colony-stimulating factor (G-CSF).7 Common pathological mechanisms for the maturation arrest of myeloid development in these patients include the lack of myeloid-specific transcription factors such as LEF-1 (lymphoid enhancer-binding factor 1) and C/EBPα (CCAAT/enhancer binding protein α), and defective G-CSF signaling.8
CN is a preleukemic syndrome with a cumulative incidence of leukemia of >20% after 20 years.9 Approximately 70% to 80% of CN patients who develop acute myeloid leukemia (AML) or myelodysplastic syndrome (MDS) acquire heterozygous G-CSF receptor (CSF3R) mutations, independent of the genetic subtype, suggesting that these mutations are involved in leukemogenesis.10-12 This pattern is distinct from de novo childhood AML in which CSFR3 mutations are very rare. Current evidence indicates that CSF3R gene mutations are not sufficient for leukemic transformation. As Welch et al13 reported, in many cases of myeloid leukemias, only 1 or 2 cooperating mutations are needed to generate the malignant founding clone. In our study, our hypothesis is that the initial driver mutations are the CSF3R mutations, and cell clones harboring CSF3R mutations have a growth advantage and acquire additional cooperating mutations (in the majority of patients RUNX1 mutations) that contribute to AML initiation and disease progression.
Point mutations in RUNX1 (runt-related transcription factor 1) have been described in de novo AML or AML secondary to MDS, radiation exposure, or chemotherapy at frequencies of 6% to 33%.14-18 High incidence of RUNX1 mutations has been associated with monosomy 7, trisomy 21, or trisomy 13.17,19,20 Most of the RUNX1 mutations are acquired heterozygous point mutations, predominantly located in the Runt homology/DNA binding (RHD) or transactivation (TAD) domains. They are associated with poor prognosis in AML.14-17,19-21 RUNX1 germline mutations, with or without acquired RUNX1 mutations, have also been described in a familial platelet disorder with a predisposition to AML.22,23
The goal of the present study was to identify the steps of leukemia progression in CN by analyzing genomic profiles of patients during the course of the disease using next-generation DNA deep-sequencing. Specifically, we sought to determine if CN patients acquired leukemia-associated mutations in the course of the development of myeloid leukemia and to assess the clinical significance of these findings.
Materials and methods
Patients
Thirty-one patients with CN who developed leukemia or MDS were analyzed. All patients had suffered from neutropenia and recurrent infections from birth and were subsequently started on treatment with G-CSF. They developed MDS or leukemia at 2 to 38 years of age. The mutation subtypes were as follows: ELANE (n = 18), HAX1 (n = 6), WAS (n = 4), ELANE plus GFI1 (n = 2), and G6PT/SLC37A4 (n = 1). Two patients were negative for ELANE, HAX1, G6PC3, GFI1, and WAS mutations. We collected bone marrow or blood samples in association with annual follow-up as recommended by the Severe Chronic Neutropenia International Registry and French Severe Chronic Neutropenia Registry.24 This study was performed with the informed consent of all subjects obtained through the respective institutional review boards and was approved by the Hannover Medical School Institutional Review Board. This study was conducted in accordance with the Declaration of Helsinki.
Pediatric AML samples were obtained from the Dutch Childhood Oncology Group (The Hague, The Netherlands), the AML-BFM Study Group (Hannover, Germany), as well as from the University Hospital in Prague (Czech Republic) and the St. Louis Hospital (Paris, France). Clinical and centrally reviewed cytogenetic and other cell-biological characteristics were made available by these cooperative groups/hospitals. All recurrent cytogenetic groups known in pediatric AML were represented among the 307 de novo pediatric AML patients (Table 2).
Mutation screening
Mutation analyses of the known leukemia-associated genes (RUNX1, NPM1, FLT3-ITD, FLT3-TKD, CEBPA, NRAS, KRAS, CBL, TET2, IDH1, IDH2, DNMT3A, SUZ12, EP300, and CSF3R) were performed using a sensitive next-generation amplicon deep-sequencing assay (454 Life Sciences, Branford, CT) or a SeqCap EZ library (Roche Nimblegen, Madison, WI) followed by sequencing on the Hisequation 2000 sequencing system (Illumina, San Diego, CA). Minimum coverage was at least 467-fold, and sequences were confirmed by ABI Sanger sequencing when supporting reads of mutated alleles were in excess of 20%. For sequential analyses of the time-course of occurrence of RUNX1 and CSF3R mutations, mutated regions of RUNX1 and CSF3R genes were amplified using polymerase chain reaction (PCR), and PCR products were sequenced using a SOLiD 5500XL ligation-based sequencing system or the SeqCap EZ library (Roche Nimblegen) followed by sequencing on the Hisequation 2000 system (Illumina). In the 302 de novo AML-samples, the protein coding sequence of the RUNX1 gene (exon 3-8) (RUNX1-002, NM_001754.4) was PCR-amplified using specific primers and subsequently sequenced in forward and reverse direction. Purified PCR products were directly sequenced from both strands. Primer sequences are available upon request. The sequence data were analyzed using CLC Workbench, version 3.5.1 (CLC Bio, Aarhus, Denmark). In case of a suspected mutation, the fragment was re-amplified and sequenced in both directions.
