Key Points
NRASG12V maintains leukemia self-renewal in a genetically engineered murine model of AML.
NRASG12V differentially regulates transcription and signaling among leukemic subpopulations.
Abstract
Mutant RAS oncoproteins activate signaling molecules that drive oncogenesis in multiple human tumors including acute myelogenous leukemia (AML). However, the specific functions of these pathways in AML are unclear, thwarting the rational application of targeted therapeutics. To elucidate the downstream functions of activated NRAS in AML, we used a murine model that harbors Mll-AF9 and a tetracycline-repressible, activated NRAS (NRASG12V). Using computational approaches to explore our gene-expression data sets, we found that NRASG12V enforced the leukemia self-renewal gene-expression signature and was required to maintain an MLL-AF9– and Myb-dependent leukemia self-renewal gene-expression program. NRASG12V was required for leukemia self-renewal independent of its effects on growth and survival. Analysis of the gene-expression patterns of leukemic subpopulations revealed that the NRASG12V-mediated leukemia self-renewal signature is preferentially expressed in the leukemia stem cell–enriched subpopulation. In a multiplexed analysis of RAS-dependent signaling, Mac-1Low cells, which harbor leukemia stem cells, were preferentially sensitive to NRASG12V withdrawal. NRASG12V maintained leukemia self-renewal through mTOR and MEK pathway activation, implicating these pathways as potential targets for cancer stem cell–specific therapies. Together, these experimental results define a RAS oncogene–driven function that is critical for leukemia maintenance and represents a novel mechanism of oncogene addiction.
Introduction
Acute myeloid leukemia (AML) arises because of an accumulation of genetic insults in hematopoietic stem and progenitor cells.1 RAS oncogenes are well-recognized drivers of proliferation and survival and are mutated in approximately 25% of cancers2 and 12% of AML cases.1 RAS proteins transduce extracellular growth factor stimulation to downstream signal–transduction pathways.3
Activation of RAS proteins in hematopoietic cells leads to myeloid progenitor cell expansion, increased bone marrow reconstitution, and chronic myeloproliferative neoplasia.4-7 In addition, NRASG12D/+ causes increased proliferation and cell-cycle entry in a hematopoietic stem cell (HSC)-specific manner.8 Recent work revealed that NRASG12D increases self-renewal in nonleukemic HSCs and that this effect was mutually exclusive with the effects of NRASG12D on increasing HSC proliferation.9 These studies reveal a potential cell type–specific role of NRAS activation in hematopoietic malignancies. However, the specific role for RAS oncogenes in the genesis of leukemia is unclear.
In our studies, we used an NRASG12V/Mll-AF9 model10 to decipher which NRASG12V functions are critical for tumor maintenance in AML. MLL is an important regulator of gene-expression programs that orchestrate hematopoietic differentiation.11 MLL fusion genes, such as MLL-AF9, block differentiation and are widely thought to confer leukemia cells with the self-renewal properties of HSCs.12-14 Moreover, rearrangements involving the MLL locus are frequent in human AML.15 Myb was shown to mediate the activity of MLL-AF9 in leukemia self-renewal.16
In our studies, we sought to decipher the NRAS functions critical for tumor maintenance in AML. We used the NRASG12V/Mll-AF9 AML model10 to study the gene-expression and signaling-response profiles of myeloid leukemia cells to NRASG12V withdrawal and to RAS-pathway inhibitors.
Methods
Mice
106 primary NRASG12V/Mll-AF9 leukemia cells were infused via tail vein into SCID/Beige mice.10 Upon development of leukemia (peripheral white blood cell count >100 000/μL), mice were injected with 10 μg of doxycycline via intraperitoneal injections and were given 2 mg/mL doxycycline-infused drinking water at time 0. Mice were sacrificed at 12-hour intervals. Complete blood counts were obtained at the time of sacrifice. Spleens were harvested, weighed, and processed to make single-cell suspensions. A portion of each spleen was stored in RNAlater, a portion was snap frozen for immunoblotting, and the remainder was fixed or fixed and permeabilized for flow and mass cytometry.
Gene-expression data analysis
Data processing and normalization.
Affymetrix gene-expression microarray data (CEL files) were normalized using Bioconductor RMA, Genedata Refiner Array 7.6 (Lexington, MA). Normalized data were analyzed for quality; statistical analyses were carried out using Genedata Analyst 7.6 (Lexington, MA). Fastq data files were mapped to the mm9 genome using TopHat.17 FPKM values and differentially expressed genes were identified using Cuffdiff18 for the comparison of NRASG12V-On to NRASG12V-Off samples.
