Key Points
Boolean implications are a useful computational algorithm to mine mutation-specific methylation relationships in large cancer data sets.
Mutant WT1 is associated with DNA hypermethylation of PRC2 targets in AML, and inhibition of EZH2 induces myeloid differentiation.
Abstract
Acute myeloid leukemia (AML) is associated with deregulation of DNA methylation; however, many cases do not bear mutations in known regulators of cytosine guanine dinucleotide (CpG) methylation. We found that mutations in WT1, IDH2, and CEBPA were strongly linked to DNA hypermethylation in AML using a novel integrative analysis of The Cancer Genome Atlas data based on Boolean implications, if-then rules that identify all individual CpG sites that are hypermethylated in the presence of a mutation. Introduction of mutant WT1 (WT1mut) into wild-type AML cells induced DNA hypermethylation, confirming mutant WT1 to be causally associated with DNA hypermethylation. Methylated genes in WT1mut primary patient samples were highly enriched for polycomb repressor complex 2 (PRC2) targets, implicating PRC2 dysregulation in WT1mut leukemogenesis. We found that PRC2 target genes were aberrantly repressed in WT1mut AML, and that expression of mutant WT1 in CD34+ cord blood cells induced myeloid differentiation block. Treatment of WT1mut AML cells with short hairpin RNA or pharmacologic PRC2/enhancer of zeste homolog 2 (EZH2) inhibitors promoted myeloid differentiation, suggesting EZH2 inhibitors may be active in this AML subtype. Our results highlight a strong association between mutant WT1 and DNA hypermethylation in AML and demonstrate that Boolean implications can be used to decipher mutation-specific methylation patterns that may lead to therapeutic insights.
Introduction
Discovery of key molecular drivers of epigenetic change in cancer is essential for the development of effective epigenetic-based therapy. Aberrant changes in DNA methylation are observed in multiple cancers including acute myeloid leukemia (AML), but the molecular events responsible for perturbing methylated genomic landscapes have not been completely characterized. A major role for dysregulated DNA methylation in the evolution of cancer has been supported by the discovery of recurrent mutations in genes that can modify DNA methylation such as isocitrate dehydrogenase 1 and 2 (IDH1/2),1,2 ten-eleven translocation-2 (TET2),2,3 and DNA (cytosine-5)-methyltransferase 3 α (DNMT3A),4,5 several of which are pharmacologic targets.6 These discoveries support the notion that de novo DNA methylation patterns in AML may be partially determined by genetic mutation.7,8 Furthermore, DNA methylation profiling can separate AML into 16 prognostically significant clusters, although several of these have no obvious associated genetic lesion.9 Clonal analysis of purified stem cell populations from patients in our laboratory and others10-13 suggests that acquisition of a methylation-modifying mutation such as TET2 or IDH2 is an early step in adult leukemogenesis. Nevertheless, many cases of AML do not bear mutations in IDH1/2 or TET2, yet clearly exhibit extensive promoter DNA hypermethylation, implying that alternative mechanisms remain to be discovered.9,14
In order to find novel genetic drivers of DNA hypermethylation in AML, we inferred Boolean implications (if-then rules) between mutation and DNA methylation in The Cancer Genome Atlas (TCGA) AML data set. The distribution of points in a scatterplot of 2 variables in a Boolean implication is L-shaped instead of linear, facilitating the discovery of subset (containment) and mutual exclusion relationships15 between samples with genetic lesions and samples with hypermethylation of specific cytosine guanine dinucleotide (CpG) sites. We found mutation in the Wilms tumor 1 (WT1) gene to be strongly linked to CpG hypermethylation in AML. WT1 is a zinc finger transcription factor encoded on 11p13 that was first identified as a tumor suppressor gene in patients with Wilms tumor predisposition-aniridia-genitourinary-mental retardation16 and is required for maintenance of mesenchymal-epithelial balance and erythropoiesis in adult tissues.17 Heterozygous insertion/deletion mutations in WT1 are present in 8% to 10% of normal karyotype (NK) AML,18-20 but their mechanism of action and contribution to the leukemic phenotype is unknown. Our findings suggest that examination of mutation-specific patterns of hypermethylated CpG sites by Boolean implications is a powerful method to find novel drivers and functional pathways that may be perturbed in cancer.
Methods
Patient sample data
Primary bone marrow and peripheral blood de novo AML samples were obtained prior to treatment with informed consent according to institutional guidelines (Stanford University Institutional Review Board No. 6453) with details provided in supplemental Table 1, available on the Blood Web site. The somatic mutation, DNA methylation, and gene expression data were retrieved from the TCGA database.14 Research was conducted in accordance with the Declaration of Helsinki.
Statistical data analysis
Details on analysis using Boolean implications as well as additional data analysis such as multidimensional analysis, gene set enrichment analysis, and ENCyclopedia of DNA elements (ENCODE) data analysis are provided in the supplemental Methods.
DNA methylation studies of AML cells
Illumina HumanMethylation450 BeadChip profiling was performed for THP1 cells expressing mutant IDH2 and WT1 complementary DNAs, CTS cells, and sample SU359. Array data are deposited in Gene Expression Omnibus database (accession number GSE62929).
Primary cord blood CD34+ and AML blast differentiation assays
Cord blood was obtained from the New York Blood Center. Fresh CD34+ cells were purified using magnetic-activated cell sorting (Miltenyi) and transduced with lentivirus (pLVX EF1α-IRES-zsGreen; Clontech) overnight. Cells were washed and incubated in Myelocult H5100 (Stem Cell Technologies) with 20 μg/mL interleukin (IL)-3, stem cell factor (SCF), fms-related tyrosine kinase 3 ligand (FLT3L), and granulocyte macrophage–colony stimulating factor (GM-CSF) (Peprotech) for 6 days. Blasts were sorted for CD117+/CD34+ and cultured for 72 hours ± GSK-126 (ActivBiochem) or all-trans retinoic acid (ATRA; Sigma). Differentiation of green fluorescent protein (GFP)–positive cells or blasts was assessed by flow cytometry using anti-human CD11c-V450 (B-ly6), CD14-APC-Cy7 (MϕP9), CD15-APC (HI98), and CD11b-PE-Cy7 (ICRF44; BD Biosciences).