All RUNX1 mutated samples were also screened for CSF3R and ELANE mutations. Briefly, if cDNA was available, CSF3R cDNA was amplified and sequenced in forward and reverse direction. When only genomic DNA was available, CSF3R was amplified and sequenced to detect mutations and deletions around 2 previously reported CSF3R hotspot mutations. For ELANE, the protein coding exons (exons 1-5) were amplified and subsequently sequenced in forward and reverse direction on genomic DNA. Primer sequences are available upon request.
Results
High frequency of RUNX1 gene mutations in CN patients who developed leukemia
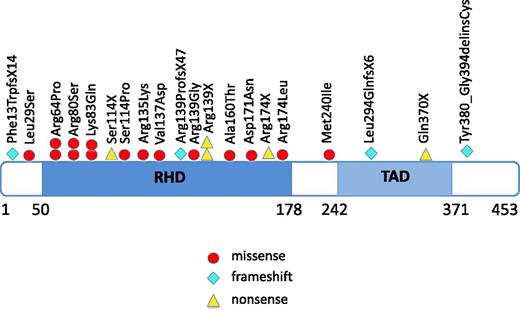
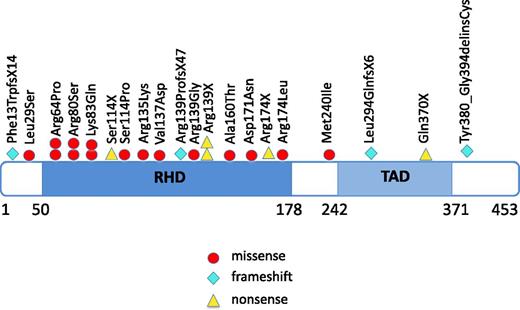
We included 31 CN patients who developed MDS or leukemia in this study (Table 1) using materials from all of the patients available to us though our international collaborations. Twenty-one patients developed leukemia and 10 patients MDS. Of the leukemic patients, 16 patients had AML, 3 had AML after MDS, 1 had biphenotypic leukemia, and one had acute lymphoblastic leukemia (ALL). Of these 31 patients, 20 (64.5%) had heterozygous RUNX1 mutations; these included missense mutations (n = 16), nonsense mutations (n = 5), frameshift mutations (n = 4), and mutations in the splice-acceptor site of intron 4 (n = 2) (Figure 1; Table 1). The most frequently affected position was Arg139, resulting in p.Arg139Gly, p.Arg139X (n = 2), and p.Arg139ProfsX47 (n = 4). Additionally, p.Arg64Pro, p.Arg80Ser, p.Lys83Gln, and Arg174 (p.Arg174X and p.Arg174Leu) mutations were identified, each in 2 patients. Of the 27 RUNX1 mutations, 18 were localized within the RHD, 4 within or proximal to the TAD and 2 in splice-sites.
Localization of RUNX1 mutations in CN patients who developed leukemia or MDS. The position of RUNX1 mutations found in CN patients, with affected amino acids numbered. Amino acid positions correspond to the RUNX1 transcript variant Q01196 (www.uniprot.org). Locations of the functionally important RHD and TAD are shown. Each symbol represents 1 patient.
Localization of RUNX1 mutations in CN patients who developed leukemia or MDS. The position of RUNX1 mutations found in CN patients, with affected amino acids numbered. Amino acid positions correspond to the RUNX1 transcript variant Q01196 (www.uniprot.org). Locations of the functionally important RHD and TAD are shown. Each symbol represents 1 patient.
Simultaneous occurrence of 2 distinct heterozygous RUNX1 mutations
In 8 patients, we detected 2 distinct heterozygous RUNX1 mutations. In one patient, 2 insertion mutations were found at the splice-acceptor site of intron 4, predicted to affect the splicing of exons 3 and 4, which encode the RHD of RUNX1. In 3 patients, 2 mutations were present solely in the RHD or were present in both RHD and TAD (1 each). In 4 patients, 1 of 2 mutations was localized to the RHD. In 2 patients, we could perform allele-specific analysis of RUNX1 mutations. Patient 10 incurred deletions leading to frame shifts on both alleles (p.Phe13TrpfsX14 and p.Arg139ProfsX47). In patient 14, 2 single missense substitutions were detected on the same allele; the first was inherited from the mother (p.Met240Ile) and is located 2 amino acids upstream of the TAD, and the second acquired mutation (p.Arg139Gly) is in the RHD of RUNX1 (supplemental Figure 1, available on the Blood Web site).