Results
Tumor burden declines after loss of oncogenic NRASG12V protein
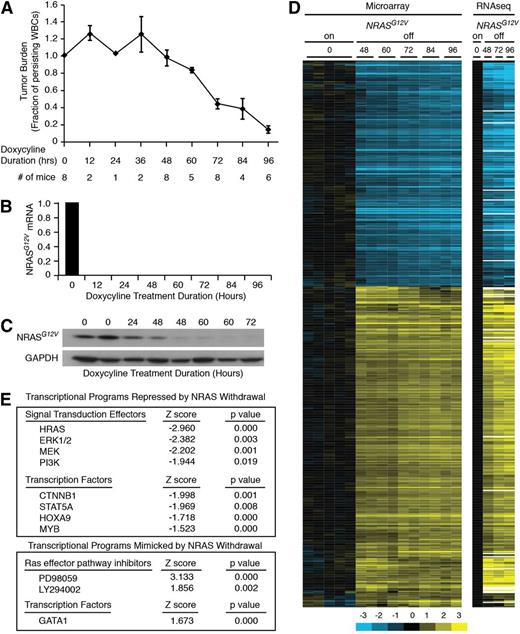
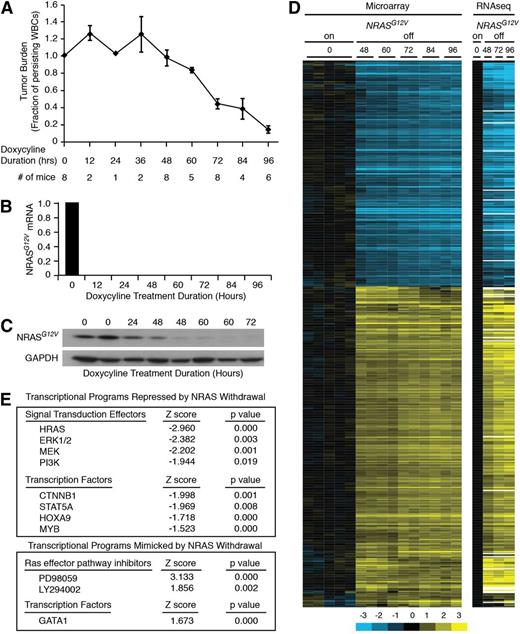
We first assessed the kinetics of the leukemia response to loss of NRASG12V by transplanting SCID/Beige mice with primary NRASG12V/Mll-AF9 AML cells. Once leukemia was established, the mice were treated with doxycycline to abolish NRASG12V expression; this transgene is under the control of a tetracycline-repressible element. Because NRASG12V is expressed as a single-allele transgene, these cells retain wild-type Nras genes at the endogenous locus and express the wild-type protein in levels comparable with the transgene.10 Mice were sacrificed at 12-hour intervals to assess early response to NRASG12V withdrawal. Tumor burden was reduced after 48 hours of doxycycline treatment, as measured by white blood counts (Figure 1A) and spleen weights (supplemental Figure 1A, available on the Blood Web site). Single-cell suspension of whole splenocytes yields leukemia cells with >90% purity (supplemental Figure 1B) that can recapitulate the leukemia in serial transplants (our unpublished observations) and were therefore used, unpurified, for these experiments. Treatment with doxycycline eliminated NRASG12V transgene expression within 12 hours as assayed by quantitative polymerase chain reaction (Figure 1B), but transgenic protein levels persisted for 48 hours (Figure 1C). Notably, tumor burden declined in parallel with protein levels after 48 hours. RNA derived from NRASG12V/Mll-AF9 AML cells from these doxycycline-treated mice was submitted for gene-expression microarray and RNA sequencing analysis. We found 679 genes whose expression was significantly altered by NRASG12V withdrawal (supplemental Table 1). Unsupervised hierarchical clustering revealed that NRASG12V removal led to gene-expression alterations that were evident by 48 hours of doxycycline treatment, before any significant decline in tumor burden, and became more pronounced at later time points (60, 72, 84, and 96 hours; Figure 1D). RNA sequencing analysis recapitulated the findings of the microarray analysis (Figure 1D). We performed Ingenuity Upstream Regulators Analysis to compare the similarity of gene-expression changes in our data set to a curated list of transcriptional regulator-directed gene-expression signatures. This analysis screens every list in Ingenuity’s Upstream Transcriptional Regulators database and confirmed that NRASG12V withdrawal led to gene-expression changes that were consistent with attenuation of known RAS-activated signaling pathways, such as MEK/ERK and PI3K. Likewise, our data set recapitulated the effects of pharmacologic RAS-pathway inhibition with MEK inhibition (PD98059) and PI3K inhibition (LY294002; Figure 1E, lower panel).
NRASG12V/Mll-AF9 AML On-Off model and gene expression. (A) Tumor burden was assayed by peripheral white blood cell (WBC) count as a fraction of the WBC count of each mouse at the time of sacrifice to the WBC count at the time of doxycycline treatment. Each point represents the mean WBC fraction of all of the animals sacrificed at that time point. Error bars represent the standard error of the mean for each time point. (B) NRASG12V transgene abundance by quantitative polymerase chain reaction. (C) NRASG12V protein levels by Western blot analysis. (D) Hierarchical clustering of gene-expression microarray and RNA sequencing data using the list of differentially expressed genes identified by microarray analysis. (E) Ingenuity Upstream Regulator Analysis identified transcriptional programs that were repressed (negative Z score, top panel) or mimicked (positive Z score, bottom panel) by NRASG12V withdrawal.
NRASG12V/Mll-AF9 AML On-Off model and gene expression. (A) Tumor burden was assayed by peripheral white blood cell (WBC) count as a fraction of the WBC count of each mouse at the time of sacrifice to the WBC count at the time of doxycycline treatment. Each point represents the mean WBC fraction of all of the animals sacrificed at that time point. Error bars represent the standard error of the mean for each time point. (B) NRASG12V transgene abundance by quantitative polymerase chain reaction. (C) NRASG12V protein levels by Western blot analysis. (D) Hierarchical clustering of gene-expression microarray and RNA sequencing data using the list of differentially expressed genes identified by microarray analysis. (E) Ingenuity Upstream Regulator Analysis identified transcriptional programs that were repressed (negative Z score, top panel) or mimicked (positive Z score, bottom panel) by NRASG12V withdrawal.
NRASG12V withdrawal leads to loss of the leukemia self-renewal gene-expression program
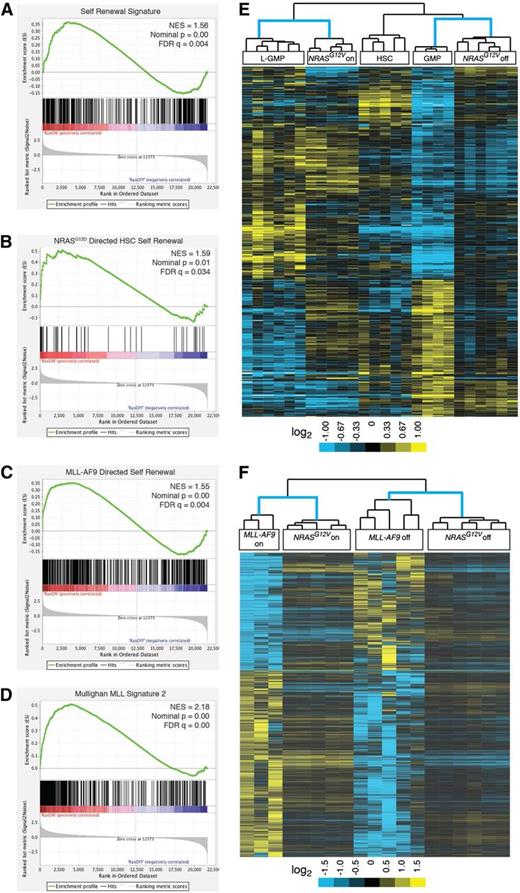
Next, we used Gene Set Enrichment Analysis (GSEA) to screen the curated gene sets in the Molecular Signatures Database v3.1 (MSigDB) for similarity to our NRASG12V-responsive gene-expression signature.19,20 In addition to the expected similarities with a number of cell cycle and survival gene sets, the top 3 enriched lists in our analyses involved hematopoiesis and cell-fate decisions (supplemental Table 2A-B). Because a recent report revealed a role for activated NRAS in HSCs and HSC self-renewal,9 and our own data suggested a role for NRASG12V in leukemia stem cell (LSC)-specific gene expression, we hypothesized that NRASG12V may endow leukemia cells with stem-cell functions. Subsequently, we used GSEA to assess previously published leukemia-specific signatures (supplemental Table 3A-B). Our NRASG12V-responsive gene-expression data set was significantly enriched in 2 independently-derived signatures of murine leukemia self-renewal14,21 (Figure 2A, supplemental Figure 2A, and supplemental Table 3A-B) and a signature of human LSCs22 (supplemental Table 3A-B). These analyses show that the NRASG12V expression signature maintains the leukemia self-renewal gene-expression program.