Results
Identification of mutation-methylation Boolean implications in AML
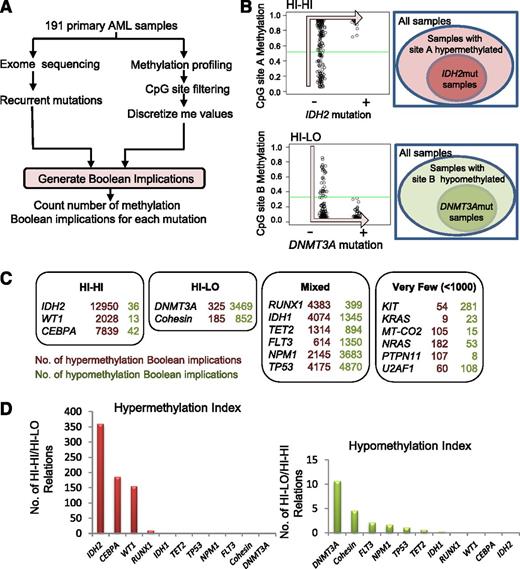
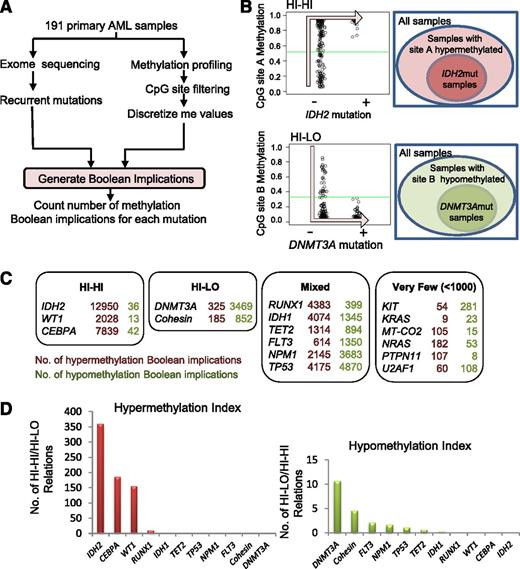
In order to identify somatic mutations in AML that are directly linked to DNA CpG methylation, we applied an algorithm based on Boolean implications to the TCGA AML patient data set consisting of 191 AML patient samples profiled in parallel for somatic gene mutations and DNA CpG methylation (Figure 1A). Boolean implications15 represent if-then rules between 2 variables, in this case between mutation in AML and DNA methylation. The distribution of points in a scatterplot of the 2 variables in a Boolean implication is L-shaped, unlike correlation, which can only find linear relationships. To be included in the analysis, a CpG site had to have available data for all patients and show a maximum methylation β value >0.2, a minimum value <0.8, and a dynamic range >0.1. To be included in the analysis, a somatic mutation had to have available methylation data from a minimum of 5 patients. Overall, 285 320 CpG sites met these criteria for inclusion in the analysis, which encompassed a total of 16 recurrently mutated genes (IDH1, IDH2, TET2, WT1, CEBPA, DNMT3A, RUNX1, FLT3, NPM1, TP53, KIT, KRAS, NRAS, MT-CO2, PTPN11, and U2AF1) plus the set of genes comprising the components of the cohesin complex. Boolean implications were then derived between these recurrent mutations and CpG sites. A HI-HI implication between a mutation m and a CpG site A states that if m is present, then A is hypermethylated. In other words, almost all samples with m are always contained within the set of patient samples that have A hypermethylated (Figure 1B). A HI-LO implication between a mutation m and a CpG site B states that if m is present, then B is hypomethylated. Thus, almost all samples with m are contained within the set of patient samples that have B hypomethylated (Figure 1B). Next, we counted the number of HI-HI (hypermethylation) and HI-LO (hypomethylation) relationships for each mutation (Figure 1C). We found a total of 57 044 HI-HI and HI-LO implications that stratified the mutations into 4 categories based on the number and type of Boolean implications with methylation: “HI-HI” or predominantly hypermethylating if the number of Boolean implications was >1000 and the hypermethylation index (ratio of HI-HI to HI-LO Boolean implications) was >150; “HI-LO” or predominantly hypomethylating if the number of Boolean implications was >1000 and the hypomethyation index (ratio of HI-LO to HI-HI Boolean implications) was >4.5; “Mixed” hyper- and hypomethylating if the total number of Boolean implications exceeded 1000, but without satisfying the criteria for the HI-HI or HI-LO categories; and mutations with “Very Few” methylation implications (<1000 total) (Figure 1C). The identity, chromosomal location, and gene annotation of all CpG sites that have Boolean implications with each recurrent mutation is provided in supplemental Table 2. Note that the number of patient samples for each mutation did not correlate with the number of Boolean implications obtained, suggesting that sample size did not confound the analysis (supplemental Figure 1).
Boolean implications identify mutations strongly linked to changes in DNA methylation. (A) Schematic of process for generating Boolean implications. (B) Examples of HI-HI (hypermethylation) and HI-LO (hypomethylation) implications between mutation and methylation. (C) Summary of the total number of HI-HI (left) and HI-LO (right) Boolean implications with CpG sites from Illumina 450K array for each recurrent mutation in the TCGA AML data set. Each mutation is categorized into 1 of 4 groups (HI-HI, HI-LO, Mixed, or Very Few) according to the predominant numbers of either HI-HI or HI-LO implications. (D) Graph of hypermethylation index (number of HI-HI/number of HI-LO) (left) or hypomethylation index (number of HI-LO/number of HI-HI) (right) for each mutation.
Boolean implications identify mutations strongly linked to changes in DNA methylation. (A) Schematic of process for generating Boolean implications. (B) Examples of HI-HI (hypermethylation) and HI-LO (hypomethylation) implications between mutation and methylation. (C) Summary of the total number of HI-HI (left) and HI-LO (right) Boolean implications with CpG sites from Illumina 450K array for each recurrent mutation in the TCGA AML data set. Each mutation is categorized into 1 of 4 groups (HI-HI, HI-LO, Mixed, or Very Few) according to the predominant numbers of either HI-HI or HI-LO implications. (D) Graph of hypermethylation index (number of HI-HI/number of HI-LO) (left) or hypomethylation index (number of HI-LO/number of HI-HI) (right) for each mutation.
Association of individual AML mutations with DNA methylation
Five of the 17 AML mutations were associated with hyper- (HI-HI) or hypo- (HI-LO) methylation (Figure 1C). The presence of an IDH2 mutation exhibited the largest number of HI-HI implications (12 950) with virtually no HI-LO implications (36), meaning that there were 12 950 CpG sites that displayed increased methylation when the IDH2 mutation was present. This is consistent with the established role of IDH2 mutations in causing DNA hypermethylation through inhibition of TET.2 On the other hand, we found that mutations in DNMT3A were linked to the largest number of HI-LO implications (3469) compared with far fewer HI-HI relationships (325), consistent with its known loss of DNA methyltransferase activity and its hypomethylating effect.21,22 These IDH2 and DNMT3A results validate this implication-based approach to the identification of mutational drivers of aberrant methylation in AML.
Both IDH1 and TET2 mutations have been directly implicated in DNA CpG hypermethylation in AML.2 Our implication-based analysis indicated that these mutations were associated with both HI-HI and HI-LO implications, placing them in the “Mixed” category, although both were skewed toward more HI-HI relationships (Figure 1C). A number of genes involved in proliferative signal transduction are recurrently mutated in AML including FLT3, KIT, NRAS, KRAS, and PTPN11. Notably, none of these mutations were associated with CpG methylation, with most having very few Boolean implications.
The implication-based analysis revealed several novel associations between gene mutations and DNA methylation. First, mutations in the cohesin complex were predominantly linked to hypomethylation with 852 HI-LO implications. Interestingly, we noted that both WT1 and CEBPA mutations were predominantly associated with HI-HI Boolean implications, similar to IDH2. We identified 2028 HI-HI implications for WT1 mutations but only 13 HI-LO, suggesting the presence of a WT1 mutation (WT1mut) is linked to DNA hypermethylation in AML (for examples of Boolean implications between WT1mut and DNA methylation, see supplemental Figure 2A). Likewise, CEBPA mutations were associated with 7839 HI-HI relationships and only 42 HI-LO. Mutations in IDH2, CEBPA, and WT1 were strongly associated with hypermethylation with an index >150 (Figure 1D); similarly, mutations in DNMT3A and the cohesin group had the strongest hypomethylation index (Figure 1D).
Mutant WT1 induces DNA hypermethylation in AML
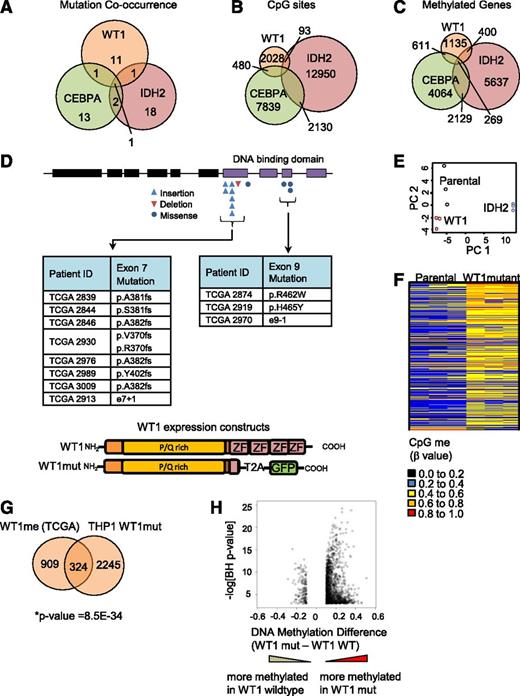
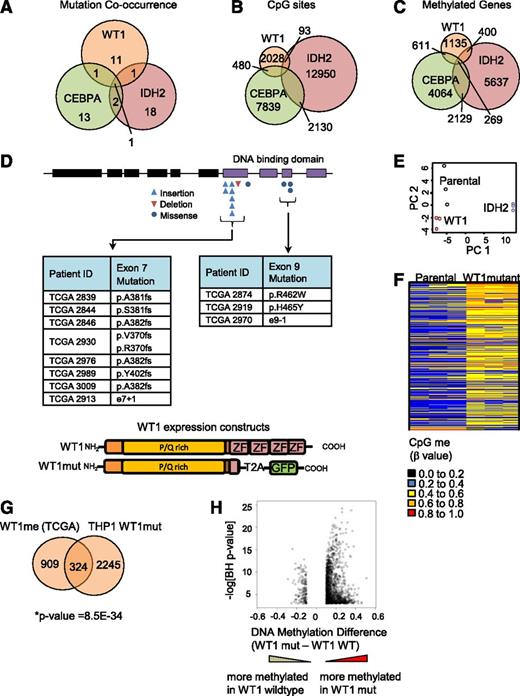
Although a link between mutation in CEBPA or IDH2 and DNA hypermethylation in AML has been reported,2,9 an association between mutation in WT1 and DNA hypermethylation has not been described. Further investigation of the TCGA data set revealed that WT1mut is almost always mutually exclusive with mutation in IDH2 or CEBPA (Figure 2A), as also reported by Eastern Cooperative Oncology Group investigators.23 Moreover, there is only a partial overlap in the methylated CpG sites in the HI-HI Boolean implications for these mutations (Figure 2B) and genes associated with the CpG sites (Figure 2C; supplemental Tables 3 and 4), suggesting that each mutation is associated with distinct DNA methylation regulatory mechanisms.