Association between RUNX1 mutations, clinical characteristics, and cytogenetics in CN patients who developed leukemia or MDS
No correlation was seen between age at progression to leukemia and RUNX1 mutations (13.8 years in RUNX1 mutated vs 10.2 years in RUNX1 wild-type, (WT) groups). No gender correlation was found; 10 male and 10 female patients acquired RUNX1 mutations, as opposed to 5 males and 6 females without RUNX1 mutations. Among 20 patients carrying mutated RUNX1, one developed AML FAB M0, 4 developed M1, 2 developed M2, 1 developed M4, and 1 developed M5. A cytogenetic analysis revealed that RUNX1 mutations were associated with monosomsy 7 in 6 patients (30%), with trisomy 21 in 6 patients (30%), and with monosomy 7/trisomy 21 in 2 patients (Table 1).
Distribution of RUNX1 mutations based on CN-specific germline mutations
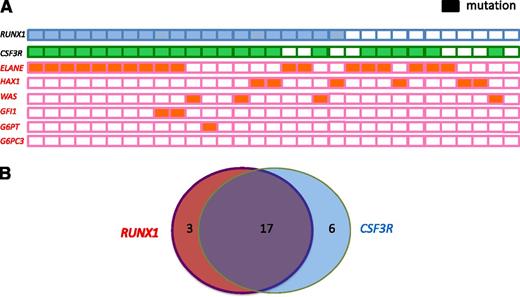
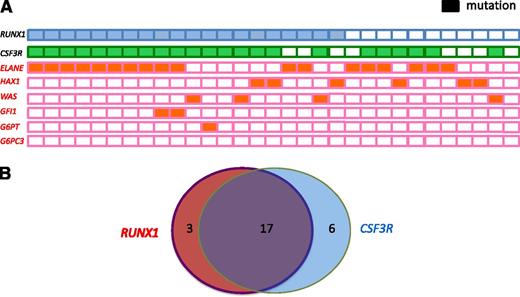
Of the 20 patients with RUNX1 mutations, 19 were positive for known CN-causing mutations: ELANE (n = 12 patients, 66%), WAS (n = 3, 75%), HAX1 (n = 3, 50%), both ELANE and GFI1 (n = 1), and G6PT/SLC37A4 (n = 1). Conversely, 5 of 17 patients harboring ELANE mutations, 3 of 6 patients with HAX1 mutations, and 1 of 4 patients with WAS mutations tested negative for RUNX1 mutations (Table 1; Figure 2A).
Frequency and distribution of acquired RUNX1 and CSF3R mutations and inherited ELANE, WAS, GFI1, GPT, and HAX1 mutations within a cohort of CN patients who developed leukemia or MDS. (A) Each patient with a RUNX1 mutation is represented by a blue rectangle, and each patient with a CSF3R mutation is represented by a green rectangle. Patients with inherited mutations are represented by orange rectangles. Open rectangles correspond to patients without mutations. (B) Venn diagram illustrating the relationship among RUNX1 and CSF3R mutations in CN patients who developed leukemia (n = 31). Diameters of each circle are roughly proportional to the number of mutations.
Frequency and distribution of acquired RUNX1 and CSF3R mutations and inherited ELANE, WAS, GFI1, GPT, and HAX1 mutations within a cohort of CN patients who developed leukemia or MDS. (A) Each patient with a RUNX1 mutation is represented by a blue rectangle, and each patient with a CSF3R mutation is represented by a green rectangle. Patients with inherited mutations are represented by orange rectangles. Open rectangles correspond to patients without mutations. (B) Venn diagram illustrating the relationship among RUNX1 and CSF3R mutations in CN patients who developed leukemia (n = 31). Diameters of each circle are roughly proportional to the number of mutations.
High frequency of cooperating RUNX1 and CSF3R mutations in CN patients who developed leukemia or MDS
To evaluate possible cooperating effects of “leukemogenic” gene mutations in combination with RUNX1 mutations, we performed targeted deep-sequencing of candidate genes known to be mutated in de novo AML, namely NPM1, FLT3, CEBPA, NRAS, KRAS, CBL, TET2, IDH1, IDH2, DNMT3A, SUZ12, EP300, and CSF3R. In 19 of 20 patients, we detected cooperating RUNX1 mutations. Intriguingly, 17 of 20 patients (80.5%) exhibited both RUNX1 and CSF3R mutations (Figure 2B), 3 had both a RUNX1 and an EP300 mutation, 2 had a RUNX1 mutation and FLT3, and 1 had a RUNX1 and a CBL mutation. Six patients with acquired CSF3R mutations had WT RUNX1. One patient with unaffected RUNX1, ELANE, or CSF3R had an activating NRAS mutation in codon 61. This may represent a unique type of CN/MDS and could suggest that NRAS and CSF3R, or RUNX1, mutations are mutually exclusive (Table 1). No mutations were found in CEBPA, DNMT3A, IDH1, IDH2, NPM1, or TET2. All CSF3R mutations were C-terminal truncated mutations, which are localized between amino acids 680 and 780.