Oncogene withdrawal results in loss of leukemia self-renewal gene-expression signatures. (A-D) GSEA analysis reveals loss of previously published gene-expression signatures. (A) The Krivtsov et al leukemia self-renewal gene-expression signature which was derived by identifying a set of genes common to self-renewing hematopoietic subsets (L-GMPs and HSCs).14 (B) The NRASG12D-directed nonleukemic HSC self-renewal signature.9 (C) The MLL-AF9–directed leukemia self-renewal gene-expression program.16 (D) The MLL fusion protein signature of primary human leukemias.23 (E) NRASG12V-On and -off gene-expression data from our study and self-renewing (HSC and L-GMP) and non–self-renewing (GMP) expression data from the Krivtsov et al14 study were compared. Each individual data set was independently transformed relative to the average value for the data set and both data sets were then merged. The probe sets of the self-renewing gene-expression signature defined by Krivtsov et al14 were used for unsupervised, 2-dimensional, hierarchical clustering of these merged data sets. (F) NRASG12V-On and -Off samples from our study and MLL-AF9-On and -Off samples from Zuber et al16 were independently transformed relative to the oncogene-on state in each data set and both data sets were then merged. The gene list from the MLL-AF9–directed leukemia self-renewal signature was used for unsupervised, 2-dimensional, hierarchical clustering of these merged data sets.
Oncogene withdrawal results in loss of leukemia self-renewal gene-expression signatures. (A-D) GSEA analysis reveals loss of previously published gene-expression signatures. (A) The Krivtsov et al leukemia self-renewal gene-expression signature which was derived by identifying a set of genes common to self-renewing hematopoietic subsets (L-GMPs and HSCs).14 (B) The NRASG12D-directed nonleukemic HSC self-renewal signature.9 (C) The MLL-AF9–directed leukemia self-renewal gene-expression program.16 (D) The MLL fusion protein signature of primary human leukemias.23 (E) NRASG12V-On and -off gene-expression data from our study and self-renewing (HSC and L-GMP) and non–self-renewing (GMP) expression data from the Krivtsov et al14 study were compared. Each individual data set was independently transformed relative to the average value for the data set and both data sets were then merged. The probe sets of the self-renewing gene-expression signature defined by Krivtsov et al14 were used for unsupervised, 2-dimensional, hierarchical clustering of these merged data sets. (F) NRASG12V-On and -Off samples from our study and MLL-AF9-On and -Off samples from Zuber et al16 were independently transformed relative to the oncogene-on state in each data set and both data sets were then merged. The gene list from the MLL-AF9–directed leukemia self-renewal signature was used for unsupervised, 2-dimensional, hierarchical clustering of these merged data sets.
Recent studies identified a role for activated NRAS in increasing self-renewal in nonleukemic HSCs.9 The gene-expression profiles of NRASG12D expressing self-renewing and non–self-renewing HSCs were reported in that study. We used data from that study to derive the NRASG12D-directed HSC self-renewal gene-expression signature by identifying the differentially expressed genes between these 2 groups. Using GSEA, we found that this NRASG12D-directed HSC self-renewal signature is enriched in our NRASG12V-expressing samples (Figure 2B and supplemental Table 3A-B), suggesting that, as in HSCs, activated NRAS can also direct self-renewal in leukemia cells.
Similarly, the NRASG12V-dependent signature was enriched in the MLL-AF9–directed leukemia self-renewal signature16 (Figure 2C). The MLL-AF9 signature is mediated by MYB,16 and this MYB-specific signature was lost in our NRASG12V-Off samples as well (supplemental Table 3A-B). Our MSigDB screen revealed highly significant similarities with human MLL-fusion gene leukemia signatures obtained from primary patient samples in multiple, independently performed studies23,24 (Figure 2D, supplemental Figure 1B-C, and supplemental Tables 2A-B and 3A-B) and other published MYB-directed signatures25 (supplemental Table 3A-B). Notably, NRASG12V withdrawal did not affect MLL-AF9 transcript levels consistent with a role for NRASG12V in mediating the activity of MYB (downstream of Mll-AF9).
Consistent with our GSEA results, Ingenuity Pathway Upstream Transcriptional Regulators Analysis (IPA) revealed that loss of NRASG12V mimicked the loss of MYB-directed and HOXA9-directed gene expression (Figure 1E and supplemental Table 4A-B). HOXA9 is a well-known mediator of the MLL-AF9 gene-expression program.26 IPA also revealed that NRASG12V loss mimicked the loss of the CTNNB1 signature (Figure 1E) without affecting CTNNB1 mRNA transcript levels. These analyses suggest that NRASG12V modulates β-catenin (the protein encoded by CTNNB1) activity at the protein level. β-catenin has been strongly implicated in LSC survival in both chronic myeloid leukemia (CML)27,28 and MLL-associated AML.29,30 In contrast, NRASG12V withdrawal directed a gene-expression pattern consistent with the activation of known hematopoietic differentiation factors, such as GATA1. These analyses reveal a consistent pattern of NRASG12V-driven gene-expression changes mimicking the transcriptional program of the orchestrators of leukemia self-renewal.