Mutant WT1 induces DNA hypermethylation in AML. (A) Venn diagram showing cooccurrence between the 3 mutations most linked to DNA hypermethylation (WT1, IDH2, and CEBPA) from 191 sequenced patients in the TCGA cohort. (B) Venn diagram showing overlap of hypermethylated CpG sites identified by Boolean implications for mutation in WT1, IDH2, and CEBPA. (C) Venn diagram showing overlap of all identified protein-coding genes associated with the hypermethylated CpG sites using the annotation file provided by Illumina. (D) Schematic showing the domain structure of the WT1 protein and the location of insertion/deletion mutations for the WT1 mutations in the TCGA AML cohort. Shown subsequently is the mutated form of WT1 used in methylation validation studies truncated at the first zinc finger domain (amino acid position 306) and linked to self-cleaving T2A peptide and GFP in a lentiviral vector. P/Q rich, proline/glutamine rich domain; ZF, zinc finger domain. (E) Principal component analysis of all probes with a dynamic β value range >0.2 assayed by Illumina HumanMethylation450 BeadChip array in THP1 AML cells transduced with either mutant WT1 or mutant IDH2 R172W in triplicate and compared with parental cells after 10 passages in culture. (F) Heatmap showing differential methylation between parental and WT1-mutant–expressing THP1 cells. Increased β values indicate increased methylation. (G) Venn diagram showing the overlap of genes methylated upon expression of mutant WT1 in THP1 cells and genes identified to be methylated in the TCGA data set by Boolean implications; *P = 8.5E-34, Fisher’s exact test, right tailed. A gene could only be included in this analysis if at least 1 of its associated CpG sites appeared in the Boolean implication-based methylome analysis of TCGA patients and in the differential methylome analysis of the THP1-derived cell lines. The total number (universe) of genes used in Fisher’s exact test calculation was 13 018. (H) Volcano plot showing the difference in methylation for WT1 mutated vs WT1 wild-type samples in the TCGA data set. The y-axis shows the negative log of Benjamini-Hochberg corrected P value of a t test for differential methylation, and the x-axis shows the magnitude change in methylation (difference in average β value between WT1-mutant and WT1 wild-type samples). Each dot represents the mean β value difference and the P value of a given CpG site in a t test between WT1-mutated vs WT1 wild-type AML.
Mutant WT1 induces DNA hypermethylation in AML. (A) Venn diagram showing cooccurrence between the 3 mutations most linked to DNA hypermethylation (WT1, IDH2, and CEBPA) from 191 sequenced patients in the TCGA cohort. (B) Venn diagram showing overlap of hypermethylated CpG sites identified by Boolean implications for mutation in WT1, IDH2, and CEBPA. (C) Venn diagram showing overlap of all identified protein-coding genes associated with the hypermethylated CpG sites using the annotation file provided by Illumina. (D) Schematic showing the domain structure of the WT1 protein and the location of insertion/deletion mutations for the WT1 mutations in the TCGA AML cohort. Shown subsequently is the mutated form of WT1 used in methylation validation studies truncated at the first zinc finger domain (amino acid position 306) and linked to self-cleaving T2A peptide and GFP in a lentiviral vector. P/Q rich, proline/glutamine rich domain; ZF, zinc finger domain. (E) Principal component analysis of all probes with a dynamic β value range >0.2 assayed by Illumina HumanMethylation450 BeadChip array in THP1 AML cells transduced with either mutant WT1 or mutant IDH2 R172W in triplicate and compared with parental cells after 10 passages in culture. (F) Heatmap showing differential methylation between parental and WT1-mutant–expressing THP1 cells. Increased β values indicate increased methylation. (G) Venn diagram showing the overlap of genes methylated upon expression of mutant WT1 in THP1 cells and genes identified to be methylated in the TCGA data set by Boolean implications; *P = 8.5E-34, Fisher’s exact test, right tailed. A gene could only be included in this analysis if at least 1 of its associated CpG sites appeared in the Boolean implication-based methylome analysis of TCGA patients and in the differential methylome analysis of the THP1-derived cell lines. The total number (universe) of genes used in Fisher’s exact test calculation was 13 018. (H) Volcano plot showing the difference in methylation for WT1 mutated vs WT1 wild-type samples in the TCGA data set. The y-axis shows the negative log of Benjamini-Hochberg corrected P value of a t test for differential methylation, and the x-axis shows the magnitude change in methylation (difference in average β value between WT1-mutant and WT1 wild-type samples). Each dot represents the mean β value difference and the P value of a given CpG site in a t test between WT1-mutated vs WT1 wild-type AML.
In order to test the hypothesis that mutation in WT1 induces DNA hypermethylation, we expressed a mutant form of the WT1 protein lacking all 4 zinc finger domains, typical of that found in AML, in the THP1 AML cell line (confirmed to be WT1 wild-type) using a lentiviral approach (Figure 2D; supplemental Figure 2B). Methylation profiling of stable cell lines was performed using Illumina HumanMethylation450 BeadChip arrays, as in the TCGA effort. THP1 cells engineered to express mutant IDH2 R172W were separately generated and analyzed in parallel as a mutation known to strongly drive DNA hypermethylation. Principal component analysis (Figure 2E) revealed that the DNA methylation profile was different in the WT1- and IDH2-mutant cells compared with parental cells and to each other, consistent with our analysis of the TCGA data (Figure 2A-C).
Next, we sought to characterize the methylome changes in the WT1- and IDH2-mutant THP1 cells. A total of 5060 CpG sites were differentially methylated in the mutant WT1-expressing cells compared with parental THP1, of which nearly all (4960) were hypermethylated (Figure 2F). In the mutant IDH2-expressing cells, 23 169 differentially methylated CpG sites were identified, of which 11 505 were hypermethylated (supplemental Figure 2C). Comparison with the TCGA primary AML data demonstrated a significant overlap between the genes corresponding to the hypermethylated CpG sites in the WT1-mutant TCGA samples (designated WT1me) and the mutant WT1-expressing THP1 cells (Figure 2G). Moreover, we observed that the distribution of CpG sites that are hypermethylated in the mutant WT1-expressing THP1 cells and WT1-mutant TCGA samples across distinct genomic regions (promoter, ±5 kb of the transcription start site) is similar (supplemental Figure 2E), and that there is significant overlap (Fisher’s exact test, P value = 1.17e-11) between the genes with hypermethylated CpG sites in promoter regions. Taken together, these results suggest that the engineered stable cell line recapitulates the DNA hypermethylation observed in patients. Finally, a volcano plot examining the methylation difference vs the statistical significance for CpG sites between the WT1-mutant and WT1 wild-type TCGA samples showed a similar asymmetric profile to that obtained for IDH2, with a skewing toward DNA hypermethylation (Figure 2H and supplemental Figure 2D). Collectively, these results suggest that WT1 mutations induce DNA hypermethylation at multiple genomic loci that are distinct from those regulated by IDH2 mutations.