Intriguingly, all 6 patients who were negative for RUNX1 and CSF3R mutations developed MDS only, whereas in the group of patients with cooperative RUNX1 and CSF3R mutations, 11 developed AML and only 6 MDS/AML.
Molecular karyotyping by array-CGH (comparative genomic hybridization)
To elucidate the underlying molecular mechanisms of cancer susceptibility and progression in secondary acute leukemia or MDS, we conducted a comprehensive genome-wide characterization of genomic aberrations in the transformed cells from several patients with inherited bone marrow syndromes. When available, samples at different time points of disease progression were analyzed. Thirty-one CN patients were analyzed. A summary of the genomic aberrations identified by array-CGH is given in supplemental Table 1. Large genomic alterations, namely, monosomy 7/-7q, +21q, or +3q, were associated with leukemia. Apart from common copy number variants, such as UGT2B, GSTT1, and HEATR4, no microdeletions or microduplications were detected in primary or secondary diseases.
RUNX1 mutations in de novo pediatric AML
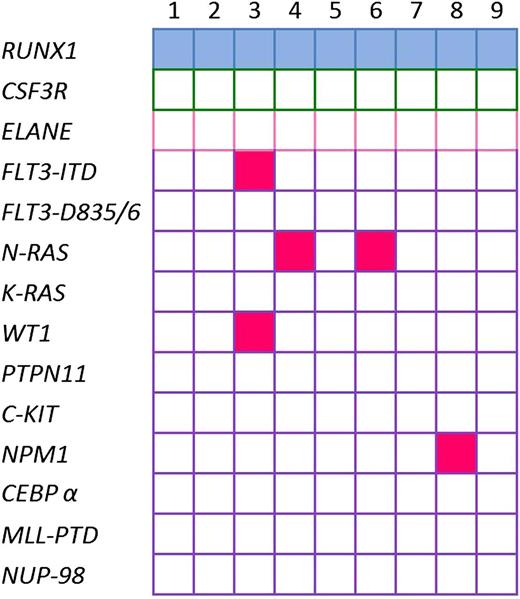
To evaluate whether de novo pediatric AML also has high frequency of RUNX1 mutations, we sequenced RUNX1 in de novo pediatric AML samples. We detected RUNX1 mutations in 9 of 307 (2.9%) pediatric patients with de novo AML (Table 2): one deletion, 4 insertions, and 4 missense mutations (single-nucleotide substitutions) for which single-nucleotide polymorphisms have not been described. We found 6 N-terminal RUNX1 mutations, 1 C-terminal mutation, and 2 mutations outside the RHD and TAD. No co-occurrence of RUNX1 mutations with CSF3R or ELANE mutations was found in these patients (Figure 3).
Mutational status of leukemia in de novo pediatric AML patients with RUNX1 aberrations. Identified gene mutations in 9 de novo pediatric AML patients with RUNX1 aberrations; each mutation is depicted by a blue rectangle. Patients with inherited mutations are represented by pink rectangles. Open rectangles correspond to patients without mutations.
Mutational status of leukemia in de novo pediatric AML patients with RUNX1 aberrations. Identified gene mutations in 9 de novo pediatric AML patients with RUNX1 aberrations; each mutation is depicted by a blue rectangle. Patients with inherited mutations are represented by pink rectangles. Open rectangles correspond to patients without mutations.
The median age was the same for patients with mutated and wild-type RUNX1. The frequency of immature phenotype (FAB M1) was higher in patients with mutated RUNX1 (56%) than in patients with WT RUNX1 (10%) (Fischer's exact test P = .0019). Interestingly, mutations were not found in patients without MLL-rearrangements, t,8,21 inv,16 t7,12 or t,15,17 whereas concomitant mutations in FLT3-ITD, NRAS, NPM1, and WT1 were found in 5 of 9 patients (supplemental Figure 2).
RUNX1 mutations were mainly found in pediatric AML patients with an adverse prognosis. Although 4 patients received allogeneic stem cell therapy, none survived. Further, we did not find a strong gene expression signature, suggestive of specific driver alterations, in RUNX1-mutated pediatric AMLs (data not shown).