To further confirm the role of NRASG12V in mediating self-renewal, we performed a third, unrelated statistical analysis by combining our data set with that of Krivtsov et al.14 We performed 2-dimensional, unsupervised hierarchical clustering on these combined data using the genes identified in the leukemia self-renewal signature of Krivtsov et al. NRASG12V-expressing samples clustered with the self-renewing Krivtsov et al samples (leukemic self-renewing cells [leukemic granulocytic-myeloid precursors (L-GMPs)]), whereas NRASG12V-Off samples clustered with the non–self-renewing samples (nonleukemic GMPs; Figure 2E). Similarly, we combined our data set with the MLL-AF9–dependent data set16 and found that NRASG12V-expressing cells clustered with MLL-AF9-expressing cells, whereas NRASG12V-Off cells clustered with MLL-AF9-Off cells (Figure 2F).
Taken together, our analyses using these 3 independent approaches (IPA, GSEA, and hierarchical clustering) reveal highly significant relationships among the NRASG12V–gene-expression signature, MLL-AF9 signature, and leukemia self-renewal signatures. These data implicate a novel function for NRASG12V in facilitating the leukemogenic activity of MLL-AF9 and in maintaining leukemia self-renewal.
Transient loss of NRASG12V abrogates leukemia self-renewal
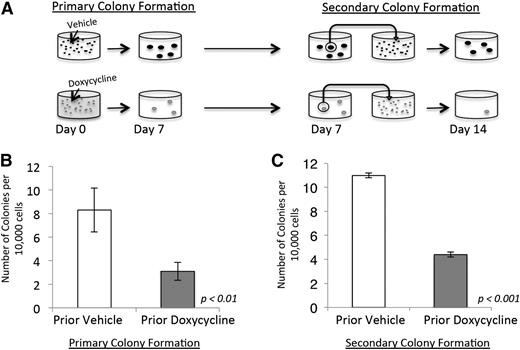
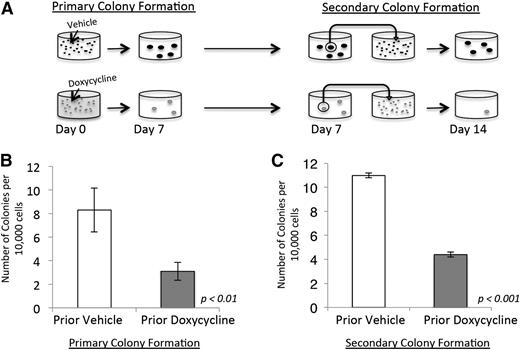
Based on our gene-expression data, we next sought to functionally validate the role of NRASG12V in maintaining leukemia self-renewal. We performed serial colony-forming assays (CFAs) on primary NRASG12V/Mll-AF9 AML cells. Initially, cells were treated with doxycycline or vehicle and plated in methylcellulose (Figure 3A, left panel). The assays were performed with minimal growth factor support (1 ng/mL granulocyte macrophage colony-stimulating factor) because higher doses of growth factor can replace the dependence on NRASG12V in CFAs (our unpublished observations) and potentially allow for the outgrowth of nonleukemic myeloid colonies. Doxycycline-treated cells developed significantly fewer colonies (3.1 colonies/10 000 plated cells) than vehicle-treated cells (8.3 colonies/10 000 plated cells, P < .01; Figure 3B). To separate the effects of NRASG12V withdrawal on proliferation and survival from its effects on leukemia self-renewal, individual colonies from both the doxycycline- and vehicle-treated groups were dispersed in liquid media and replated in secondary CFAs in methylcellulose with no further doxycycline treatment (Figure 3A, right panel). This experimental design reveals the colony-forming capacity of cells after transient loss of NRASG12V. Furthermore, this experiment tests the colony-forming ability of cells that retained survival and proliferation capacity after doxycycline treatment and, therefore, tests specifically for self-renewal capacity. Cells initially treated with doxycycline at the first plating grew significantly fewer colonies upon secondary plating (4.4 colonies/10 000 plated cells) than vehicle-treated cells (11 colonies/10 000 plated cells, P < .001; Figure 3C), even though no additional doxycycline was used in the secondary plating. These experiments show that cells that retain survival and proliferation capacity (and are able to form primary colonies) are deficient in self-renewal capacity, and functionally validate our gene-expression findings and show that transient loss of NRASG12V expression abolishes the capacity for self-renewal in AML cells. Recent work has shown that activated NRAS increases self-renewal capacity in a subset of normal HSCs.9 Our data show that activated NRAS also confers self-renewal capacity on leukemia cells.
NRASG12V withdrawal abrogates self-renewal capacity. (A) Colony-forming assay (CFA) schematic. Doxycycline was administered on day 0. Colonies that grew by day 7 were extracted, dispersed, and plated in methylcellulose for a secondary CFA with no further treatment (doxycycline was not administered after day 0). (B) Primary colony formation of primary leukemia cells treated with doxycycline or vehicle on the day of plating. Each bar represents the average value for 13 independent experiments. (C) Secondary colony formation of replated colonies. Each bar represents the average value for 3 independent experiments. Error bars represent standard error of the mean.
NRASG12V withdrawal abrogates self-renewal capacity. (A) Colony-forming assay (CFA) schematic. Doxycycline was administered on day 0. Colonies that grew by day 7 were extracted, dispersed, and plated in methylcellulose for a secondary CFA with no further treatment (doxycycline was not administered after day 0). (B) Primary colony formation of primary leukemia cells treated with doxycycline or vehicle on the day of plating. Each bar represents the average value for 13 independent experiments. (C) Secondary colony formation of replated colonies. Each bar represents the average value for 3 independent experiments. Error bars represent standard error of the mean.