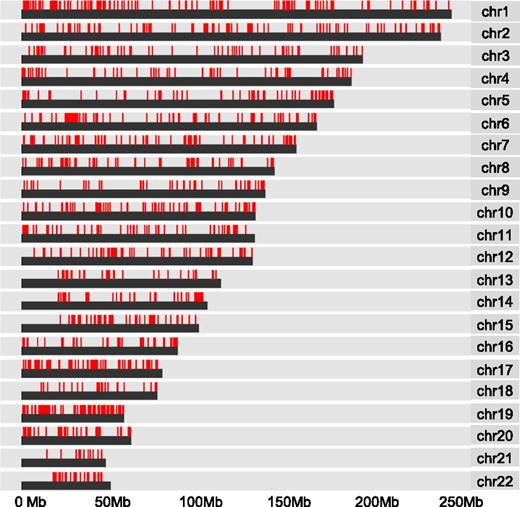
DNA hypermethylation associated with WT1 mutation is widely distributed
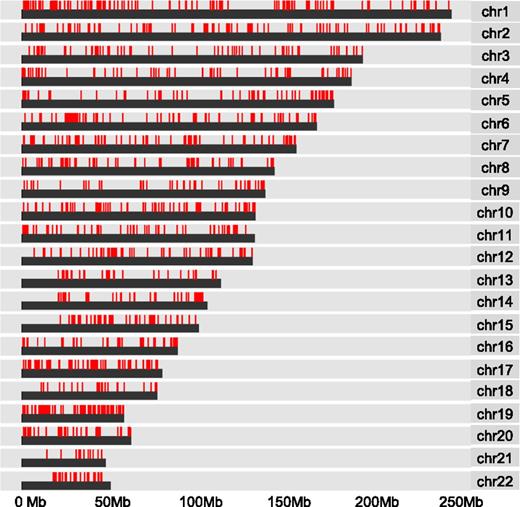
To further examine the observed pattern of hypermethylation, we mapped the distribution of CpG methylation associated with WT1 mutation across human chromosomes. We found widespread dispersal of hypermethylated sites throughout all chromosome regions rather than clustering to more restricted loci (Figure 3). In addition, the induction of hypermethylation in the mutant WT1-expressing THP1 cells suggests that the observed pattern of hypermethylation is de novo. To examine which DNA methyltransferases could be responsible, we compared the mRNA expression levels of all isoforms of the known mammalian enzymes DNMT3A, DNMT3B, and DNMT1 in WT1-mutated samples and NK AML (supplemental Figure 3). DNMT1 was expressed but not differentially upregulated in WT1-mutated AML and is generally considered unlikely to play a role in de novo methylation.24 Significantly, both active isoforms of DNMT3A (A isoform, P < .02; B isoform, P < .008), but not DNMT3B, were upregulated in the WT1-mutated samples, indicating DNMT3A rather than DNMT3B is the predominant de novo DNA methyltransferase expressed in WT1mut AML.
DNA hypermethylation associated with WT1 mutation is widely distributed. Chromosome distribution map of all CpG sites linked to mutation in WT1 by Boolean implications in the TCGA data set. Red lines represent methylated CpG sites.
DNA hypermethylation associated with WT1 mutation is widely distributed. Chromosome distribution map of all CpG sites linked to mutation in WT1 by Boolean implications in the TCGA data set. Red lines represent methylated CpG sites.
WT1 mutation induces DNA hypermethylation of PRC2 target genes
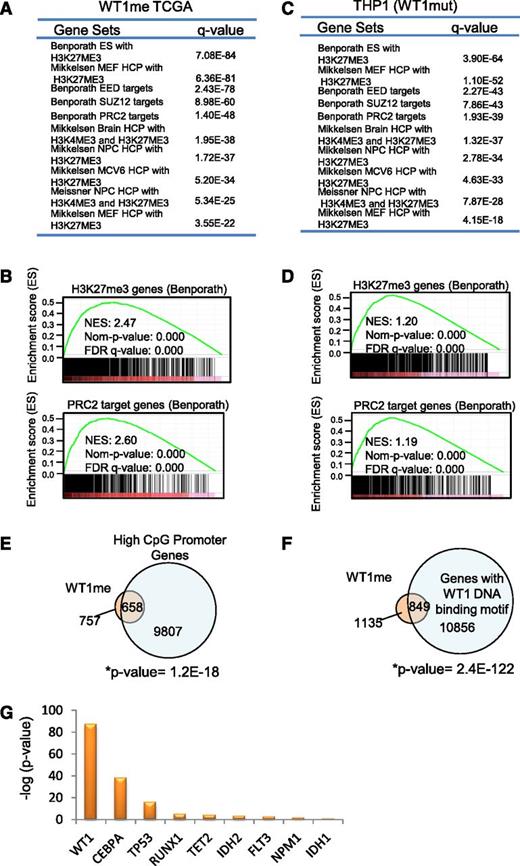
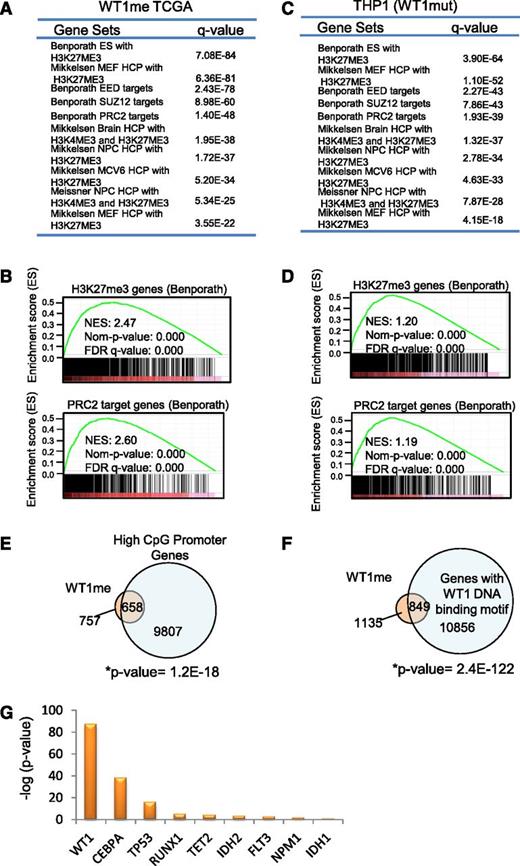
In order to further understand the origin of hypermethylation associated with mutation in WT1, we conducted gene set enrichment analysis25 of the methylated genes identified by Boolean implications. Intriguingly, WT1me genes were overrepresented in multiple gene sets of polycomb repressor complex 2 (PRC2) targets and genes associated with histone H3 lysine 27 (H3K27) trimethylated (me3) histone marks in early human development (P < .0E-25 for multiple sets)26,27 (Figure 4A-B). This was not the case for the mutant IDH2 HI-HI genes (IDH2me), which showed enrichment for a range of gene sets including metabolic syndrome, mitogen-activated protein kinase, RUNX, p53, as well as bivalent histone H3 lysine 4/H3K27 trimethyl targets (supplemental Figure 4A). Enrichment of PRC2/H3K27me3-associated gene sets was also observed for the hypermethylated genes induced by mutant WT1 in THP1 cells (P < 9.66E-21 for multiple sets) (Figure 4C-D). The PRC2 complex is known to preferentially associate with CpG rich promoters that are characteristically hypomethylated in early human development.28 Consistent with this, we found significant overlap between WT1me genes and high CpG-containing promoters as defined by Brutlag29 (P = 1.2E-18) (Figure 4E). Moreover, we also determined that the sequences within 500 bp of the transcription start site of WT1me genes were enriched for the WT1 DNA binding motif GCG-GGG-NGN indicating that mutations that disrupt normal DNA binding by this transcription factor may promote DNA hypermethylation of WT1 transcriptional target genes, in addition to PRC2 targets (Figure 4F). Interestingly, all WT1-mutated samples with measured hypermethylation in the TCGA cohort (Figure 2D) had frameshift or point mutations in either exon 7 or exon 9 of WT1 that disrupt the DNA binding domains.