RUNX1 mutations are late events in the leukemic transformation of CN
To evaluate the time points and sequence of acquisition of RUNX1 and CSF3R mutations, we performed a consecutive analysis of both mutations in 10 CN/AML patients. DNA from bone marrow cells collected at different time points prior to leukemia was analyzed. In 6 of 10 patients, a CSF3R mutation occurred prior to RUNX1 mutations (Figure 4A and supplemental Figure 3), consistent with previous data demonstrating that acquisition of CSF3R mutations is an early event in the leukemic transformation in CN.11,12 In 2 patients, both RUNX1 and CSF3R mutations were detected in the earliest available DNA samples prior to leukemia/MDS. Two patients had no CSF3R mutations and acquired RUNX1 mutations 2 months prior to AML or at the time point of AML progression. Interestingly, monosomy 7 or trisomy 21 appeared after acquisition of RUNX1 mutations.
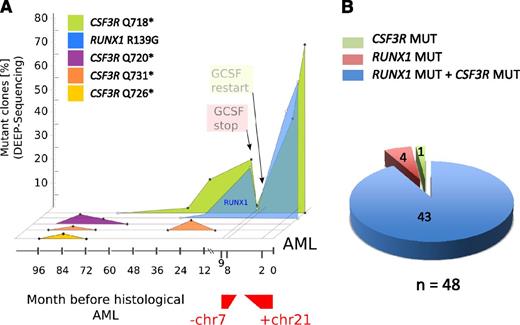
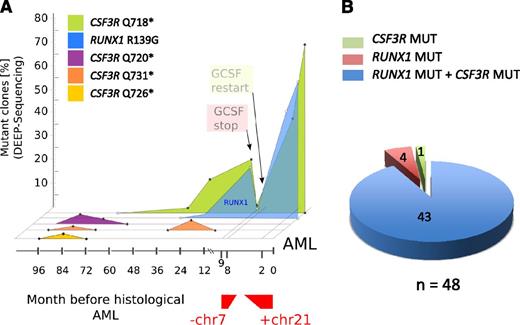
Cooccurrence of RUNX1 and CSF3R mutations in combination with monosomy 7 and trisomy 21 in a leukemic clone of a CN/AML patient. (A) Graphic presentation of a mutational analysis of deep-sequencing data for RUNX1 and CSF3R genes in CN/AML patient 14. Results of sequencing DNA samples from different time points prior to overt AML (x-axis) and the percentage of mutant clones (y-axis) are presented. (B) Diagram of the distribution of RUNX1 and CSF3R mutations in DNA isolated from single clones (n = 48) of AML samples from CN patient 14.
Cooccurrence of RUNX1 and CSF3R mutations in combination with monosomy 7 and trisomy 21 in a leukemic clone of a CN/AML patient. (A) Graphic presentation of a mutational analysis of deep-sequencing data for RUNX1 and CSF3R genes in CN/AML patient 14. Results of sequencing DNA samples from different time points prior to overt AML (x-axis) and the percentage of mutant clones (y-axis) are presented. (B) Diagram of the distribution of RUNX1 and CSF3R mutations in DNA isolated from single clones (n = 48) of AML samples from CN patient 14.
The time course of mutational events in patient 14 is shown in Figure 4A. The transient appearance of cell clones with different CSF3R mutations was detected as early as 8 years prior to leukemia. Three different CSF3R mutations (p.Q720X, p.Q726X, and p.Q731X) were detected at the age of 12 years (8 years prior to leukemia). All 3 clones fell below the detection level within 2-3 years, and one clone (p.Q731X) reappeared for a short period of time 3 years prior to leukemia. One additional clone with a p.Q718X mutation was detected 5 years prior to leukemia at the age of 15 years. Intriguingly, this clone acquired an additional RUNX1 mutation 2 years prior to leukemia. The mutations in RUNX1 and CSF3R preceded monosomy 7, which was observed nine months prior to leukemia. After acquisition of monosomy 7, treatment with G-CSF was discontinued and, intriguingly, the clone with mutated RUNX1 and CSF3R was no longer detectable. Because of the life-threatening infection status of the patient, G-CSF therapy was restarted, which was rapidly followed by expansion of the cell clone with RUNX1 and CSF3R mutations, an additional trisomy 21, and a bone marrow morphology that revealed overt AML.
To evaluate whether RUNX1 and CSF3R mutations were present in the same transformed cell clone or whether 2 different clones carried RUNX1 and CSF3R mutations, we performed colony-forming assays using leukemia blasts and CD34+/CD33+ bone marrow cells isolated 7 years prior to leukemia development of one CN/AML patient. We also isolated DNA from single colonies and analyzed samples for the presence of RUNX1 and CSF3R mutations by Sanger sequencing. Both RUNX1 and CSF3R p.Q718X mutations were detected in 43 of 48 leukemic colonies, whereas no mutations were found in cells isolated 7 years prior leukemia (Figure 4B).