NRASG12V-mediated self-renewal gene-expression signature is preferentially expressed in the leukemia subpopulations with stem-cell capacity
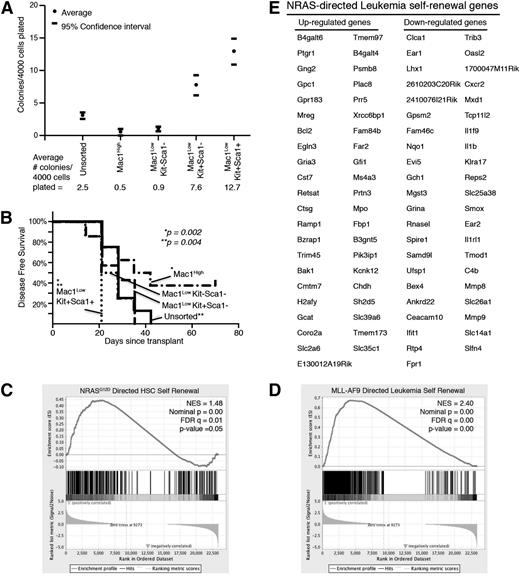
Leukemia cells from our model were characterized according to commonly used myeloid and stem-cell markers (Mac1, c-Kit, and Sca1).31 Unlike the normal hematopoietic hierarchy, we found that virtually all of our leukemia cells express the lineage marker Mac1 (supplemental Figure 1B). Some of these cells also coexpress the stem-cell markers c-Kit and Sca1 (supplemental Figure 3A-B). To identify the immunophenotype of the leukemia subpopulation enriched with LSCs, we harvested the spleens of leukemic mice and sorted cells into immunophenotypic subgroups (Mac1High, Mac1LowKit–Sca-1–, Mac1LowKit+Sca-1–, and Mac1LowKit+Sca-1+; supplemental Figure 3A) and plated cells from each of these groups in equal numbers in methylcellulose CFAs. We found that the Mac1LowKit+Sca-1– and Mac1LowKit+Sca-1+ groups were significantly enriched in colony-forming capacity in 7 independent experiments (Figure 4A and supplemental Figure 3A). We also transplanted each of these groups, or unsorted cells, into SCID/Beige recipients and found that the Mac1LowKit+Sca-1+ group was significantly more efficient at inducing AML in vivo (7-8 mice/group: 1-2 mice/group in 4 independent sorting experiments; Figure 4B). Although these data do not define the details of the immunophenotypic hierarchy of our leukemia model, these data identify the Mac1LowKit+Sca-1+ group as enriched with LSCs compared with the other groups, particularly the Mac1High group that accounts for the majority of the leukemia population. The groups with less LSC enrichment showed variable results between the CFAs and the in vivo assays, possibly reflecting the ability of these cells to change immunophenotype and stem-cell capacity in response to in vivo signals. Both of these modalities identify the Mac-1High group as the group with the least stem-cell capacity.
LSC-enriched population expresses self-renewal gene-expression signature. Primary leukemia cells harvested from spleens were sorted into immunophenotypic subpopulations (Mac-1High, Mac-1LowKit–Sca-1–, Mac-1LowKit+Sca-1–, and Mac-1LowKit+Sca-1+). (A) Sorted cells were plated in methylcellulose CFAs. The number of colonies that developed per 4000 cells plated is noted. Numbers represent the average of 7 independent experiments. 95% confidence intervals are indicated. (B) 105 sorted cells were intravenously injected into SCID/Beige mice. Disease-free survival is indicated. Each group consisted of 7 or 8 mice. P values noted reflect comparisons between the (*) Mac1LowKit+Sca1+ vs Mac1High groups and (**) Mac1LowKit+Sca1+ and unsorted groups. (C-E) Cells from each sorted subpopulation were submitted for RNA sequencing. (C) The NRASG12D-directed gene-expression signature specific to self-renewing HSCs9 is enriched in the LSC-enriched subpopulation (Mac-1LowKit+Sca-1+). (D) The MLL-AF9-mediated self-renewal gene-expression signature is enriched in the LSC-enriched subpopulation (Mac-1LowKit+Sca-1+). (E) The NRASG12V leukemia self-renewal signature was derived by identifying the NRASG12V-mediated gene-expression profile that is also present in the LSC-enriched subpopulation.
LSC-enriched population expresses self-renewal gene-expression signature. Primary leukemia cells harvested from spleens were sorted into immunophenotypic subpopulations (Mac-1High, Mac-1LowKit–Sca-1–, Mac-1LowKit+Sca-1–, and Mac-1LowKit+Sca-1+). (A) Sorted cells were plated in methylcellulose CFAs. The number of colonies that developed per 4000 cells plated is noted. Numbers represent the average of 7 independent experiments. 95% confidence intervals are indicated. (B) 105 sorted cells were intravenously injected into SCID/Beige mice. Disease-free survival is indicated. Each group consisted of 7 or 8 mice. P values noted reflect comparisons between the (*) Mac1LowKit+Sca1+ vs Mac1High groups and (**) Mac1LowKit+Sca1+ and unsorted groups. (C-E) Cells from each sorted subpopulation were submitted for RNA sequencing. (C) The NRASG12D-directed gene-expression signature specific to self-renewing HSCs9 is enriched in the LSC-enriched subpopulation (Mac-1LowKit+Sca-1+). (D) The MLL-AF9-mediated self-renewal gene-expression signature is enriched in the LSC-enriched subpopulation (Mac-1LowKit+Sca-1+). (E) The NRASG12V leukemia self-renewal signature was derived by identifying the NRASG12V-mediated gene-expression profile that is also present in the LSC-enriched subpopulation.
To determine whether the NRASG12V-mediated self-renewal signature was differentially expressed among leukemia subpopulations, we submitted cells from each sorted group for RNA sequencing and derived normalized gene-expression values. Using GSEA, we showed that the Mac1LowKit+Sca-1+ group was enriched in the NRASG12D-directed HSC self-renewal gene-expression signature and the Mll-AF9–directed leukemia self-renewal signature (Figure 4C-D and supplemental Table 5), as well as several of the other leukemia self-renewal signatures that are NRASG12V-dependent in our model (supplemental Table 5). By identifying the list of genes that are differentially regulated in response to NRASG12V withdrawal and are concordantly differentially regulated between the Mac-1High and Mac-1LowKit+Sca-1+ samples, we derive the NRASG12V-mediated leukemia self-renewal gene-expression signature (Figure 4E). Ingenuity Pathway Analysis shows these genes are involved in hematologic development and function, consistent with a role in activating the stem cell self-renewal program.