WT1 mutation induces DNA hypermethylation of PRC2 target genes. (A-B) Summary and examples of gene sets enriched in the set of WT1mut-associated hypermethylated genes (WT1me) derived from Boolean implications from the TCGA data. Gene sets were obtained from the MSigDB. Hypergeometric test P values are indicated, and enrichment plots for selected gene sets are illustrated. (C-D) Summary and examples of gene sets enriched in the set of genes hypermethylated in THP1 cells engineered to express mutant WT1 (as in Figure 2F). Hypergeometric test P values are indicated and enrichment plots for selected gene sets are illustrated. (E) Venn diagram showing overlap of the WT1me genes from the TCGA data set and genes with high CpG content in their promoters. Overlap of these gene sets was measured by Fisher’s exact test (right tailed). The universe of genes in the contingency table is equal to 15 880, which is the set of genes that were studied and classified as having high or low CpG content.29 (F) Venn diagram showing overlap of the WT1me genes from the TCGA data set and genes with WT1 DNA binding motifs in their promoters. Overlap of these gene sets was measured by Fisher’s exact test. (G) Comparison of PRC2 target gene enrichment among hypermethylated genes associated with individual AML mutations from the TCGA data set. The y-axis shows the negative log of the P value of the cooccurrence between the H3K27me3 Benporath gene set and methylated genes derived from HI-HI Boolean implications for each recurrent mutation. Overlap of the gene sets was measured by Fisher’s exact test (right tailed).
WT1 mutation induces DNA hypermethylation of PRC2 target genes. (A-B) Summary and examples of gene sets enriched in the set of WT1mut-associated hypermethylated genes (WT1me) derived from Boolean implications from the TCGA data. Gene sets were obtained from the MSigDB. Hypergeometric test P values are indicated, and enrichment plots for selected gene sets are illustrated. (C-D) Summary and examples of gene sets enriched in the set of genes hypermethylated in THP1 cells engineered to express mutant WT1 (as in Figure 2F). Hypergeometric test P values are indicated and enrichment plots for selected gene sets are illustrated. (E) Venn diagram showing overlap of the WT1me genes from the TCGA data set and genes with high CpG content in their promoters. Overlap of these gene sets was measured by Fisher’s exact test (right tailed). The universe of genes in the contingency table is equal to 15 880, which is the set of genes that were studied and classified as having high or low CpG content.29 (F) Venn diagram showing overlap of the WT1me genes from the TCGA data set and genes with WT1 DNA binding motifs in their promoters. Overlap of these gene sets was measured by Fisher’s exact test. (G) Comparison of PRC2 target gene enrichment among hypermethylated genes associated with individual AML mutations from the TCGA data set. The y-axis shows the negative log of the P value of the cooccurrence between the H3K27me3 Benporath gene set and methylated genes derived from HI-HI Boolean implications for each recurrent mutation. Overlap of the gene sets was measured by Fisher’s exact test (right tailed).
Notably, WT1me genes showed the largest statistically significant overlap with the embryonic stem cell H3K27me3 gene set30 compared with all other AML mutations and their associated hypermethylated genes (Figure 4G and supplemental Table 5). In order to test whether the association between WT1me genes and H3K27me3-marked genes was also apparent in hematopoietic cells, we identified genes whose transcription start sites are within ±5 kb of the H3K27me3 peaks in the AML K562 cell line data from the ENCODE project.31 The WT1me genes had a large overlap with these H3K27me3 marked genes in K562 (supplemental Figure 4B), consistent with a link between aberrant DNA methylation and H3K27 modification by PRC2 in WT1mut AML.
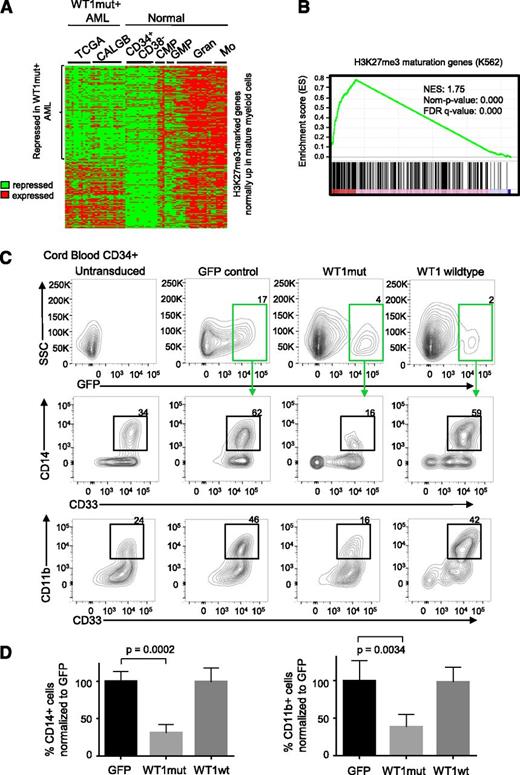
WT1 mutation is associated with aberrant repression of H3K27me3 marked genes in AML
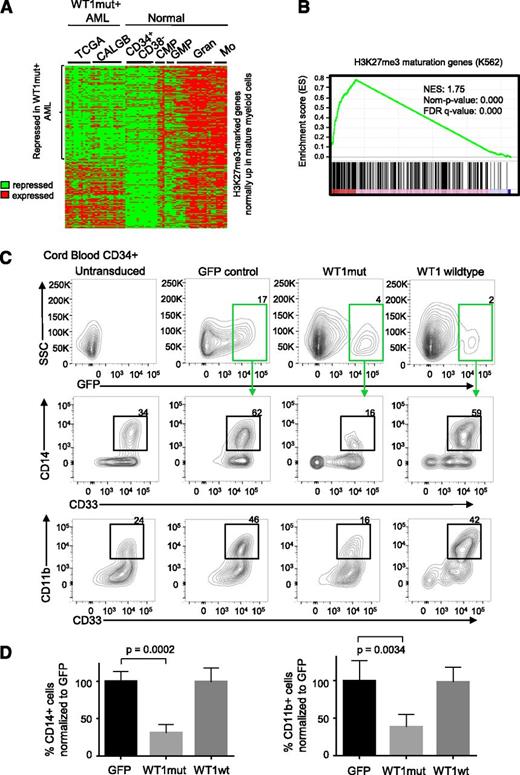
In order to examine whether mutation in WT1 may be specifically interfering with the regulation of PRC2 target genes in AML, we analyzed gene expression data from DNA microarrays of WT1-mutant TCGA samples, WT1-mutant Cancer and Leukemia Group B (CALGB) samples,32 and normal hematopoietic populations.33 We compiled a list of H3K27me3-marked genes in K562 that are upregulated at least 1.5-fold in normal mature myeloid cells (granulocytes and monocytes) compared with normal progenitor cells (Figure 5A). Interestingly, we found a high degree of repression of these genes in WT1mut AML (Figure 5A), with 58% of genes repressed. Furthermore, we identified genes that are differentially expressed in WT1-mutant AML compared with WT1 wild-type AML by pooling gene expression data for NK AML samples from TCGA and CALGB. We noted a significant enrichment for the H3K27me3-marked myeloid maturation genes (Figure 5A) among the genes that were preferentially downregulated in the WT1-mutant samples (Figure 5B). In addition, we also sought to determine the degree of repression of mature myeloid H3K27me3-marked genes in the different mutant populations within each patient cohort. The H3K27me3 genes were analyzed by gene set enrichment analysis25 using repression scores for each gene in the WT1mut AML subgroup within the TCGA cohort and the CALGB cohort. This analysis identified a large enrichment for maturation genes (supplemental Figure 5A-B) with a positive enrichment score for repression in WT1mut AML in the TCGA (normalised enrichment score (NES) = 2.47, false discovery rate q value = 0) and in CALGB (NES = 1.88, false discovery rate q value = 0), indicating repression of H3K27me3-marked genes that become expressed during normal myeloid differentiation. The IDH2-mutant and NPM1-mutant subgroups within the TCGA did not show the same degree of repression with negative enrichment scores for the H3K27me3 maturation genes (supplemental Figures 5C-D).
WT1 mutation is associated with repression of hematopoietic PRC2 targets and blocks myeloid differentiation. (A) Heatmap showing normalized U133A microarray expression of H3K27me3-marked genes (identified from the K562 H3K27me3 chromatin immunoprecipitation ENCODE data) that are normally upregulated in maturing myeloid cells in WT1mut AML (total of 26 patient samples, 11 from TCGA and 15 from Cancer and Leukemia Group B data sets) and normal CD34+CD38− hematopoietic stem and progenitor cells, common myeloid progenitors (CMP), granulocyte-macrophage progenitors (GMP), mature granulocytes (Gran), and mature monocytes (Mo). (B) Gene set enrichment plot assessing the degree of PRC2 target gene repression in the WT1mut AML subgroup with respect to other NK AML subtypes (TCGA and CALBG). Genes were ranked according to the degree of downregulation in WT1mut AML compared with rest of NK AML based on the t test. A positive enrichment score (normalised enrichment score = 1.75, nominal P value = 0) indicates repression of H3K27me3 genes that are normally upregulated during myeloid differentiation in WT1mut AML when compared with other NK AML. (C) Human CD34+ cord blood stem/progenitor cells were transduced with lentiviruses encoding WT1mut–IRES-GFP, WT1 wild type–IRES-GFP, or GFP empty vector and cultured in myeloid differentiation media supplemented with IL-3, SCF, FLT3L, and GM-CSF. At day 6, GFP-positive cells were analyzed for expression of myeloid markers CD33, CD11b, and CD14. Flow cytometry analysis shows the percentage of CD14- and CD11b-positive cells within the GFP-positive populations, as well as untransduced cells from 1 representative experiment out of 3 independent experiments. (D) Summary of data from 3 independent experiments showing the percentage of CD33+/CD14+ and CD33+/CD11b+ cells compared with GFP control. Unpaired Student t test was used to determine statistical significance between GFP and WT1mut populations.