Analysis of familial CN/AML cases
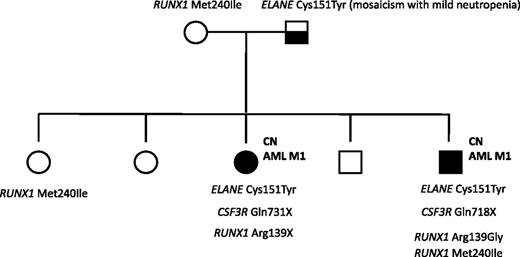
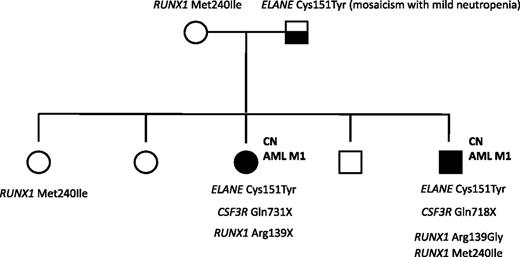
A mutation analysis of RUNX1 was also performed in a family in which 2 siblings with ELANE-CN developed AML M1. The father was mosaic for the ELANE mutation and had a mild, asymptomatic neutropenia. One of the affected siblings had a germline RUNX1 variation (p.Met240Ile) that was inherited from the healthy mother and which has not been previously reported as a common single-nucleotide polymorphism. A healthy sister also carried this p.Met240Ile RUNX1 alteration. Both affected siblings had acquired distinct RUNX1 mutations in Arg139: a p.Arg139Gly missense mutation in the brother and a p.Arg139X nonsense mutation in the sister. Both affected siblings also had CSF3R nonsense mutations: p.Q718X and p.Q731X in the brother and sister, respectively. None of the healthy siblings had acquired RUNX1 or CSF3R mutations, or inherited ELANE mutations (Figure 5).
Mutation analysis of a CN/AML pedigree. Squares indicate males and circles indicate females. Open symbols represent unaffected persons; the half-filled square represent the father with a mosaicism for ELANE mutation and mild neutropenia; closed symbols represent 2 persons affected by CN, both of whom also developed AML-M1. Inherited mutations in ELANE (p.Cys151Tyr) and RUNX1 (p.Met240Ile) as well as acquired RUNX1 (p.Arg139Gly; p.Arg139*) and CSF3R (p.Gln718X; p.Gln731*) mutations are indicated for each affected person.
Mutation analysis of a CN/AML pedigree. Squares indicate males and circles indicate females. Open symbols represent unaffected persons; the half-filled square represent the father with a mosaicism for ELANE mutation and mild neutropenia; closed symbols represent 2 persons affected by CN, both of whom also developed AML-M1. Inherited mutations in ELANE (p.Cys151Tyr) and RUNX1 (p.Met240Ile) as well as acquired RUNX1 (p.Arg139Gly; p.Arg139*) and CSF3R (p.Gln718X; p.Gln731*) mutations are indicated for each affected person.
Enhanced proliferation of CD34+ cells cotransduced with mutated RUNX1 and mutated CSF3R
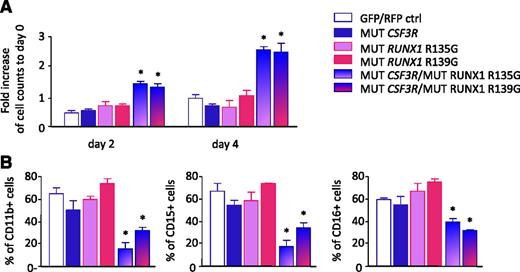
To test the effects of coexpression of RUNX1 and CSF3R mutations, we cotransduced CD34+ hematopoietic cells with cDNAs encoding the truncated CSF3R mutant (d715) or RUNX1 variants carrying missense mutations within the RHD (Arg135Gly or Arg139Gly). We treated transduced cells with G-CSF and evaluated proliferation and myeloid differentiation. Proliferation of cells expressing with the CSF3R mutant in combination with either RUNX1 Arg135Gly or RUNX1 Arg139Gly mutants was elevated relative to cells transduced with control vectors, the CSF3R mutant alone, or RUNX1 mutants alone (Figure 6A). In parallel, we observed diminished myeloid differentiation of cells transduced with mutated CSF3R and RUNX1, as assessed by surface expression of the myeloid-specific surface markers, CD11b, CD15 and CD16 (Figure 6B; supplemental Figure 4).