NRASG12V withdrawal leads to stem cell–specific effects on leukemic signaling behavior
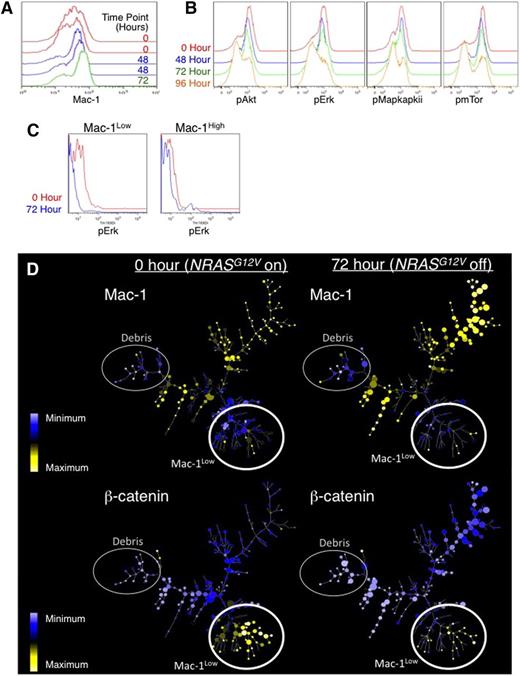
To determine the effects of NRASG12V withdrawal on the LSC frequency, cells from doxycycline-treated leukemic mice were analyzed by flow cytometry. We found a selective depletion of Mac1Low cells (Figure 5A and supplemental Figure 3B-C) that reflected the loss of the population enriched in c-Kit+Sca-1+ cells (Population A in supplemental Figure 3D-E). As we demonstrated in Figure 4, these Mac1Lowc-Kit+Sca-1+ cells are enriched in LSCs, revealing that NRASG12V withdrawal leads to a preferential loss of cells with LSC potential.
Intracellular flow cytometry and mass cytometry reveals loss of the LSC population in response to NRASG12V withdrawal. Primary AML cells were harvested from the spleens of leukemic mice at 0, 48, 72, and 96 hours after doxycycline treatment to abolish NRASG12V expression. Flow cytometry was used to assay for levels of (A) Mac1 (myeloid lineage–commitment surface marker). (B) Phospho-AKT, -ERK1/2, -MAPAPKII, and -mTOR. (C) Using mass cytometry, Mac1High and Mac1Low populations were gated separately, and the levels of pERK were assessed in each group in each sample. Because mass cytometry data lack autofluorescence (as seen with traditional flow cytometry), these histograms abut the y-axis. (D) SPADE trees reveal loss of Mac-1Low cells in response to NRASG12V withdrawal. These cells were also submitted for mass cytometry to simultaneously evaluate the levels of 26 surface markers and intracellular signaling molecules. SPADE analysis displaying the patterns of Mac-1Low (top 2 panels) and β-catenin are displayed. Expression values represent arcsinh difference: for Mac-1 the range is 1.15 to 6.13 and for β-catenin the range is −0.14 to 2.6. The smallest node corresponds to 1 cell and the largest node corresponds to 150 cells.
Intracellular flow cytometry and mass cytometry reveals loss of the LSC population in response to NRASG12V withdrawal. Primary AML cells were harvested from the spleens of leukemic mice at 0, 48, 72, and 96 hours after doxycycline treatment to abolish NRASG12V expression. Flow cytometry was used to assay for levels of (A) Mac1 (myeloid lineage–commitment surface marker). (B) Phospho-AKT, -ERK1/2, -MAPAPKII, and -mTOR. (C) Using mass cytometry, Mac1High and Mac1Low populations were gated separately, and the levels of pERK were assessed in each group in each sample. Because mass cytometry data lack autofluorescence (as seen with traditional flow cytometry), these histograms abut the y-axis. (D) SPADE trees reveal loss of Mac-1Low cells in response to NRASG12V withdrawal. These cells were also submitted for mass cytometry to simultaneously evaluate the levels of 26 surface markers and intracellular signaling molecules. SPADE analysis displaying the patterns of Mac-1Low (top 2 panels) and β-catenin are displayed. Expression values represent arcsinh difference: for Mac-1 the range is 1.15 to 6.13 and for β-catenin the range is −0.14 to 2.6. The smallest node corresponds to 1 cell and the largest node corresponds to 150 cells.
Intracellular phosphospecific flow cytometry showed that NRASG12V withdrawal led to loss of phosphorylated RAS-effector molecules including pERK, pAKT, and pmTOR (Figure 5B). Because we found cell type–specific effects of NRASG12V on gene expression, we used mass cytometry to investigate whether NRASG12V-mediated signaling events were cell-type specific as well. We found that the effects of NRASG12V withdrawal on phospho-ERK1/2 (pERK) were significantly more pronounced in Mac1Low cells than in Mac1High cells (Figure 5C).
To further understand the dynamics of this cell type–specific RAS-induced signaling network, we used high-dimensional mass cytometry to measure the levels and activities of 26 signaling molecules simultaneously on a single-cell basis.32 We applied spanning-tree progression analysis of density-normalized events (SPADE33 ) to our mass cytometry data to organize the data according to clusters of similarities of the levels of all the molecules we measured (Figure 5D). Mac1Low clusters are enriched for LSCs (Figure 5D, top panels). In contrast to the cell type–specific response of pERK, NRASG12V withdrawal uniformly reduced p4EBP1 levels in clusters of all levels of Mac1 (supplemental Figure 4A). β-catenin levels were high only in Mac1Low cells; loss of NRASG12V caused a preferential loss of β-catenin–expressing, Mac1Low cells (Figure 5D, bottom panel, and supplemental Figure 4C). NF-κB levels displayed a similar pattern and showed Mac1Low-specific reductions in abundance (supplemental Figure 4B). Notably, this corresponds to our finding that loss of NRASG12V led to loss of the β-catenin–driven gene-expression pattern (Figure 1E). Like the gene-expression data, these findings suggest that NRASG12V-activated signaling mediates unique functions in LSCs.