WT1 mutation is associated with repression of hematopoietic PRC2 targets and blocks myeloid differentiation. (A) Heatmap showing normalized U133A microarray expression of H3K27me3-marked genes (identified from the K562 H3K27me3 chromatin immunoprecipitation ENCODE data) that are normally upregulated in maturing myeloid cells in WT1mut AML (total of 26 patient samples, 11 from TCGA and 15 from Cancer and Leukemia Group B data sets) and normal CD34+CD38− hematopoietic stem and progenitor cells, common myeloid progenitors (CMP), granulocyte-macrophage progenitors (GMP), mature granulocytes (Gran), and mature monocytes (Mo). (B) Gene set enrichment plot assessing the degree of PRC2 target gene repression in the WT1mut AML subgroup with respect to other NK AML subtypes (TCGA and CALBG). Genes were ranked according to the degree of downregulation in WT1mut AML compared with rest of NK AML based on the t test. A positive enrichment score (normalised enrichment score = 1.75, nominal P value = 0) indicates repression of H3K27me3 genes that are normally upregulated during myeloid differentiation in WT1mut AML when compared with other NK AML. (C) Human CD34+ cord blood stem/progenitor cells were transduced with lentiviruses encoding WT1mut–IRES-GFP, WT1 wild type–IRES-GFP, or GFP empty vector and cultured in myeloid differentiation media supplemented with IL-3, SCF, FLT3L, and GM-CSF. At day 6, GFP-positive cells were analyzed for expression of myeloid markers CD33, CD11b, and CD14. Flow cytometry analysis shows the percentage of CD14- and CD11b-positive cells within the GFP-positive populations, as well as untransduced cells from 1 representative experiment out of 3 independent experiments. (D) Summary of data from 3 independent experiments showing the percentage of CD33+/CD14+ and CD33+/CD11b+ cells compared with GFP control. Unpaired Student t test was used to determine statistical significance between GFP and WT1mut populations.
Mutant WT1 blocks myeloid differentiation in CD34+ cord blood stem cells
The previous analysis suggests that mutation in WT1 can interfere with normal myeloid differentiation. To test this hypothesis, we investigated the function of WT1mut in a well-established model of myeloid differentiation, erythropoietin (EPO)–induced fetal hemoglobin expression in TF1 cells. We generated TF1 cells stably expressing mutant WT1 using lentiviral transduction. TF1 cells require GM-CSF for proliferation and will undergo differentiation down the erythroid lineage upon exposure to EPO. However, with expression of mutant WT1, TF1 cells failed to upregulate fetal hemoglobin upon exposure to EPO (supplemental Figure 5E), indicating that expression of mutant WT1 induces a block in myeloid differentiation. To further explore our hypothesis, we expressed wild-type or mutant WT1, or GFP empty vector control in purified CD34+ cord blood hematopoietic stem/progenitor cells using lentiviral transduction. After 6 days in myeloid-differentiation–promoting liquid culture with SCF, IL-3, FLT3L, and GM-CSF, we noted that mutant WT1 induced a block in myeloid differentiation as indicated by fewer cells expressing mature myeloid markers CD11b, CD14, and CD33 (Figure 5C-D). This differentiation block was not observed in cells expressing wild-type WT1 or in untransduced or GFP controls.
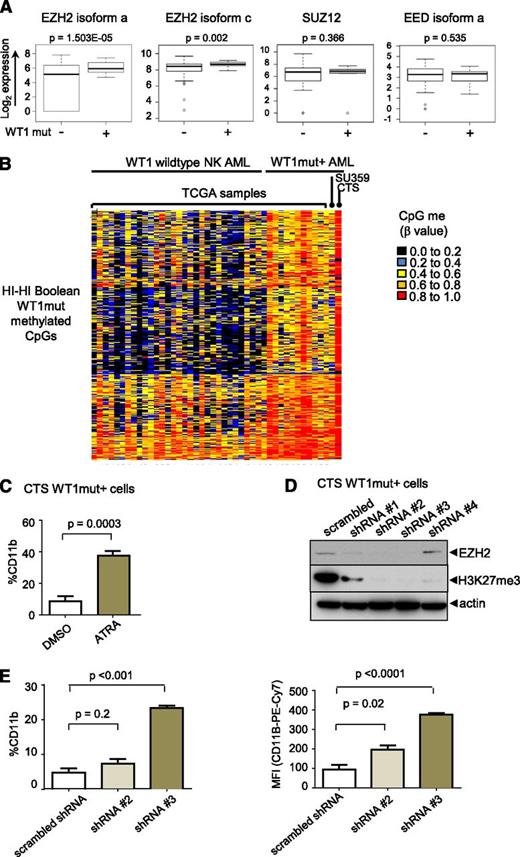
Enhancer of zeste homolog 2 (EZH2) is upregulated in WT1-mutated AML and contributes to myeloid differentiation block
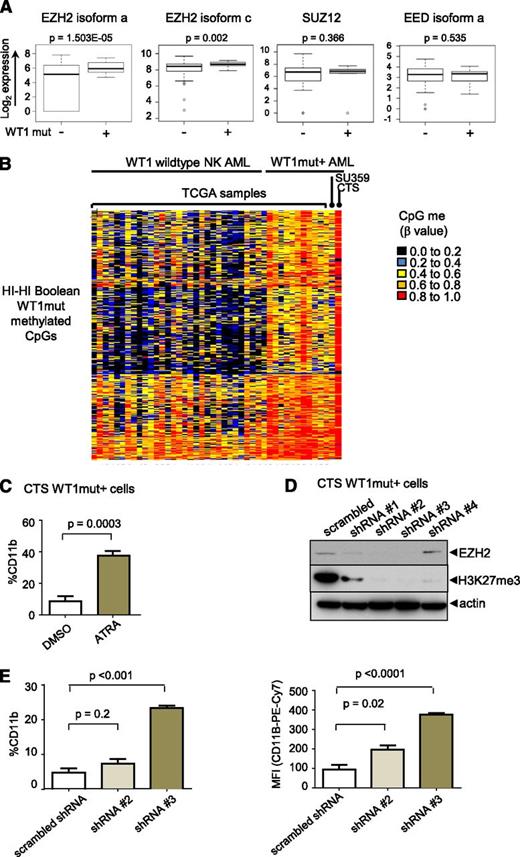
We examined the expression of PRC2 complex components and found that the catalytic subunit, EZH2,18 but not EED or SUZ12, is significantly overexpressed in WT1mut AML compared with NK AML (fold change in means is 4.0 and 1.45 for EZH2 isoforms a and c, respectively) (Figure 6A). To test the hypothesis that the differentiation block in WT1mut leukemia is mediated through PRC2/EZH2, we employed the CTS AML cell line that harbors an endogenous WT1 exon 7 mutation.18 Methylome analysis of CTS cells revealed hypermethylation of the WT1me CpG sites identified by Boolean analysis in patient samples with WT1 mutations and, in contrast to other NK AML samples (Figure 6B), with strong enrichment for PRC2 targets in the methylated genes (supplemental Figure 6A). Treatment of CTS cells with ATRA induced differentiation as indicated by increased expression of CD11b (Figure 6C). We transduced CTS cells with lentiviruses expressing several different short hairpin RNAs (shRNAs) targeting EZH2. Markedly decreased EZH2 protein expression and loss of H3K27 trimethylation after 5 days in culture was confirmed on sorted red fluorescent protein (RFP)–positive virally transduced cells for 2 of the 4 shRNAs (Figure 6D). In cells expressing these shRNAs, CTS cells upregulated expression of the mature myeloid marker CD11b (Figure 6E), consistent with release of the myeloid differentiation block.