Enhanced proliferation and diminished myeloid differentiation of hematopoietic cells. CD34+ bone marrow cells from healthy individuals were transduced with lentivirus-based green fluorescent protein (GFP)-tagged RUNX1 mutants (p.Arg135Gly or p.Arg139Gly) or red fluorescent protein (RFP)-tagged CSF3R mutant (d715) alone, cotransduced with RUNX1 and CSF3R mutants, or cotransduced with control GFP- and RFP-tagged lentiviral constructs. Transduced cells were treated with 10 ng/mL of G-CSF. (A) Cell number was evaluated on days 2 and 4 of culture by estimation of RFP+/GFP+ cells using fluorescence-activated cell sorter. (B) G-CSF–triggered myeloid differentiation was evaluated on day 8 of culture. Data represent means ± standard deviations and are derived from 2 independent experiments, each in triplicate (*P < .05).
Enhanced proliferation and diminished myeloid differentiation of hematopoietic cells. CD34+ bone marrow cells from healthy individuals were transduced with lentivirus-based green fluorescent protein (GFP)-tagged RUNX1 mutants (p.Arg135Gly or p.Arg139Gly) or red fluorescent protein (RFP)-tagged CSF3R mutant (d715) alone, cotransduced with RUNX1 and CSF3R mutants, or cotransduced with control GFP- and RFP-tagged lentiviral constructs. Transduced cells were treated with 10 ng/mL of G-CSF. (A) Cell number was evaluated on days 2 and 4 of culture by estimation of RFP+/GFP+ cells using fluorescence-activated cell sorter. (B) G-CSF–triggered myeloid differentiation was evaluated on day 8 of culture. Data represent means ± standard deviations and are derived from 2 independent experiments, each in triplicate (*P < .05).
Discussion
CN is a preleukemic bone marrow failure syndrome with a high risk of progression to AML or MDS.9 Rare cases of ALL associated with CN have also been described.27 The genetic changes involved in the evolution of CN to leukemia are still largely unknown. The frequency of acquired CSF3R mutations in CN/AML patients is ∼80%, substantially higher than that in patients who have not yet progressed to leukemia (∼20%), suggesting that CSF3R mutations are drivers of leukemogenesis.10,11 Previous studies have demonstrated a clonogenic proliferative advantage of hematopoietic cells carrying a truncated CSF3R.40,41 However, CSF3R mutations alone are not sufficient for leukemia development, and additional genetic events are required.28-30 Moreover, the first case of leukemia in a CN patient was described in 1969, many years before G-CSF was used for the treatment of CN.31,32 This suggests that G-CSF treatment is not causative for leukemic transformation.
Here, we provide the first report of the high frequency of RUNX1 mutations in CN/AML patients. It is important to mention that there is no other clinical entity with this high frequency of RUNX1 mutations. In contrast, we found that RUNX1 mutations are a rare event in de novo childhood AML (frequency <3%) and are not associated with the acquisition of CSF3R mutations. Interestingly, RUNX1 mutations were found in pediatric AML patients mutually exclusive of MLL-rearrangements, t(8,21), inv(16), t(7,12), or t(15,17) whereas concomitant mutations in the FLT3, NRAS, NPM1, and WT1 genes were found in 5 of 9 patients. Intriguingly, patients with CN associated with glycogen storage disease type Ib, or those harboring ELANE, HAX1, WAS, or GFI1 mutations who developed secondary leukemia, acquired RUNX1 mutations. This observation demonstrates that RUNX1 is a crucial driver of leukemogenesis, independent of the underlying diverse inherited aberrations. The vast majority of RUNX1 mutations in CN/AML and de novo AML patients impair RUNX1 function (loss of DNA binding or transactivation capacity) and would be predicted to have dominant negative effects or to generate a null allele (no protein or extremely truncated protein).33-37
The high frequency of combined RUNX1 and CSF3R mutations and the presence of both mutations in the same leukemic cell clones clearly suggest that these 2 mutations cooperate as drivers in leukemogenesis, Our hypothesis is that the initial driver mutations are the CSF3R mutations and cell clones harboring CSF3R mutations having a growth advantage acquire in the majority of patients RUNX1 mutations that contribute to AML initiation and disease progression.