MEK and mTOR mediate NRASG12V-dependent leukemia self-renewal
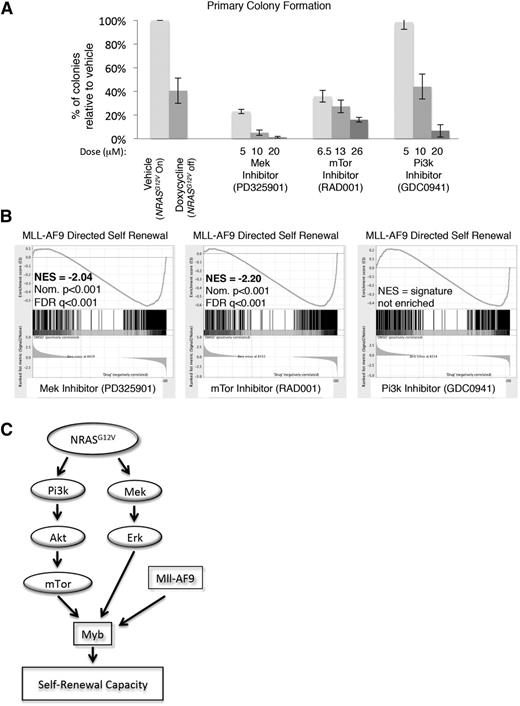
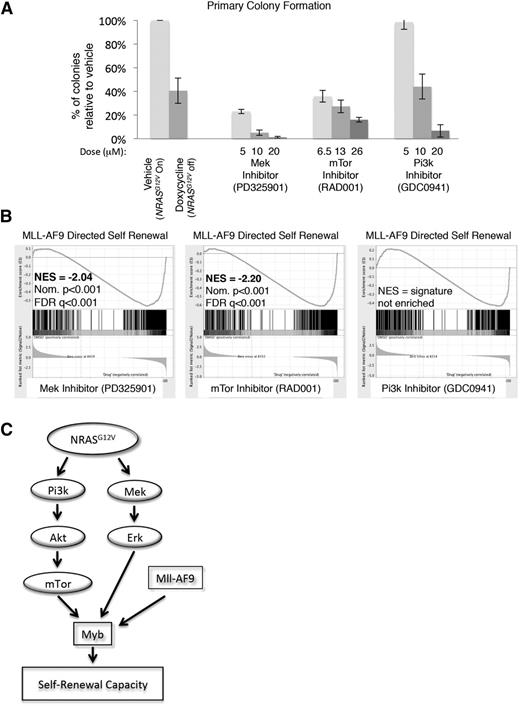
To delineate the specific NRASG12V-mediated signals maintaining self-renewal, primary NRASG12V/Mll-AF9 AML cells were treated with RAS-pathway inhibitors PD325901 (MEK inhibitor), GDC-0941 (PI3K inhibitor), and RAD001 (mTOR inhibitor). These agents effected specific pathway inhibition without evidence of parallel pathway activation, indicating that these agents do not lead to feedback upregulation of the alternate pathways we tested, as described in other settings33 (supplemental Figure 5). Each of these inhibitors diminished colony formation in methylcellulose relative to vehicle control, although the lowest dose of the PI3K inhibitor had no effect on colony formation (Figure 6A).
mTOR and MEK inhibition recapitulate the effects of NRASG12V withdrawal on leukemia self-renewal capacity. (A) Primary leukemia cells were treated with RAS-pathway inhibitors and plated in CFAs. Colonies were scored as the number of colonies that developed per 10 000 cells plated. The percentage of colonies that developed from drug-treated cells, relative to vehicle-treated cells from the same experiment, is shown. Each bar represents the average value of 4 to 13 experiments (doxycycline treatment was performed 13 times, RAD001 and PD325901 treatments were performed 4 times, and GDC0941 treatment was performed 8 times; vehicle treatment was performed at each experiment). Error bars represent the standard error of the mean. (B) RNA was extracted from primary leukemia cells after 24-hour in vitro treatment with RAS-pathway inhibitors and submitted for RNA sequencing in triplicate. Doses used were PD325901 20 μM, RAD001 26 μM, and GDC0941 20 μM. Normalized transcript expression levels were used for GSEA, comparing our data set with leukemia-specific self-renewal gene lists, and showed a loss of the MLL-AF9–directed signature in MEK and mTOR-inhibited cells. (C) Schematic figure indicating the NRASG12V-driven pathways that mediate leukemic self-renewal based on our findings.
mTOR and MEK inhibition recapitulate the effects of NRASG12V withdrawal on leukemia self-renewal capacity. (A) Primary leukemia cells were treated with RAS-pathway inhibitors and plated in CFAs. Colonies were scored as the number of colonies that developed per 10 000 cells plated. The percentage of colonies that developed from drug-treated cells, relative to vehicle-treated cells from the same experiment, is shown. Each bar represents the average value of 4 to 13 experiments (doxycycline treatment was performed 13 times, RAD001 and PD325901 treatments were performed 4 times, and GDC0941 treatment was performed 8 times; vehicle treatment was performed at each experiment). Error bars represent the standard error of the mean. (B) RNA was extracted from primary leukemia cells after 24-hour in vitro treatment with RAS-pathway inhibitors and submitted for RNA sequencing in triplicate. Doses used were PD325901 20 μM, RAD001 26 μM, and GDC0941 20 μM. Normalized transcript expression levels were used for GSEA, comparing our data set with leukemia-specific self-renewal gene lists, and showed a loss of the MLL-AF9–directed signature in MEK and mTOR-inhibited cells. (C) Schematic figure indicating the NRASG12V-driven pathways that mediate leukemic self-renewal based on our findings.
To assess which RAS-activated signaling pathways mediate the NRASG12V-directed self-renewal gene-expression pattern, primary NRASG12V/Mll-AF9 leukemia spleen cells were incubated with each inhibitor in vitro for 24 hours and submitted for RNA sequencing. Each condition was performed in triplicate. The reads were mapped using TopHat,17 and normalized expression values (FPKMs) were derived using CuffDiff.18 We used GSEA to compare the gene-expression profiles of inhibitor-treated cells with leukemia-specific self-renewal signatures and found that either mTOR or MEK inhibition, but not PI3K inhibition, led to a significant loss of the MYB and MLL-AF9–directed leukemia self-renewal gene-expression patterns16 (Figure 6B and supplemental Table 6). PI3K activates a number of downstream mediators, including mTOR. In our model, PI3K and mTOR inhibition are not equivalent. Both of these agents inhibit p4EBP1 phosphorylation, but only the PI3K inhibitor targets pAKT (supplemental Figure 5B-C), suggesting that PI3K inhibition may activate feedback loops or alternate pathways, which we did not measure, and these pathways overcome effects on p4EBP1. To confirm these findings, we examined the expression of 33 individual genes from the MLL-AF9 and MYB self-renewal signatures and found that inhibition of either mTOR or MEK most closely recapitulated the effects of NRASG12V withdrawal on these genes (supplemental Figure 6). These data implicate mTOR and MEK as mediators of NRASG12V-directed leukemia self-renewal gene expression and functional capacity.
Discussion
Our study demonstrates a novel role for NRASG12V in maintaining leukemia self-renewal. By using computational approaches to explore our data sets and comparing them with the published literature, we found that NRASG12V enforced the leukemia self-renewal gene-expression signature and was required for leukemia self-renewal independent of its effects on growth and survival. NRASG12V was also required for MLL-AF9– and MYB-dependent leukemia self-renewal gene expression. We also showed that these effects of NRASG12V are specific to the leukemic subpopulation enriched for LSCs and that this subpopulation is preferentially lost upon NRASG12V withdrawal. Using RAS-pathway inhibitors, we found NRASG12V maintained this gene-expression program and leukemia self-renewal through mTOR and MEK pathway activation. These experiments demonstrate that RAS oncogene–driven signaling is critical for leukemia maintenance and explain a mechanism of oncogene addiction.