EZH2 is upregulated in WT1 mutated AML and contributes to myeloid differentiation block. (A) Box-and-whisker plots showing log2-normalized RNA Seq expression data for EZH2, SUZ12, and EED in NK AML with and without mutation in WT1 from TCGA patient data. Bar represents Student t test was used to determined statistical significance. (B) Heatmap of β methylation values of WT1me CpG sites in primary AML sample SU359, CTS cells, 11 AML TCGA patients with WT1 mutation, and NK AML TCGA patients without WT1 mutations. (C) CTS cells with endogenous mutation in WT1 were treated with ATRA for 72 hours, and CD11b expression was measured by flow cytometry. Cumulative data from 3 independent experiments is presented with statistical significance determined by unpaired 2-tailed Student t test. (D) CTS cells were transduced with scrambled or EZH2-specific shRNAs and RFP using lentivirus. RFP-positive cells were sorted after 5 days and whole cell lysates were blotted with anti-EZH2, anti-H3K27me3, and anti-actin–specific antibodies. (E) Sorted RFP-positive CTS cells expressing scrambled or EZH2 targeting shRNAs #2 or #3 were passaged for 7 days and stained with anti-CD11b antibody. The percentage of CD11b-positive cells compared with isotype control (left); geometric mean fluorescence intensity (right). Bars indicate standard deviations of a representative experiment performed in triplicate. **P < .001, Student t test unpaired, 2-tailed.
EZH2 is upregulated in WT1 mutated AML and contributes to myeloid differentiation block. (A) Box-and-whisker plots showing log2-normalized RNA Seq expression data for EZH2, SUZ12, and EED in NK AML with and without mutation in WT1 from TCGA patient data. Bar represents Student t test was used to determined statistical significance. (B) Heatmap of β methylation values of WT1me CpG sites in primary AML sample SU359, CTS cells, 11 AML TCGA patients with WT1 mutation, and NK AML TCGA patients without WT1 mutations. (C) CTS cells with endogenous mutation in WT1 were treated with ATRA for 72 hours, and CD11b expression was measured by flow cytometry. Cumulative data from 3 independent experiments is presented with statistical significance determined by unpaired 2-tailed Student t test. (D) CTS cells were transduced with scrambled or EZH2-specific shRNAs and RFP using lentivirus. RFP-positive cells were sorted after 5 days and whole cell lysates were blotted with anti-EZH2, anti-H3K27me3, and anti-actin–specific antibodies. (E) Sorted RFP-positive CTS cells expressing scrambled or EZH2 targeting shRNAs #2 or #3 were passaged for 7 days and stained with anti-CD11b antibody. The percentage of CD11b-positive cells compared with isotype control (left); geometric mean fluorescence intensity (right). Bars indicate standard deviations of a representative experiment performed in triplicate. **P < .001, Student t test unpaired, 2-tailed.
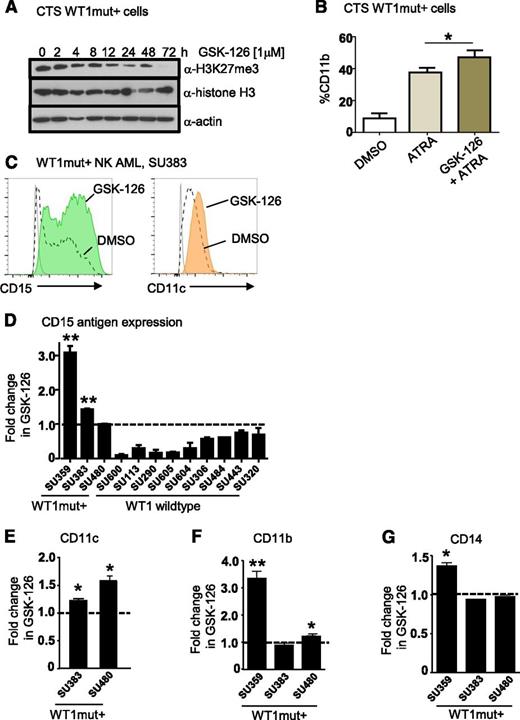
Pharmacologic inhibition of EZH2 in WT1-mutated AML promotes myeloid differentiation
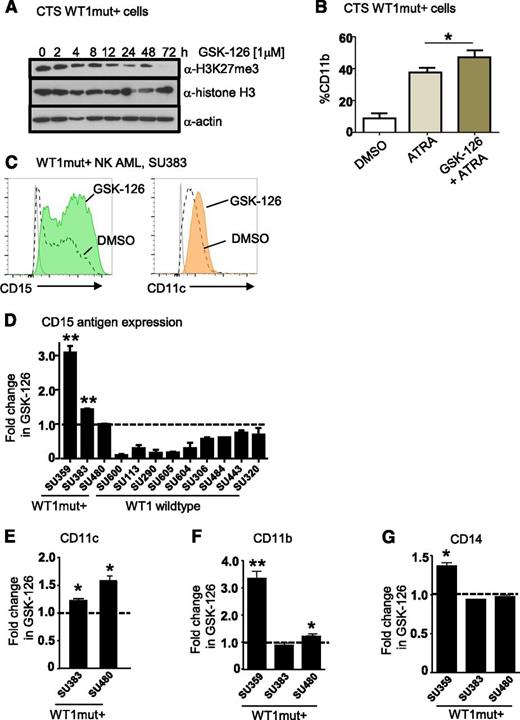
Small-molecule inhibitors of EZH2 have been developed and are currently in early phase clinical trials for other cancer indications.34 To assess their effects on myeloid differentiation of WT1mut AML, we treated CTS cells with the EZH2 inhibitor GSK-12634 and measured expression of mature myeloid markers by flow cytometry. Inhibition of H3K27 trimethylation at 1 μM GSK-126 was confirmed by flow cytometry and western blotting (Figure 7A and supplemental Figure 6B). Treatment of CTS cells with ATRA and GSK-126 induced CD11b expression beyond that of ATRA alone (Figure 7B). Next, we identified our own primary AML sample SU359 heterozygous for a WT1 exon 7 frameshift mutation (1301 ins 1 bp) (supplemental Figure 6C-D) that exhibited increased methylation of the WT1me CpG sites (Figure 6B). We treated purified CD34+/CD117+ leukemic blasts with GSK-126 for 72 hours and observed increased expression of the myeloid maturation markers CD11b, CD15, and CD14 compared with vehicle control (supplemental Figure 6E). Similarly, we observed upregulation of at least 2 mature myeloid markers (CD15, CD11c, CD11b, or CD14) in purified blasts from 2 additional primary de novo NK AML samples with WT1 exon 7 mutations (Figure 7C-G). For comparison, we treated primary acute promyelocytic leukemia blasts lacking WT1 mutation with GSK-126 and ATRA (supplemental Figure 6F). Although we observed increased CD11b expression in response to ATRA in primary acute promyelocytic leukemia cells, GSK-126 treatment did not promote upregulation of mature myeloid markers (supplemental Figure 6F), suggesting that it did not have a global effect. We then tested a further 8 WT1 wild-type de novo NK AML samples with GSK-126, and although we saw heterogeneity in response to GSK-126 with some WT1 wild-type samples exhibiting evidence of upregulation of a single marker, none of these samples showed upregulation of at least 2 myeloid markers (supplemental Figure 6G-J). Thus, WT1mut primary AMLs exhibit more pronounced differentiation responses with multiple myeloid markers than WT1 wild-type cases (Fisher’s exact test, P = .004).