In contrast, the association between RUNX1 and CSF3R mutations was not found in de novo pediatric AML, suggesting a distinct and specific mechanism in CN. Only 2 patients with a RUNX1 mutation had no CSF3R mutation; conversely, 6 patients with a CSF3R mutation had no RUNX1 mutations. Intriguingly, 5 patients who were negative for CSF3R, and RUNX1 mutations had MDS only and no overt AML, suggesting that CSF3R/RUNX1 mutations are rather a feature of AML and less frequent of MDS. There was no evidence from a copy number analysis that the chromosomal region 21q22.1 containing RUNX1 was lost in these latter 6 cases, but the loss of RUNX1 expression (eg, through DNA or histone methylation) cannot be excluded. CN patients require lifelong treatment with high therapeutic doses of G-CSF.1 Most of the CSF3R mutations described in CN patients lead to a truncated CSF3R protein lacking the intracellular domain responsible for the termination of proliferative signals.10-12,38,39 Moreover, we demonstrated the transitory appearance of clones carrying different CSF3R mutations in CN patients many years prior to AML (Figure 4A). These data are in line with our previously published findings showing more than one mutation in CSF3R in a CN patient who developed AML.42 Our data suggest that as soon as a cell clone carrying a CSF3R mutation is hit by an additional mutation in RUNX1, the inevitable fate of this clone is transformation into leukemic cells. It has been reported that a RUNX1 loss-of-function mutation per se is not sufficient to cause leukemia but does block myeloid differentiation.43 Our patient data taken together with in vitro studies strongly support the hypothesis that, by markedly elevating proliferation and diminishing myeloid differentiation of hematopoietic cells, CSF3R and RUNX1 mutations are the major drivers of leukemogenesis in patients carrying both mutated proteins.
The importance of RUNX1 mutations in leukemic transformation was substantially strengthened by the analysis of a unique family with 2 siblings suffering from CN that subsequently transformed into AML. In both children, cooperating RUNX1 and CSF3R mutations were detected that were not present in healthy family members. Inherited RUNX1 mutations have been described in familial platelet disorder-AML, another inherited preleukemic bone marrow failure syndrome with a high propensity to develop into AML.22,23 Cryptic RUNX1 lesions (translocations, deletions, or mutations) have also been observed in Fanconi anemia patients who developed AML or MDS.44 These data, together with our observations, suggest that RUNX1 mutations could represent an important common event in triggering secondary leukemia in patients with bone marrow failure syndromes.
In conclusion, our findings may improve monitoring of the possible leukemic transformation in CN patients in which CSF3R mutations are already present. We recommend yearly bone marrow examinations. Patients with both RUNX1 and CSF3R mutations appear to be at high risk for rapid evolution to leukemia and should be considered candidates for stem cell transplantation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank A. Müller Brechlin, M. Reuter, A.-L. Hagemann, N. Bruckhoff, and F. Grundstedt for the excellent technical assistance. We also thank the physicians cooperating with the Severe Chronic Neutropenia International Registry and French Severe Chronic Neutropenia Registry for providing patient material. We thank Dr J. Stary, Department of Pediatric Hematology and Oncology, University Hospital Motol, Prague, Czech Republic and Dr A. Baruchel from Paris. We thank the patients for their cooperation in providing their bone marrow cells. Computational support and infrastructure were provided by the North-German Supercomputing Alliance.
This work was partially supported by Madeleine-Schickedanz Kinderkrebsstiftung, Deutsche Krebshilfe, Josè-Carreras Leukemie Stiftung, and the Federal Ministry of Education and Research (German Network on Congenital Bone Marrow Failure Syndromes).
Authorship
Contribution: J.S. and D.S. collected and analyzed data, supervised experiments, and wrote the manuscript; J.S. also provided financial support; J.E.K.-K., J.O., C.M.Z., D.R., and M.M.v.d.H.-E. collected and analyzed data on de novo pediatric AML patients and wrote the part of the manuscript describing de novo AML patients data; C.Z. provided patient materials; O.K. performed in vitro experiments with CD34+ cells; M.K. performed DNA sample preparation, analysis of sequenced data, sequencing of single CFU clones, and allele-specific PCR; M.Ü. and S.K. analyzed deep-seq data of CSF3R; R.B., K.B., and C.S. made lentiviral constructs with WT and MUT RUNX1; S.S. and A.K. performed deep-seq of RUNX1 and other leukemia-associated mutations and wrote manuscript; M.G.V., R.H., and I.P.T. performed RUNX1 and CSF3R analysis of some CN/AML patients; I.P.T. wrote the manuscript; V.M., D.C.D., P.V., and J.D. provided some patient materials; D.C.D. wrote the manuscript; G.G. and B.S. provided some patient materials and cytogenetic analysis; M.S. wrote the manuscript; and K.W. provided patients material, supervised experiments, analyzed data, wrote the manuscript, and provided financial support.
Conflict-of-interest disclosure: D.C.D. is a consultant and receives research support from Amgen, the manufacturer of G-CSF mentioned in the paper and used to treat cyclic and CN. The remaining authors declare no competing financial interests.
Correspondence: Karl Welte, Department of Molecular Hematopoiesis, Hannover Medical School, Carl-Neuberg Str. 1, 30625 Hannover, Germany; e-mail: Welte.Karl.H@mh-hannover.de; and Julia Skokowa, Department of Molecular Hematopoiesis, Hannover Medical School, Carl-Neuberg Str. 1, 30625 Hannover, Germany; e-mail: skokowa.julia@mh-hannover.de.
References
Author notes
J.S. and D.S. contributed equally to the work.