Both our gene-expression analyses and multidimensional signaling studies hint at a possible role for β-catenin in NRASG12V-mediated self-renewal. Inhibition of β-catenin has been shown to be critical for CML stem-cell targeting.27,35,36 Likewise, β-catenin levels have been shown to correlate with the acquisition of the leukemic phenotype in MLL-AF9–expressing cells,6 indicating that MLL-AF9 is insufficient to induce leukemia and that other signals, supplied by β-catenin, are required. Our data support the notion that RAS-pathway activation may supply these additional signals. Indeed, RAS activation is known to activate the PI3K-AKT pathway, which inhibits GSKβ, a member of the β-catenin destruction complex.37
Our data suggest that the activity of MLL-AF9, a well-known transcriptional regulator of the self-renewal phenotype in leukemia, may rely on additional signals from a cooperating oncogene to exert these effects. Our data show that NRASG12V can facilitate the self-renewal effects of MLL-AF9. Previous reports have shown the MLL-AF9 transcriptional program is regulated by MYB,16 whose function is known to be posttranscriptionally regulated.38 We therefore propose that NRASG12V-dependent signals may posttranscriptionally activate MYB, allowing it to facilitate MLL-AF9 activity (Figure 5C). Interestingly, reduced MYB levels have been associated with improved clinical outcomes in chronic myelomonocytic leukemia (CMML),39 a disease in which RAS-pathway activation is common.2 This observation leads us to speculate that the RAS-MYB relationship may be an important feature of myeloid malignancies.
A recent phase 1b clinical trial incorporating everolimus (RAD001, the mTOR inhibitor used in our study) with standard chemotherapy for relapsed AML patients, showed promising results, with a 68% complete remission rate in these historically refractory patients.40 In this study, relapsed patients who received higher doses of everolimus achieved an 85% complete remission rate. Thus, mTOR signaling may be a critical RAS-effector pathway in maintaining leukemia self-renewal.
The similarity between our data set and several human MLL and MYB data sets (Figure 2 and supplemental Tables 2A-B and 3A-B) suggests that activated NRAS may facilitate self-renewal in human leukemia as well. Accordingly, several reports have identified the central role of NRAS in the development of human leukemia. MLL-translocated human AML harbor the fewest number of mutations of any other subtype of leukemia,1 yet these translocations commonly co-occur with NRAS mutations.41 Single-cell profiling of human myeloproliferative neoplasms that commonly harbor NRAS mutations (M4/M5 AML, CMML, and juvenile myelomonocytic leukemia) reveal a common NRAS-associated signaling architecture in these diseases.42 Whole-genome sequencing of early T-cell precursor acute lymphoblastic leukemia revealed activating mutations of RAS and cytokine receptor signaling in 67% of cases. Notably, this rare, aggressive subtype of ALL expressed a transcriptional profile more consistent with myeloid than lymphoblastic LSCs.43 These studies highlight the importance of NRAS in human leukemia and support our findings implicating NRAS as an important contributor to the development of human AML.
Previous studies have investigated the effects of RAS inhibition in various tumor models. Although numerous reports have explored and validated the role of RAS oncoproteins in cancer cell proliferation and survival, a role in leukemia self-renewal has not been previously described in leukemia and has only recently been described in nonleukemic HSCs.9 We speculate that many experimental systems are not designed to distinguish between proliferation and self-renewal and would be expected to overlook this feature. Unbiased exploration of the gene-expression signature of RAS withdrawal has identified the role of Kras in metabolic reprogramming of pancreatic tumors.44 Similar to our study, the pancreatic cancer study began with a GSEA screen of gene lists in the database. The Ying et al study, along with ours, highlights the value of unbiased screens to uncover unexpected biological behaviors in well-studied proteins such as Ras.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Arkady Khodursky for his assistance with microarray data analysis; Michael Franklin, a scientific writing editor supported by the Division of Hematology, Oncology, and Transplantation, for his editorial assistance; and Carol Taubert for her assistance with figures.
D.A.L. and K.M.S. are American Cancer Society Research Professors.
This work used resources of Minnesota Supercomputing Institute, University of Minnesota Genomics Center, and Masonic Cancer Center (which is supported by National Institutes of Health [NIH] National Cancer Institute; [P30 CA77598]); and was supported in part by an NIH National Heart, Lung, and Blood Institute training grant (T32HL007062) (Z.S.); funds from the Division of Hematology, Oncology, and Transplantation, Department of Medicine, University of Minnesota (Z.S.); a Leukemia and Lymphoma Society Specialized Center of Research grant (D.A.L.); an NIH National Cancer Institute grant (R37CA72614) (K.M.S); a Leukemia and Lymphoma Society Specialized Center of Research Award LLS (7019-04) (K.M.S); a Department of Defense (CDMRP) Ovarian Cancer Teal Innovator Award (G.P.N); the National Heart, Lung and Blood Institute, NIH (N01-HV-00242) (G.P.N.); and the National Cancer Institute, NIH (1R01CA130826) (G.P.N.).
Authorship
Contribution: Z.S. designed the study, performed experiments, analyzed data and wrote the manuscript; R.S.L., K.S., S.K.R., and A.L.S. analyzed data; H.T.N., K.E.N., N.A.M.H., E.D.-F., S.C.B., N.A.H., and M.D.D. performed experiments; G.P.N. and K.M.S. provided guidance; and D.A.L. designed the study and provided guidance.
Conflict-of-interest disclosure: S.C.B. is a consultant for DVS sciences, the supplier of some of the reagents and instrumentation used in this study. G.P.N. is Chairman of the SAB for DVS Sciences and Nodality, Inc., and a consultant for Cell Signaling Technologies and Becton Dickinson, Inc.— holder of patents or manufacturer of the instruments and/or reagents used in this study. The remaining authors declare no competing financial interests
Correspondence: Zohar Sachs, Mayo Mail Code 480, 420 Delaware St SE, Minneapolis, MN 55455; e-mail:sachs038@umn.edu.