Pharmacologic inhibition of EZH2 in WT1 mutated AML promotes myeloid differentiation. (A) Time course of inhibition of H3K27 trimethylation by GSK-126 in WT1 mutation–positive CTS cells. Western blots were performed on CTS cells after treatment with 1 μM GSK-126 for the time intervals as shown. Whole cell extracts were blotted for anti-H3K27me3, anti-histone H3 (total), and anti-actin. (B) CTS cells were treated with dimethylsulfoxide (DMSO), 1 μM ATRA, or 10 μM GSK-126 and 1 μM ATRA for 72 hours, after which the percentage of cells expressing CD11b was determined by flow cytometry. Bars represent mean of 3 replicates plus standard deviation. A representative of 3 independent experiments in shown. *P < .05, Student t test, 2-tailed. (C) Flow plots showing upregulation of CD15 and CD11c after 72 hours of GSK-126 treatment. Primary AML blasts were obtained from an adult patient (SU383) with a 2-bp insertion mutation in exon 7 of WT1 at P381 and sorted for blasts expressing CD117 and CD34 and cultured in either DMSO or 1 μM GSK-126 for 72 hours. (D) Summary of fold increase in CD15 antigen expression after GSK-126 treatment in vitro compared with DMSO for WT1mut+ AMLs SU359, SU383, and SU480, and 9 WT1 wild-type AMLs as shown, including 1 acute promyelocytic leukemia, SU600. *P < .05; **P < .001, Student t test unpaired, 2-tailed. Statistical significance is only indicated for AMLs that responded with upregulation of CD15 after treatment. (E-G) The fold increase in CD11c (E), CD11b (F), and CD14 antigen expression (G) as measured by flow cytometry in cultured primary AML WT1mut+ blasts after 72 hours of GSK-126 treatment is presented. *P < .05; **P < .001, Student t test unpaired, 2-tailed.
Pharmacologic inhibition of EZH2 in WT1 mutated AML promotes myeloid differentiation. (A) Time course of inhibition of H3K27 trimethylation by GSK-126 in WT1 mutation–positive CTS cells. Western blots were performed on CTS cells after treatment with 1 μM GSK-126 for the time intervals as shown. Whole cell extracts were blotted for anti-H3K27me3, anti-histone H3 (total), and anti-actin. (B) CTS cells were treated with dimethylsulfoxide (DMSO), 1 μM ATRA, or 10 μM GSK-126 and 1 μM ATRA for 72 hours, after which the percentage of cells expressing CD11b was determined by flow cytometry. Bars represent mean of 3 replicates plus standard deviation. A representative of 3 independent experiments in shown. *P < .05, Student t test, 2-tailed. (C) Flow plots showing upregulation of CD15 and CD11c after 72 hours of GSK-126 treatment. Primary AML blasts were obtained from an adult patient (SU383) with a 2-bp insertion mutation in exon 7 of WT1 at P381 and sorted for blasts expressing CD117 and CD34 and cultured in either DMSO or 1 μM GSK-126 for 72 hours. (D) Summary of fold increase in CD15 antigen expression after GSK-126 treatment in vitro compared with DMSO for WT1mut+ AMLs SU359, SU383, and SU480, and 9 WT1 wild-type AMLs as shown, including 1 acute promyelocytic leukemia, SU600. *P < .05; **P < .001, Student t test unpaired, 2-tailed. Statistical significance is only indicated for AMLs that responded with upregulation of CD15 after treatment. (E-G) The fold increase in CD11c (E), CD11b (F), and CD14 antigen expression (G) as measured by flow cytometry in cultured primary AML WT1mut+ blasts after 72 hours of GSK-126 treatment is presented. *P < .05; **P < .001, Student t test unpaired, 2-tailed.
Discussion
Identifying key mutations that contribute to aberrant DNA methylation in cancer is a complex and challenging problem.35 Here, we demonstrate a novel application of Boolean implications (strict subset and mutual exclusion rules) to uncover relationships between somatic mutations and DNA methylation. Boolean implications are if-then rules that provide a conceptually simple, uniform, and highly scalable way to find associations between pairs of random variables. The distribution of points in a scatterplot of 2 variables in a Boolean implication is L-shaped instead of linear (Figure 1B), thereby allowing it to discover many relationships missed by other methods for mining associations such as correlation or t test.36 Applying this method to the TCGA AML data set, we found that mutation in WT1 is strongly associated with CpG hypermethylation. Furthermore, introduction of the mutant WT1 gene into AML cells also induced DNA hypermethylation acting on a similar set of genes, suggesting a causal, although not necessarily direct, effect conferred by the mutant protein. Intriguingly, the WT1mut-associated hypermethylated genes were enriched for PRC2 gene targets marked by H3K27 trimethylation in both embryonic stem cells and K562 hematopoietic cells, revealing an unexpected link between DNA hypermethylation and dysregulation of histone epigenetic marks in this AML subgroup. This pattern was not observed to the same extent for the other recurrent mutations linked to hypermethylation by Boolean implications (Figure 4G). Consistent with this model, AML cells with mutation in WT1 exhibited marked repression of PRC2 gene targets in 2 independent clinical cohorts, and mutant WT1 led to myeloid differentiation block when introduced into CD34+ cord blood cells. Finally, shRNA or pharmacologic inhibition of EZH2 with the GSK-126 inhibitor resulted in myeloid differentiation of WT1-mutant cells, suggesting that EZH2 inhibitors may be clinically active in WT1-mutant AML.
The observed mutation-specific methylation patterns described here may provide clues to the nature of upstream-acting programs that are dysregulated in cancer7,8,37 and provide leads for therapeutic intervention. Besides identifying the total number of hyper- and hypomethylated CpG sites associated with each recurrent mutation, the implication-based method we used also facilitates direct identification of each CpG site that is consistently aberrantly methylated. Knowledge of these precise sites of aberrant methylation may be useful in interpreting mutation-specific methylomes. Here, for WT1mut AML, the pattern of hypermethylated CpGs revealed enrichment of PRC2 targets in both patient samples and WT1mut-transduced cells (Figure 4A-D). Although the molecular mechanisms responsible for PRC2 target DNA hypermethylation in WT1mut AML remain unknown, we note a number of key findings. Curiously, WT1mut AML has a high degree of polycomb target gene repression in comparison with normal maturing myeloid cells (Figure 5A-B and supplemental Figure 5A-B) and also when compared with AML with IDH2 and NPM1 mutations (supplemental Figure 5C-D). Members of the polycomb group complex are critical for developmental stage-specific repression of key master transcriptional regulators of differentiation, and it has been suggested that dysregulation of polycomb-mediated repression may contribute to precancerous states. Consistent with this, our findings of aberrant PRC2 gene repression and DNA methylation in WT1mut AML imply that polycomb-mediated silencing may be critically required for WT1mut-induced leukemogenesis. Furthermore, we find that small molecule-mediated inhibition of EZH2 can promote differentiation of AML cells that harbor endogenous WT1 exon 7 mutations (Figure 7). This may indicate that ongoing EZH2 histone methyltransferase activity is required to maintain the differentiation block in WT1mut AML and is consistent with recent reports showing EZH2 knockdown induces differentiation to macrophage-like cells in mouse mixed lineage leukemia models.38,39 Our methods also identified genetic drivers of hypomethylation including mutation in DNMT3A and the cohesin complex, which will be an interesting avenue for further study (Figure 1C).
Here, we report a strong link between mutation in WT1 and DNA hypermethylation in association with aberrant PRC2-mediated gene repression. This results in a block in myeloid differentiation that can be relieved by EZH2 inhibition. Further preclinical testing of EZH2 inhibitors should be conducted in WT1-mutant AML and possibly brought forward into clinical trials.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE62929).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Feifei Zhao for laboratory management and Sylvia Plevritis for leadership of the Stanford Center for Cancer Systems Biology. The authors acknowledge the Stanford Hematology Division Tissue Bank and the patients for donating their samples.
This work was supported by a Stanford Center for Cancer Systems Biology grant (National Institutes of Health, National Cancer Institute U54CA149145). D.T. is supported by an Australian National Health and Medical Research Council Overseas Biomedical Research Fellowship. R.M. holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund and is a New York Stem Cell Foundation Robertson Investigator.
Authorship
Contribution: S.S. and D.T. designed research, performed research, analyzed data, and wrote the manuscript; A.J.G. analyzed data; N.J., A.R., M.R.C.-Z., S.M.C., and L.Y. performed experiments; A.P.F. analyzed data and edited the manuscript; D.L.D. and S.S. designed computational methods for applying Boolean analysis to methylome data; and R.M. and D.L.D. designed research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David L. Dill, Department of Computer Science, Gates Building 3A, Stanford, CA 94305; e-mail: dill@cs.stanford.edu; and Ravindra Majeti, Stanford Institute for Stem Cell Biology and Regenerative Medicine, Lokey Stem Cell Building, 265 Campus Dr, Stanford, CA 94305; e-mail: rmajeti@stanford.edu.
References
Author notes
S.S., D.T., D.L.D., and R.M. contributed equally to this study.